Abstract
Protein Peeling 2 (PP2) is a web server for the automatic identification of protein units (PUs) given the 3D coordinates of a protein. PUs are an intermediate level of protein structure description between protein domains and secondary structures. It is a new tool to better understand and analyze the organization of protein structures. PP2 uses only the matrices of protein contact probabilities and cuts the protein structures optimally using Matthews' coefficient correlation. An index assesses the compactness quality of each PU. Results are given both textually and graphically using JMol and PyMol softwares. The server can be accessed from http://www.ebgm.jussieu.fr/~gelly/index.html.
INTRODUCTION
Even with no relationship with the folding process, folded state is often described in a hierarchical way. The top is frequently associated with domains, i.e. autonomous folding units (1), and the bottom to secondary structures (2,3). In the middle, supersecondary structures are frequently identified. Such descriptions are mainly based on the frequency of similar motifs, described at different levels of 3D complexity. Many experiments and the recent theory of protein folding suggest that the 3D folded state dictates the folding process. Yet, few approaches aim at identifying folding features through the analysis of the folded state. In a pioneer work, Wetlaufer was the first to examine the organization of known structures and suggested that the early stages of 3D structure formation, i.e. nucleation, occur independently in separate parts of these molecules (4). He referred to these domains as folding units (5) and suggested that they could fold independently during the folding process, creating structural modules which are assembled to give the native structure (6). Early analyses were often performed manually and on a limited number of proteins (4,7–9).
Since then, different strategies have been designed for extracting such folding units. The procedures have differed in many aspects, e.g. the measures and the criteria used. Gō (10) based his description on the Cα–Cα distance map. Janin and Wodak defined putative compact globular units as units with minimal interface area (11). Rose (12,13) identified a disclosing plane that cut the protein chain into compact continuous segments. Subsequently, Zehfus (14), extending earlier work by Zehfus and Rose (15), reported an algorithm that identified compact structures and located discontinuous domains in four globular proteins. Sowdhamini and Blundell's approach was based on Cα–Cα distances and secondary structures (16). Tsai and Nussinov described a scoring function, based on compactness, hydrophobicity and isolatedness, that estimates the stability of these units (12,17). These different automatic approaches define a hierarchical organization of the protein in compact units (11,12,16–18). However, few servers are accessible to the scientific community at this time. Moreover, they focus mainly on the top level of organization, namely, protein domains [e.g. Protein Domain Parser (19)]. DIAL, another web tool, focuses on automatic identification of structural compact domains given the 3D coordinates of a protein; it extends the detection of other hierarchical levels of 3D organization of protein structure (20) compared with the previous one.
Here, we propose a new web server, called Protein Peeling 2 (PP2), that aims at describing different levels of organization of 3D protein structures, depending on the user choices. PP2 is based on a new methodology able to decompose the 3D protein structure from secondary structures to domains. The procedure may yield an intermediate level of organization, through what we have named protein units (PUs). A PU is defined as a compact subregion of the 3D structure corresponding to one sequence fragment, defined by a high number of intra-PU contacts and a low number of inter-PU contacts. PP2 works from the Cα-contact matrix translated into contact probabilities (21). An optimization procedure, based on the Matthews' coefficient correlation (MCC) (22) between contact submatrices, defines optimal cutting points that separate generally into two or three PUs, the region examined. The process is iterated until the compactness of the resulting PUs reaches a given limit, fixed by the user. The PU compactness is quantified by an index, CI (compaction index). This index is based on a correlation coefficient R between the mutual entropy of the contact submatrices (23–25). The procedure leads the 3D protein structure being cut into a limited set of PUs. Thus, it defines a series of successive nested partitions, i.e. a dendogram showing the successive splitting of the PUs into sub-PUs.
PEELING SERVER
The web server allows the user to work with one structure (or a structural model). Using default parameters, the user uploads a Protein Data Bank (PDB) file. The contact matrix is then computed and transformed into a probabilities contact matrix. The procedure performs the splitting of protein structures into PUs. Various tools are used to show the results: (i) a dendogram showing the successive splitting of the PUs into sub-PUs, (ii) a contact matrix of the PUs, (iii) a 3D representation of the PUs and (iv) a summary of the different PUs.
IMPLEMENTATION
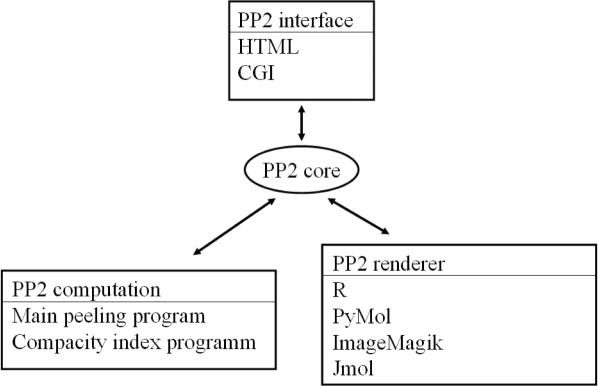
Figure 1 shows the flowchart representation of the PP2 web server. The interface component consists of a web page (HTML) and common gateway interface (CGI). This interface allows the retrieval of values given by the user (parameters and PDB file) and their transmission to the perl core instance. The core component is a perl module that embeds all the information necessary for two other components. The first component consists of programs that perform the protein peeling process and compute CI. The second component consists of different rendering programs. Thus, R (26) is used to visualize (i) the hierarchical peeling of the protein structure, (ii) the probabilities contact matrix and (iii) the final splitting of the protein structure contents into secondary structures. PP2 also relies on PyMol (27) (http://www.pymol.org). The communication between the PP2 core instance and the graphical viewer is based on the generation of a script. This component manages the post-rendering of the pictures. The conversion is based on the conversion program of the ImageMagick suite (http://www.imagemagick.org). We have also added the possibility with a Java Virtual Machine to use JMol. JMol allows the 3D visualization of the final cutting into PUs (www.jmol.org). It is possible, using this design, to trigger the rendering using a mechanism other than the CGI (e.g. interactively using a command line) and to generate a databank of PUs from a non-redundant databank. Further analyses may therefore be carried outeasily.
Figure 1.
Flowchart of the PP2 web server.
PEELING SERVER FEATURES
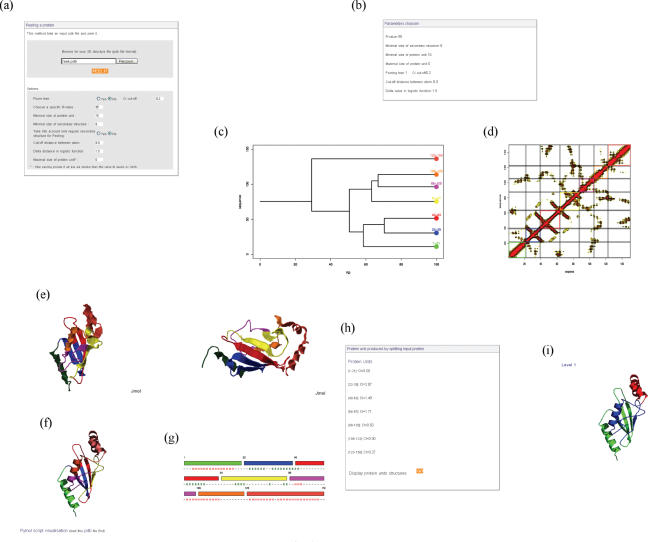
Figure 2 presents snapshots of the different information and results given by the PP2 web server.
Figure 2.
PP2 web server. (a) PP2 interface. (b) Summary of the parameters used. (c) Tree showing the protein peeling process with the different final PUs and their positions. (d) Probability contact matrix with the cutting of the PUs. (e) JMol and (f) PyMol representation of the protein colored by PU. (g) Linear representation of the PUs with their secondary structure assignments. (h) Sequence positions of the PUs with their CI values. (i) Previous level of protein peeling represented using PyMol.
Data input
PP2 supports only the PDB format.
Adjustable parameters
See Figure 2a. PUs are split using the Cα distance matrix translated in terms of probabilities. This version of our web server allows the Cα distance threshold and the curvature of the probability function to be changed. In the same way, it is also possible to use only the regular secondary structures in the creation of the PUs. The minimal size for secondary structures and PUs can also be changed by the user. To stop the peeling process, the R-value threshold can be modified. Moreover, a pruning of the final dendogram is proposed. It permits the discarding of any PU that presents a low number of inter-contacts; i.e. only pertinent cutting will be done.
Representation of the results
During the submission process, an automatic image generated using Rasmol (28) and representing the protein structure is shown. After the protein peeling, each PU is characterized by its position in the protein sequence and is associated with a fixed color for its representation.
First, a summary of the different parameters used is shown (Figure 2b). A dendogram representation of the peeling process details the different events (Figure 2c). The contact matrix is also represented, colored according to the PUs (Figure 2d).
We have also added a schematic representation of the results with a description of PUs with their contents in secondary structures and a 3D visualization using the JMol applet (http://jmol.sourceforge.net/) (Figure 2e). This viewer allows the entire colored protein to be viewed in terms of PUs. The user can easily interact with such a description. A classical static representation is also generated using PyMol (27) (http://www.pymol.org). The corresponding script can be downloaded locally (Figure 2f). A linear representation of the PUs along the sequence is given in the corresponding colors (Figure 2g). The precise position of the different PUs is given in text form with the corresponding CI, i.e. the index that quantifies the compactness of the PU (Figure 2h). For clarity, only the final level of cutting is given. However, all the other levels are available, with the corresponding CI of each intermediate PU (Figure 2i).
Non-redundant databank
An updated non-redundant databank taken from the PDB (29) is also available. This non-redundant set of protein structures includes 2309 elements from crystallographic experiments with >2 Å resolution from the PISCES server (30). The proteins share no more than 30% sequence identity. All these structures have been dissected with the protein peeling procedure. The results have been stored in a flat file database, and these pre-cut proteins can be easily accessed through a form or by selecting the protein from a list.
PERSPECTIVES
The PU provides a new view of the protein folded state. It offers an original and rapid way to analyze interesting regions in the structure. The PP2 web server allows the composition in terms of PUs of protein structures to be obtained. The PP2 server is thus a useful tool to examine in an original way, the 3D structure of proteins. The different parameters can be easily controlled and the subsequent PUs graphically analyzed. Results are given both textually and visually. In the future, we would like to analyze the Pus' distribution across protein families (31) and perform prediction from the sequence.
Acknowledgments
This paper is dedicated to the memory of Prof. Serge Hazout. This work was supported in part by ACI Action Bioinformatique 2003–2004. Authors thank the Institut National de la Recherche Médicale (INSERM), Université Paris 7—Denis Diderot, Centre National de la Recherche Scientifique (CNRS) and the Ministère de l'Education Nationale de l'Enseignement Supérieur et de la Recherche for financial and infrastructural supports. Funding to pay the Open Access publication charges for this article was provided by the Institut National de la Santé et de la Recherche Médicale (INSERM).
Conflict of interest statement. None declared.
REFERENCES
- 1.Richardson J.S. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 2.Pauling L., Corey R.B. The pleated sheet, a new layer configuration of polypeptide chains. Proc. Natl Acad. Sci. USA. 1951;37:251–256. doi: 10.1073/pnas.37.5.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pauling L., Corey R.B. Atomic coordinates and structure factors for two helical configurations of polypeptide chains. Proc. Natl Acad. Sci. USA. 1951;37:235–240. doi: 10.1073/pnas.37.5.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wetlaufer D.B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc. Natl Acad. Sci. USA. 1973;70:697–701. doi: 10.1073/pnas.70.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wetlaufer D.B. Folding of protein fragments. Adv. Protein Chem. 1981;34:61–92. doi: 10.1016/s0065-3233(08)60518-5. [DOI] [PubMed] [Google Scholar]
- 6.Chothia C. Principles that determine the structure of proteins. Annu. Rev. Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- 7.Drenth J., Jansonius J.N., Koekoek R., Swen H.M., Wolthers B.G. Structure of papain. Nature. 1968;218:929–932. doi: 10.1038/218929a0. [DOI] [PubMed] [Google Scholar]
- 8.Phillips D.C. The three-dimensional structure of an enzyme molecule. Sci. Am. 1966;215:78–90. doi: 10.1038/scientificamerican1166-78. [DOI] [PubMed] [Google Scholar]
- 9.Janin J., Wodak S.J. Structural domains in proteins and their role in the dynamics of protein function. Prog. Biophys. Mol. Biol. 1983;42:21–78. doi: 10.1016/0079-6107(83)90003-2. [DOI] [PubMed] [Google Scholar]
- 10.Go M. Correlation of DNA exonic regions with protein structural units in haemoglobin. Nature. 1981;291:90–92. doi: 10.1038/291090a0. [DOI] [PubMed] [Google Scholar]
- 11.Wodak S.J., Janin J. Location of structural domains in protein. Biochemistry. 1981;20:6544–6552. doi: 10.1021/bi00526a005. [DOI] [PubMed] [Google Scholar]
- 12.Rose G.D. Hierarchic organization of domains in globular proteins. J. Mol. Biol. 1979;134:447–470. doi: 10.1016/0022-2836(79)90363-2. [DOI] [PubMed] [Google Scholar]
- 13.Lesk A.M., Rose G.D. Folding units in globular proteins. Proc. Natl Acad. Sci. USA. 1981;78:4304–4308. doi: 10.1073/pnas.78.7.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zehfus M.H. Binary discontinuous compact protein domains. Protein Eng. 1994;7:335–340. doi: 10.1093/protein/7.3.335. [DOI] [PubMed] [Google Scholar]
- 15.Zehfus M.H., Rose G.D. Compact units in proteins. Biochemistry. 1986;25:5759–5765. doi: 10.1021/bi00367a062. [DOI] [PubMed] [Google Scholar]
- 16.Sowdhamini R., Blundell T.L. An automatic method involving cluster analysis of secondary structures for the identification of domains in proteins. Protein Sci. 1995;4:506–520. doi: 10.1002/pro.5560040317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai C.J., Nussinov R. Hydrophobic folding units derived from dissimilar monomer structures and their interactions. Protein Sci. 1997;6:24–42. doi: 10.1002/pro.5560060104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crippen G.M. The tree structural organization of proteins. J. Mol. Biol. 1978;126:315–332. doi: 10.1016/0022-2836(78)90043-8. [DOI] [PubMed] [Google Scholar]
- 19.Alexandrov N., Shindyalov I. PDP: protein domain parser. Bioinformatics. 2003;19:429–430. doi: 10.1093/bioinformatics/btg006. [DOI] [PubMed] [Google Scholar]
- 20.Pugalenthi G., Archunan G., Sowdhamini R. DIAL: a web-based server for the automatic identification of structural domains in proteins. Nucleic Acids Res. 2005;33:W130–W132. doi: 10.1093/nar/gki427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelly J.-C., de Brevern A.G., Hazout S. ‘Protein Peeling’: an approach for splitting a 3D protein structure into compact fragments. Bioinformatics. 2006;22:129–133. doi: 10.1093/bioinformatics/bti773. [DOI] [PubMed] [Google Scholar]
- 22.Matthews B.W. X-ray crystallographic studies of proteins. Annu. Rev. Phys. Chem. 1976;27:493–523. [Google Scholar]
- 23.de Brevern A.G., Etchebest C., Hazout S. Bayesian probabilistic approach for predicting backbone structures in terms of protein blocks. Proteins. 2000;41:271–287. doi: 10.1002/1097-0134(20001115)41:3<271::aid-prot10>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 24.de Brevern A.G., Hazout S. ‘Hybrid protein model’ for optimally defining 3D protein structure fragments. Bioinformatics. 2003;19:345–353. doi: 10.1093/bioinformatics/btf859. [DOI] [PubMed] [Google Scholar]
- 25.Etchebest C., Benros C., Hazout S., de Brevern A.G. A structural alphabet for local protein structures: improved prediction methods. Proteins. 2005;59:810–827. doi: 10.1002/prot.20458. [DOI] [PubMed] [Google Scholar]
- 26.Ihaka R., Gentleman R. R: a language for data analysis and graphics. J. Comput. Graph Stat. 1996;5:299–314. [Google Scholar]
- 27.DeLano W.L.T. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 28.Sayle R.A., Milner-White E.J. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 29.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G., Dunbrack R.L., Jr PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 31.Efimov A.V. Structural trees for protein superfamilies. Proteins. 1997;28:241–260. doi: 10.1002/(sici)1097-0134(199706)28:2<241::aid-prot12>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]