Abstract
SABBAC is an on-line service devoted to protein backbone reconstruction from alpha-carbon trace. It is based on the assembly of fragments taken from a library of reduced size, selected from the encoding of the protein trace in a hidden Markov model-derived structural alphabet. The assembly of the fragments is achieved by a greedy algorithm, using an energy-based scoring. Alpha-carbon coordinates remain unaffected. SABBAC simply positions the missing backbone atoms, no further refinement is performed. From our tests, SABBAC performs equal or better than other similar on-line approach and is robust to deviations on the alpha-carbon coordinates. It can be accessed at http://bioserv.rpbs.jussieu.fr/SABBAC.html.
INTRODUCTION
There are various fields where the structure of proteins is expressed using a coarse grained model that needs to be expanded in an all atom model. Such preoccupation occurs for instance in fields such as ab-initio or de novo protein fold generation, comparative modelling or the refinement of experimental data obtained at low resolution. It might also occur for structures generated from normal modes simulations. Starting from a coarse grained model of a protein structure, which might provide only some information about where the residues are roughly located in 3D space, the full protein structure generation is often decomposed in a two-part process: (i) generation of the backbone coordinates and (ii) side chain positioning. The alpha-carbon reconstruction process is one way to address the former process, and several studies have tackled this problem of the protein all-atom peptide reconstruction from alpha-carbon trace (1–19).
Among the different approaches that have been proposed, some explore the conformational space of the peptidic units to produce a complete backbone. For instance, the approach developed by Payne (16) attempts to identify the optimal rotation of peptide units using a potential of mean force depending on adjacent residues. Most approaches, such as Ref. (17,19), rely on the assembly of fragments extracted from libraries derived from known structures. Such fragments are then assembled to produce a peptidic chain fitting as best as possible the alpha-carbon trace, usually using energy or geometry criteria to drive the search. Fragment based approaches are in general confronted with the limitation that, in order to improve the accuracy of the reconstruction, it is necessary to maintain large and up to date collections of fragments. The MaxSprout server (17) has been one of the first proposed. It can still be considered as a reference. It is based on a fragment library regularly updated from the protein structures available.
Here, we introduce SABBAC, a procedure that relies on a new approach to fragment selection—among a reduced set—and assembly. It uses the encoding of the alpha-carbon trace using a hidden Markov model derived structural alphabet (20) to select at each position in the structure a small set of candidates among a complete set of only 155 candidates fragments describing all the letters of the structural alphabet—i.e. to describe all the conformations of all protein structures. It then assembles the fragments using a greedy algorithm, searching for the ‘optimal’ combination of fragments compatible with the alpha-carbon trace and produce a full-protein backbone reconstruction.
METHODS
Structure dependent fragment library
To select fragments that will be used for the structure generation we encode the alpha-carbon trace in the Hidden Markov Model derived SA-27 structural alphabet described in Ref. (20). Here, we use the optimal trajectory as produced by the viterbi algorithm. Then at each position, we retain as candidate fragments that represent the letter of the structural alphabet. As described in Ref. (20–22), we have only a number of 155 fragments to describe the 27 letters of the alphabet. Having selected only one letter to describe the conformation of each fragment of four residue length, the average number of fragments used, is only on the order of 5–6 per position.
Peptide units coordinates generation
To quickly generate atoms coordinates for the N, HN, C′ and O atoms of the peptide units, we follow a procedure described previously by Milik et al. (18). We use coordinates pre-computed in a local reference defined from three consecutive alpha carbons. Since SA-27 describes the conformations of fragments of four residue length, two different sets of atoms can be used to define such local reference. For each prototype fragment associated to each letter of SA-27, we have pre-computed the atom coordinates at the first, second and third peptidic bond in these two possible references. We have assessed the stability and accuracy of using each set of coordinate (data not shown). Here, we use the reference associated with the last three alpha-carbons of the fragments of four residue length to position the atoms between the second and third alpha-carbon. For the extremities of the polypeptidic chain only, we consider the first and third peptidic bonds of the fragments.
Search for an optimal combination of fragments
Given the collection of candidate fragments at each position of the structure, we use a greedy algorithm, as described in Ref. (21,22) to search for their optimal combination. At each position, given a collection of reconstruction up to the previous position, we generate all the new possible assemblies of size increased by one residue. Each of them is scored (see below), and we retain only a limited subset of assemblies to iterate to the next position (heap size). Here, we use a heap size of only 10, a number learnt from the reconstruction of series of structures. Compared with Ref. (21,22), we use a simpler strategy: the rebuild process is only achieved by growing from N- towards C-terminal. The process is not iterated.
Energy evaluation
To drive the search, we use an energy criterion combining some of the OPEP force field (23) with some terms assessing the quality of the geometry of the alpha-carbons.
where ESC,SC describes side chain–side chain interactions; EHB, hydrogen bonds, EVdW the Van der Waals backbone–backbone and backbone–side chain interactions, EPhiP the phi positive contribution, EBB the alpha carbon valence angle distortion and ETrans is a pseudo energy term related to the transitions between consecutive fragments. Backbone is described using an all-atom representation (N, HN, CA, C, O). Side chains are represented using a single particle, described by a centroid and a radius for each residue type. Centroid coordinates and ray values have been computed and optimized on a non-redundant Protein Data Bank (PDB) of <30% identity (J. Manupetit, P. Tufféry and P. Derreumaux, manuscript in preparation). All these terms are computed as described in Ref. (23) except for the phi positive term, that is set to a constant of +0.5 kcal/mol if the phi angle value is positive, 0 otherwise, except for glycines (−0.3 kcal/mol for phi positive values). For EBB, we simply use the square of the deviation to the canonical angle value of 110°. For ETrans, we use
where p(i → i + 1) is the probability that the fragment selected at position i + 1 follows the fragment at position i. p(i → i + 1) have been evaluated from a non-redundant set of proteins of the PDB with <30% sequence identity.
PERFORMANCES
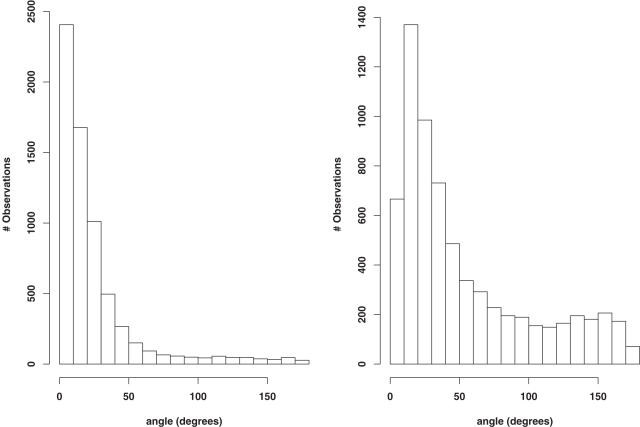
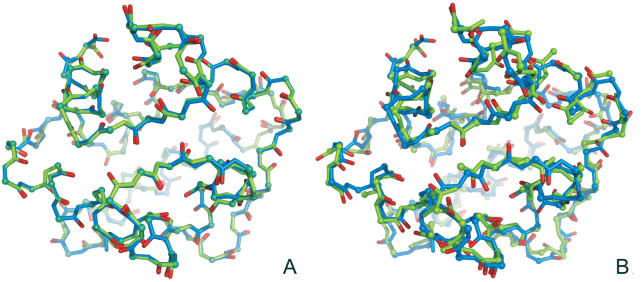
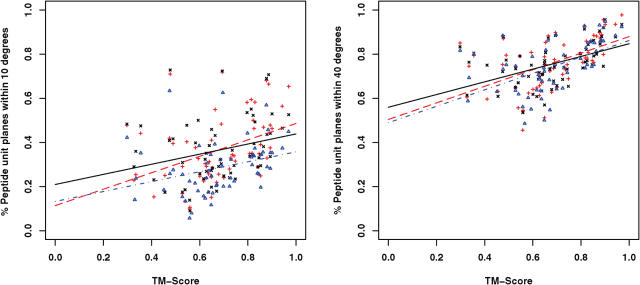
We have assessed the performances of SABBAC on various test sets. Some results are reported Table 1. Its upper section presents SABBAC results obtained for a collection of proteins discussed in Ref. (19), which provides some elements of comparison with earlier approaches. For bb, we present the results as obtained with the bb program available for download, without any minimization. For MaxSprout, we have used the on-line server. As shown, SABBAC gives on average better results than both MaxSprout and bb. The lower part reports some results obtained for recent entries of the PDB, not introduced for the identification of our collection of fragments. In this subset, the performances of SABBAC are unaffected. We have also checked how SABBAC performs for perturbed alpha-carbon trace. Figure 1 presents distributions of deviations of the peptide unit planes between native and rebuilt structures of the proteins of Table 1; for perturbed alpha-carbon traces. Even for traces randomly perturbed by over 1 Å on alpha-carbon coordinates, SABBAC results are only marginally affected. We emphasize that correct peptide unit plane orientation also implies correct side chain orientation. Figure 2 shows the rebuilt structures obtained for the oncomodulin (1OMD PDB code), from native trace (A) and from highly degenerate alpha carbons trace (B). Finally, we have also considered the reconstruction for a series of models from the CASP6 experiment (25). We have retained all the targets corresponding to complete structures (not domain-only targets) and removed targets having missing fragments. For each, we have considered the best and the rank 5 models (as classified by CASP GDT-TS). Incomplete models were discarded. This resulted in 31 targets—60 models—including homology modelling (14 targets), fold recognition (13 targets) and new fold (4 targets) categories. We have rebuilt the backbone using SABBAC and MaxSprout. For MaxSprout, we obtained results for only 57 models. Figure 3 plots, for all the models, for SABBAC and MaxSprout, the fractions of the peptide unit planes that deviate by <10° or 40° to that of the native structures. It is expressed as a function of the deviation of the model to the native structure, quantified using the TM-Score (26). A value of TM-Score of 1 is reached for perfect correspondence between modelled and native main chain. The lowest the value, the worse the model. Such measure is more relevant than the classical root mean square deviation (r.m.s.d.) since it assess the parts of the structures that correspond, not necessarily resulting in extremely low values if a domain undergoes a collective motion relative to another, for instance the alpha-carbon RMS deviations are between 1.6 and over 10 Å; for the 60 models. As can be seen, SABBAC performs overall better than MaxSprout for a 10° deviation, implying a more accurate reconstruction. The performance differences get smaller for 40°. Compared with the CASP models, SABBAC tends to propose better results for the lower TM scores, which suggest a good robustness to main chain perturbation. For values of TM score close to 1, CASP models are in general better. One must however consider that the processes of model generation and SABBAC reconstruction largely differ, SABBAC not performing any refinement.
Table 1.
Backbone reconstruction performance. Comparison with other methods
| Main chain RMSd | ||||
|---|---|---|---|---|
| PDB | n Residues | SABBAC | MaxSprout | bb |
| Adcock subset | ||||
| 111M | 154 | 0.29 | 0.42 | 0.91 |
| 1CTF | 68 | 0.43 | 0.73 | 0.85 |
| 1IGD | 61 | 0.36 | 0.44 | 0.68 |
| 1OMD | 107 | 0.35 | 0.41 | 0.77 |
| 1SEMA | 58 | 0.48 | 0.34 | 1.00 |
| 1TIMA | 247 | 0.59 | 0.60 | 0.97 |
| 1UBQ | 76 | 0.35 | 0.38 | 0.96 |
| 2CTS | 437 | 0.40 | 0.45 | 0.86 |
| 2LYM | 129 | 0.38 | 0.44 | 0.98 |
| 2MHR | 118 | 0.50 | 0.54 | 0.88 |
| 2PCY | 99 | 0.42 | 0.54 | 0.91 |
| 2WRP | 104 | 0.30 | 0.42 | 0.87 |
| 4PTI | 58 | 0.53 | 0.44 | 0.81 |
| 5NLL | 138 | 0.37 | 0.46 | 0.85 |
| Mean | 0.41 | 0.47 | 0.89 | |
| SD | 0.09 | 0.10 | 0.08 | |
| PDB newcomers subset | ||||
| 1PXZA | 346 | 0.55 | 0.54 | 0.96 |
| 1RKIA | 101 | 0.58 | 0.44 | 0.88 |
| 1S7LA | 177 | 0.29 | 0.36 | 0.86 |
| 1T70A | 255 | 0.42 | 0.50 | 0.95 |
| 1TXOA | 235 | 0.41 | 0.38 | 0.96 |
| 1V0ED | 666 | 0.48 | 0.45 | 0.89 |
| 1V7BA | 175 | 0.30 | 0.41 | 0.87 |
| 1VB5B | 255 | 0.34 | 0.42 | 0.84 |
| 1VKCA | 149 | 0.28 | 0.33 | 0.82 |
| 1VR4A | 103 | 0.47 | 0.59 | 1.00 |
| 1VR9A | 121 | 0.42 | 0.45 | 0.79 |
| 1WMHA | 83 | 0.27 | 0.28 | 0.82 |
| 1WPBG | 168 | 0.37 | 0.35 | 0.86 |
| 1WMIA | 88 | 0.41 | 0.42 | 0.81 |
| 1X6JA | 88 | 0.43 | 0.36 | 0.76 |
| 1XB9A | 108 | 0.46 | 0.51 | 0.81 |
| 1XE0B | 107 | 0.61 | 0.62 | 0.90 |
| Mean | 0.42 | 0.44 | 0.88 | |
| SD | 0.10 | 0.09 | 0.07 | |
| GLOBAL | ||||
| Mean | 0.41 | 0.45 | 0.87 | |
| SD | 0.09 | 0.10 | 0.07 | |
Backbone reconstruction performance is evaluated compared with the native structure (backbone heavy atoms r.m.s.d). Two test sets are presented, the first one is a subset of those presented by SA Adcock (19), and the second one is composed of recent newcomers of the PDB (24).
Figure 1.
Peptidic bonds angle distribution. Distribution of the peptide unit planes angular deviations between native and rebuilt structures for perturbed alpha-carbon traces. Each deviation is calculated for locally fitted backbones. Left, average alpha-carbon trace perturbation of 0.2 Å. Right, average perturbation of 1.0 Å.
Figure 2.
SABBAC rebuilding example, 1OMD. (A) Native and SABBAC rebuilt structures. (B) Native and SABBAC rebuilt structures from an alpha-carbon trace perturbed by 0.8 Å on average. The native structure is represented in blue.
Figure 3.
SABBAC and MaxSprout reconstruction performance for 31 CASP6 targets. For each of target, the best and the rank 5 models have been considered. The fraction of the peptide unit planes deviating by <10° (left) and 40° (right) are plotted as a function of the TM-Scores of the models. Lines correspond to regressions. Red + CASP6 models versus native structure; black x SABBAC reconstruction; blue triangles MaxSprout reconstruction.
IMPLEMENTATION
The SABBAC server integrates all the steps required for the complete protein reconstruction from an alpha-carbon trace specified in the PDB format. In the present version, atoms present in the PDB file other than alpha-carbons are discarded, as well as hetero groups. As output, SABBAC will return a file with all the backbone atoms, and the detail of the energy scores, for the complete structure and by residue. Since the greedy algorithm heap size is of 10, it is possible to ask for a number of models varying between 1 and 10. The models are sorted according to their scores, by decreasing order—the best is at rank 1. Side chains are not positioned by default, but it is possible to chain SABBAC to SCit (27) fast side chain positioning method in order to obtain all atom models. This fast version of SCit positions side chains by selecting the most probable side chain conformation given backbone conformation, removing side chain conformations having clashes with the backbone. Side chain–side chain clashes are then considered. Using a greedy algorithm, the calculation time depends on the size of the protein and is expected to increase linearly with the size. However, this is presently not true for the energy calculation. For small proteins, typical calculation times are in the order of few seconds. For larger size proteins, calculation time can increase up to several minutes or tens of minutes, depending on the server load.
DISCUSSION AND FUTURE WORK
In its present form, the SABBAC seems a reliable alternative to the reference MaxSprout server that pioneered the field. SABBAC provides, on average, a more accurate reconstruction for accurate alpha-carbon trace, and its performances for perturbed traces are still relevant. One strong feature of SABBAC is to provide an answer even in cases where the trace is degenerated. In such cases, the MaxSprout may return structures having missing parts, if the conformations are too far from those observed in its bank of fragments. In addition, SABBAC performances rely on a very small set of conformations that are selected using a structural alphabet encoding, and that rely much less than previous approaches on updates from new structures resolved. Finally, SABBAC performs reconstruction in reasonable time, although work is under progress to improve that point.
Future direction for the SABBAC service are in terms of introducing flexibility. Firstly, it could be of interest to implement some filter to rebuild only parts of the structure specified by the user. Secondly, we are also considering the possible automation of an interface with the side chain positioning facility using a more accurate positioning method.
Acknowledgments
The authors thank Ph. Derreumaux for his help in implementing OPEP, G. Debret and P. Fuchs for helpful discussions and the referees for their constructive remarks. Funding to pay the Open Access publication charges for this article was provided by INSERM U726.
Conflict of interest statement. None declared.
REFERENCES
- 1.Purisima E.O., Scheraga H.A. Conversion from a virtual-bond chain to a complete polypeptide backbone chain. Biopolymers. 1984;23:1207–1224. doi: 10.1002/bip.360230706. [DOI] [PubMed] [Google Scholar]
- 2.Jones T.A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986;5:819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid L.S., Thornton J.M. Rebuilding flavodoxin from C alpha coordinates: a test study. Proteins. 1989;5:170–182. doi: 10.1002/prot.340050212. [DOI] [PubMed] [Google Scholar]
- 4.Claessens M., Van Cutsem E., Lasters I., Wodak S. Modelling the polypeptide backbone with ‘spare parts’ from known protein structures. Protein Eng. 1989;2:335–345. doi: 10.1093/protein/2.5.335. [DOI] [PubMed] [Google Scholar]
- 5.Levitt M. Accurate modeling of protein conformation by automatic segment matching. J. Mol. Biol. 1992;226:507–533. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- 6.Rey A., Skolnick J. Efficient algorithm for the reconstruction of a protein backbone from the a-carbon coordinates. J. Comput. Chem. 1992;13:443–456. [Google Scholar]
- 7.Feldman H.J., Hogue C.W. A fast method to sample real protein conformational space. Proteins. 2000;39:112–131. [PubMed] [Google Scholar]
- 8.Liwo A., Pincus M.R., Wawak R.J., Rackovsky S., Scheraga H.A. Calculation of protein backbone geometry from alpha-carbon coordinates based on peptide-group dipole alignment. Protein Sci. 1993;2:1697–1714. doi: 10.1002/pro.5560021015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwata Y., Kasuya A., Miyamoto S. An efficient method for reconstructing protein backbones from alpha-carbon coordinates. J. Mol. Graph. Model. 2002;21:119–128. doi: 10.1016/s1093-3263(02)00142-0. [DOI] [PubMed] [Google Scholar]
- 10.Correa P.E. The building of protein structures from alpha-carbon coordinates. Proteins. 1990;7:366–377. doi: 10.1002/prot.340070408. [DOI] [PubMed] [Google Scholar]
- 11.van Gelder C.W., Leusen F.J., Leunissen J.A., Noordik J.H. A molecular dynamics approach for the generation of complete protein structures from limited coordinate data. Proteins. 1994;18:174–185. doi: 10.1002/prot.340180209. [DOI] [PubMed] [Google Scholar]
- 12.van Hooft P.A., Holtje H.D. Construction of a full three-dimensional model of the transpeptidase domain of Streptococcus pneumoniae PBP2x starting from its Calpha-atom coordinates. J. Comput. Aided Mol. Des. 2000;14:719–730. doi: 10.1023/a:1008164914993. [DOI] [PubMed] [Google Scholar]
- 13.Mathiowetz A.M., Goddard W.A., III Building proteins from C alpha coordinates using the dihedral probability grid Monte Carlo method. Protein Sci. 1995;4:1217–1232. doi: 10.1002/pro.5560040619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan K., Alexander P., Coxon J.M., McKinnon J.A., Worth G.A. The reconstruction of a protein backbone from Ca coordinates. Biopolymers. 1996;41:381–389. [Google Scholar]
- 15.Rajmund Kazacutemierkiewicz, Adam Liwo, Harold A. Scheraga. Energy-based reconstruction of a protein backbone from its alpha-carbon trace by a Monte-Carlo method. J. Comput. Chem. 2002;23:715–723. doi: 10.1002/jcc.10068. [DOI] [PubMed] [Google Scholar]
- 16.Payne P.W. Reconstruction of protein conformations from estimated positions of the C alpha coordinates. Protein Sci. 1993;2:315–324. doi: 10.1002/pro.5560020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holm L., Sander C. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J. Mol. Biol. 1991;218:183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- 18.Milik M., Kolinski A., Skolnick J. Algorithm for rapid reconstruction of protein backbone from alpha carbon coordinates. J. Comput. Chem. 1997;18:80–85. [Google Scholar]
- 19.Adcock S.A. Peptide backbone reconstruction using dead-end elimination and a knowledge-based forcefield. J. Comput. Chem. 2004;25:16–27. doi: 10.1002/jcc.10314. [DOI] [PubMed] [Google Scholar]
- 20.Camproux A.C., Gautier R., Tuffery P. A hidden markov model derived structural alphabet for proteins. J. Mol. Biol. 2004;339:591–605. doi: 10.1016/j.jmb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Tuffery P., Guyon F., Derreumaux P. Improved greedy algorithm for protein structure reconstruction. J. Comput. Chem. 2005;26:506–513. doi: 10.1002/jcc.20181. [DOI] [PubMed] [Google Scholar]
- 22.Tuffery P., Derreumaux P. Dependency between consecutive local conformations helps assemble protein structures from secondary structures using Go potential and greedy algorithm. Proteins. 2005;61:732–740. doi: 10.1002/prot.20698. [DOI] [PubMed] [Google Scholar]
- 23.Santini S., Wei G.H., Mousseau N., Derreumaux P. Exploring the folding pathways of proteins through energy landscape sampling: Application application to Alzheimer's beta-amyloid peptide. Internet Electron. J. Mol. Des. 2003;2:564–577. [Google Scholar]
- 24.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moult J., Fidelis K., Rost B., Hubbard T., Tramontano A. Critical assessment of methods of protein structure prediction (CASP)—Round 6. Proteins. 2005;61(S7):3–7. doi: 10.1002/prot.20716. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gautier R., Camproux A.C., Tuffery P. SCit: web tools for protein side chain conformation analysis. Nucleic Acids Res. 2004;32:W508–W11. doi: 10.1093/nar/gkh388. [DOI] [PMC free article] [PubMed] [Google Scholar]