Abstract
We describe Protogene, a server that can turn a protein multiple sequence alignment into the equivalent alignment of the original gene coding DNA. Protogene relies on a pipeline where every initial protein sequence is BLASTed against RefSeq or NR. The annotation associated with potential matches is used to identify the gene sequence. This gene sequence is then aligned with the query protein using Exonerate in order to extract a coding nucleotide sequence matching the original protein. Protogene can handle protein fragments and will return every CDS coding for a given protein, even if they occur in different genomes. Protogene is available from http://www.tcoffee.org/.
INTRODUCTION
Although they constitute the material with which primary biological databases are made of, nucleotide sequences are rarely used when it comes to analyzing proteins. The reason is that evolutionary models designed for comparing nucleic acids are often too simplistic and almost never take into account the constraints associated with the coding nature of gene sequences. In practice, biologists dealing with proteins are often encouraged to use protein databases and associated tools to build their models. However, the transposition of these results onto the bona fide nucleotide sequences is time consuming. This limitation can be an issue, especially when reconstructing phylogenetic trees of closely related species or when looking for conserved nucleotide patterns within multiple coding sequences.
In theory, the task of turning a protein multiple sequence alignment (MSA) into the associated CDS (CoDing Sequence) MSA is trivial. Yet in practice things can prove more complicated for a variety of simple reasons: unknown gene names, MSAs of domain or partial protein sequence, incomplete database annotation, and the ever faster evolution of genomic resources. Of course, each of these problems can usually be solved manually, on a case by case basis, but altogether they tend to hamper the establishment of automatic procedures for seamlessly connecting the protein and the nucleotide worlds.
When comparing protein and nucleotide sequences, efficient methods exist to either align CDSs using their coding potential (1–3) or thread CDSs onto a pre-established protein sequences (protal2dna: http://bioweb.pasteur.fr/seqanal/interfaces/protal2dna.html and pal2nal (http://www.bork.embl-heidelberg.de/pal2nal/) but all these tools require the user to preprocess the data, gather the appropriate CDSs and make sure these are compatible with any subdomain extracted from the original protein sequence. To the best of our knowledge, no tool is available online to automatically identify the nucleotide sequence (genomic or transcript) associated with a protein partial sequence (domain or fragment) and process it to replace that protein with its bona fide CDS while retaining the original alignment.
We developed a fully automated program named Protogene (PROtein TO GENE) that when given a protein sequence alignment (pairwise or multiple) returns the corresponding CDS alignment. Protogene searches RefSeq (4) and NR with BLAST (5) in order to identify the transcript or genomic sequence(s) most likely to be associated with the original protein sequences. The sequences thus identified are processed by Exonerate (6) in order to extract a portion of CDS matching perfectly the original protein sequence. This CDS is re-introduced within the MSA to replace the original protein. Although every attempt is made to identify the genuine protein CDS, a conservative post-filtering step is still needed to eliminate any sequence that may not have been correctly processed. Protogene is not a gene finding tool and depends entirely on the pre-established proteomes found in RefSeq and NR. We have put the emphasis on robustness and reliability rather than exhaustiveness. In our hands, Protogene returns a bona fide CDS for 95% of the sequences and fails on ∼5%. Whenever the missed sequences matter, it is up to the user to explore the labyrinth of nucleotide databases and discover the glitch that breaks the information chain between the protein and its CDS. Protogene is available on http://www.tcoffee.org/.
METHODS
Server pipeline
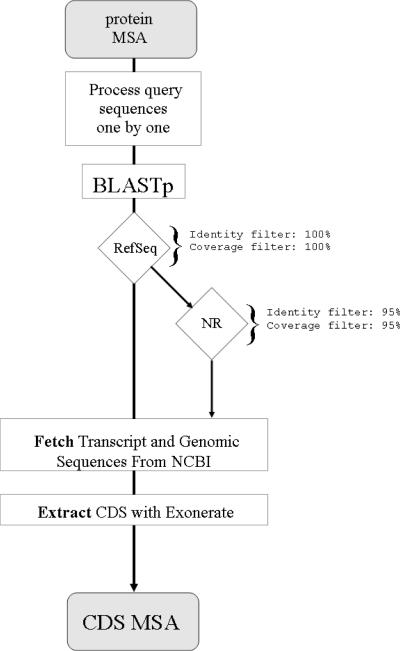
Figure 1 shows Protogene's flow chart. Each sequence within the provided MSA is treated individually. The first step is a BLASTp (5) against the RefSeq (4) protein database. Matches against RefSeq are only accepted when associated with an alignment that displays 100% identity and 100% coverage with the query sequence (i.e. 100% of the query sequence residues aligned with identical residues). Multiple hits with the same score are all kept. If no suitable hit is found in RefSeq, the query sequence is BLASTed against the NR non-redundant protein sequence database, with a lowered acceptance threshold (95% identity, 95% coverage). NCBI EFetch and EUtils utilities (http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html) are then used to fetch the nucleotide sequences associated with the query sequence. Exonerate is finally used to extract a CDS matching perfectly the original protein sequence (be it a full-length protein or just a fragment). Exonerate is able to splice introns and can handle genomic and transcript sequences alike. Mismatches between the query and the CDS are indicated with NNN codons. It is up to the user to decide whether these are sequences errors, polymorphisms or identification errors. The user may also request automatic back-translations using the IUC ambiguity code for unmatched amino acids. CDSs are returned along with some basic annotation including the nucleotide sequence accession number, the source organism and the RefSeq/NR accession number. CDSs where >5% of the nucleotides have had to be replaced with Ns are considered unreliable and discarded with an explicit mention in the output.
Figure 1.
Protogene flow chart sequences are first BLASTed against RefSeq. If no match is found, they are then BLASTed against NR. Nucleotide sequences are fetched from NCBI and processed with Exonerate to yield CDSs that perfectly match the original protein.
It is important to point out that when several proteins from different organisms have a perfect identity with the original query, each corresponding nucleotide sequence is integrated within the final alignment, leaving it to the user to remove the nucleotide sequences he is not interested in (see the Histone example in the next section). To ease this selection, the final alignment is reported in FASTA format. Nucleotide sequences gathered from the NCBI are kept in cache for 2 weeks, thus insuring faster second runs when reanalyzing a dataset with minor modifications.
Distribution
The Protogene server is available from http://www.tcoffee.org/. It relies on a collection of Perl scripts and two external programs: BLAST and Exonerate. BLAST searches against NR and RefSeq are made using the gigablaster service (http://www.igs.cnrs-mrs.fr/).
USING PROTOGENE
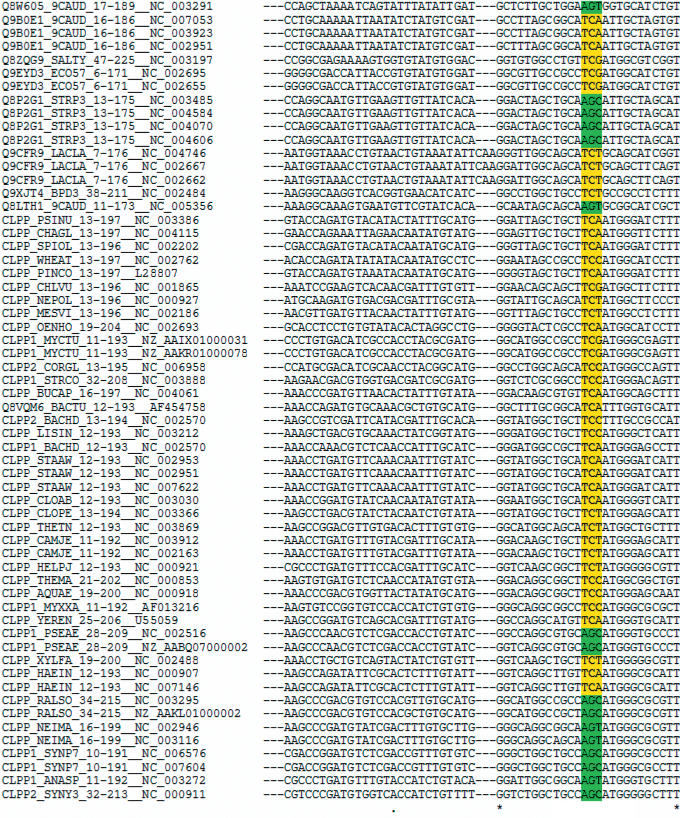
We provide three simple examples of how Protogene can be used to rapidly and efficiently ask simple questions regarding protein sequence conservation at the nucleotide level. The first one is an analysis of the CLP Serine protease family. Serine is the only amino acid coded by two sets of codons (UCN and AGY) that cannot be interconverted by a single point mutation. Class switches, appear however to be very frequent and the question whether they arise from double mutations has been the subject of intense scrutiny and debate (7). Given a collection of protein sequences, Protogene makes it straightforward to analyze the codon conservation of the serine. Figure 2 shows the output obtained after cutting and pasting the Pfam (8) seed MSA of the CLP Serine Protease family (PF00574). This alignment is not made of complete proteins but restricted to the serine protease domain. For each domain, Protogene managed to identify at least one corresponding gene and also reported identical protein sequences coming from closely related genomes. Figure 2 represents the portion of this alignment containing the conserved Serine that is part of the catalytic triad. In a second test, we evaluated Protogene's ability to process a complete eukaryotic domain sequence dataset. For that purpose we selected the 209 human trypsin like serine proteases listed in the SMART database (9) (SM00020). These are protein domains processed using a SMART Hidden Markov Model. Protogene returned the CDSs associated with 253 protein sequences found in RefSeq and NR (157 in RefSeq and 96 in NR). Out of the 209 original human sequences, 4 matched equally well a chimpanzee protein (thus prompting the return of the associated Chimpanzee CDSs) and 66 matched two distinct human entries (thus prompting the return of 66 extra Human CDSs). Of the original sequences 26 could not be associated with an acceptable CDS: 20 did not pass the BLAST step (i.e. no suitable match was found in RefSeq or NR) and the 6 remaining could not be properly processed by exonerate against the nucleotide sequences indicated by the database annotation.
Figure 2.
Protogene output on the CLP Serine Protease family. The Seed MSA of the PFAM profile entry (PFAM PF00574) was processed by Protogene. The portion of the alignment containing the Serine active site classes are indicated in yellow (UCN) and green (AGY).
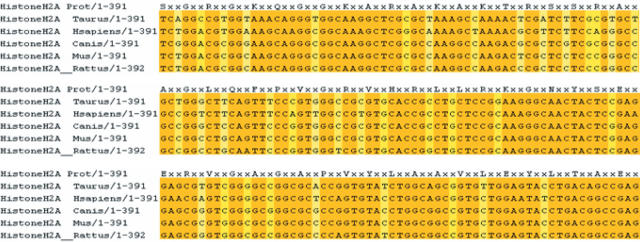
Our third example (Figure 3) addresses the question of nucleotide sequence conservation in the Histone H2A family. Histones are notoriously conserved proteins and in the present case, launching a Protogene analysis on the Human H2A sequence (SwissProt P28001) returned 5 perfect matches in RefSeq, resulting in 5 CDSs being reported: Human, Cow, Rat, Mouse and Dog. The alignment is shown of Figure 3. Such a nucleotide alignment of perfectly conserved protein sequences is ideal for phylogenetic studies or motif discoveries. It is worth pointing out that although it is identical, the Chimpanzee Histone was not reported by Protogene because it is not included in RefSeq. This finding reveals the heavy bias of Protogen toward model systems included in RefSeq. The systematic use of NR rather than RefSeq could help solve this problem, but this would come at the cost of a more complex output.
Figure 3.
Protogene output on the Human H2A Histone protein. The original protein sequence is indicated on the top. Light coloured columns are those not entirely conserved.
CONCLUSION
In this paper we describe Protogene, a web server that makes it possible to turn a protein MSA into the corresponding CDS MSA, using bona fide genomic or transcriptome data. Protogene is meant to be a simple yet powerful data exploration tool. Its purpose is to rapidly ask simple questions, with an emphasis on accuracy and robustness rather than sensitivity.
Acknowledgments
We thank Guy Slater for his advice on the use of Exonerate, NCBI team for their help with EFetch and EUtils tools. We thank Prof. Jean-Michel Claverie (head of IGS) for stimulating discussions and support. The development was supported by CNRS (Centre National de la Recherche Scientifique), Sanofi-Aventis Pharma SA., Marseille-Nice Génopole and the French National Genomic Network (RNG). Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bininda-Emonds O.R. transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinformatics. 2005;6:156. doi: 10.1186/1471-2105-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stocsits R.R., Hofacker I.L., Fried C., Stadler P.F. Multiple sequence alignments of partially coding nucleic acid sequences. BMC Bioinformatics. 2005;6:160. doi: 10.1186/1471-2105-6-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wernersson R., Pedersen A.G. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Slater G.S., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Averof M., Rokas A., Wolfe K.H., Sharp P.M. Evidence for a high frequency of simultaneous double-nucleotide substitutions. Science. 2000;287:1283–1286. doi: 10.1126/science.287.5456.1283. [DOI] [PubMed] [Google Scholar]
- 8.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letunic I., Copley R.R., Pils B., Pinkert S., Schultz J., Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]