Abstract
With the high number of sequences and structures streaming in from genomic projects, there is a need for more powerful and sophisticated annotation tools. Most problematic of the annotation efforts is predicting gene and protein function. Over the past few years there has been considerable progress in automated protein function prediction, using a diverse set of methods. Nevertheless, no single method reports all the information possible, and molecular biologists resort to ‘shopping around’ using different methods: a cumbersome and time-consuming practice. Here we present the Joined Assembly of Function Annotations, or JAFA server. JAFA queries several function prediction servers with a protein sequence and assembles the returned predictions in a legible, non-redundant format. In this manner, JAFA combines the predictions of several servers to provide a comprehensive view of what are the predicted functions of the proteins. JAFA also offers its own output, and the individual programs' predictions for further processing. JAFA is available for use from http://jafa.burnham.org.
INTRODUCTION
The huge influx of sequences from the various genome projects has brought protein function prediction to the forefront of computational molecular biology. Not only are we being overwhelmed by an exponentially increasing number of sequences, but those sequences are getting more diverse as well (1). This increase in unknown sequences has made computational function prediction a priority in bioinformatics research.
Automated protein function prediction is a difficult problem for several reasons. The first is the many aspects of the term function in a biological context. For example, if we identify a protein to be a kinase, this tells us only one aspect of its function which is the molecular aspect: an activity performed at the molecular level. This kinase may also participate in one or more intracellular signaling pathways so another aspect, the cellular process this kinase is involved in, comes into play. Additionally, there may be some organism-level physiological effect: this kinase may be a proto-oncogene, which makes for a third aspect, manifest in the whole organism. A second reason for the difficulty is partial knowledge we often have. In many cases we know only certain aspects of a protein's function. We may know that the protein is a kinase, but not in which cellular pathway or pathways it participates in. Conversely, we may know that a protein plays a role in a specific pathway, but not its molecular function. The latter case does not preclude the protein's participation in other pathways, of which we know nothing about. Also, even within a given functional aspect our knowledge may be incomplete: we may know the protein is a kinase, but we may not know what this kinase phosphorylates. In light of this situation, different computational methods have been devised to increase our knowledge of a protein's function, based on its sequence or, when available, its structure.
Probably the most common method for computational function prediction is that of homology transfer. The function of the query protein is inferred from similarity to another protein, which is well annotated. The use of sequence similarity search tools, such as the BLAST family (2) for function prediction has long been routine. Homology transfer is limited by the requirements that a sequence be found which are (i) well annotated and (ii) has a reasonably high similarity to the query sequence. Nevertheless, significant levels of mis-annotations can occur even at 60% sequence identity (3). Another method of sequence similarity based prediction is by using sequence motifs. In motif based identification, all which is required is to identify a sequence-based feature which can be associated with a function. Molecular function and cellular localization can be well determined using these methods, which are more sensitive than whole sequence homology based predictions. Other methods do not rely on sequence similarity of any kind, but require knowledge of genomic location of the sequence, and the identification of co-evolving genes (4). The rationale is that genes which are proximal on a chromosome, and/or co-evolve, are linked to the same functions. This type of prediction is better in inferring the cellular processes a gene may be associated with, but not so much the molecular function. Other methods for function prediction based on sequence or structure exist, and have been reviewed extensively (5–7).
The motivation for creating a function prediction meta-server is twofold. First, the simple concentration of many predictions together in a legible and concise fashion can be very helpful in interpreting function. Second, as shown above, different prediction methods have different strengths. Combining these strengths may produce a better prediction than any single individual method can. Yet in order to combine, compare and contrast predictions there is a need for a standardized description of function. Natural language is rife with synonyms and ambiguity, making the comparison of predictions difficult. Controlled vocabularies such as the Gene Ontology (GO) (8) and the Enzyme Commission Classification (EC) have been established to eliminate the semantic and contextual ambiguity of the term function. EC and GO standardize the vocabulary, eliminate synonymy and ambiguity. They both represent the terms describing functions in a semi-hierarchical fashion, from the general to the specific. Thus, even if a functional aspect is only partially known, the protein may still be annotated using a less specific term. EC deals only with enzymatic functions. GO covers all of EC, and many more molecular functions. Additionally, GO has ontologies for two other functional aspects: biological process and cellular location. For these reasons, many databases are incorporating GO into their annotations. Consequently, for our meta-server, we have decided to use GO for functional annotation.
JAFA INPUT
The input screen consists of a sequence upload screen, a selection of function prediction servers and their configuration parameters. The user may upload up to 10 sequences, in FASTA format for querying. The limit is set so that the queried servers will not be overloaded.
QUERIED SERVERS
At the time this manuscript is being written, Joined Assembly of Function Annotations (JAFA) queries five function prediction servers: GOFigure (9), GOtcha (10), GOblet (11), InterProScan (12) and PhydBac2 (4). The first three servers predict function using BLAST for locating homologs. The user may choose the BLAST E-value cutoff, and the scanned databases. InterProScan relies on a set of Hidden Markov Models to locate short sequence motifs and longer sequence domains in various domain and motif databases. In a sense, InterProScan is somewhat of an aggregate meta-server by itself. Finally, PhydBac2 uses co-evolution, co-localization and gene-fusion events to predict gene and gene product function. Each of the servers is queried with the input sequence or sequences, and the server output is parsed for GO terms which in turn are compiled and displayed by JAFA.
JAFA OUTPUT
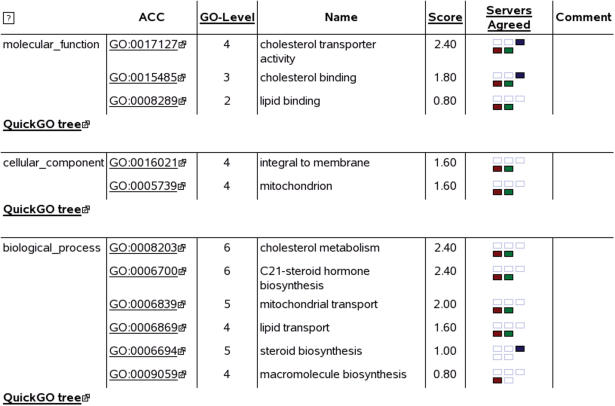
An example output screen is shown in Figure 1. Three tables are given, one for each GO aspect. Each table lists the GO terms, their accession numbers and their level in the GO graph, which is the number of edges along the minimal path from the root node to the term in question. The higher the GO level number, the more specific the prediction. Another column reports the servers agreeing on a particular GO term in color code boxes. Each box is linked to the original server's results, for further study of that server's result. Each table is linked to EBI's QuickGO, which presents the predicted GO terms within the context of the GO hierarchy. An example of terms progressing from a low level (less specific) to a high level (more specific) would be ‘catalytic activity’→‘transferase activity’→‘transferase activity, transferring phosphorous-containg groups’→‘kinase activity’→‘protein kinase activity’→‘protein threonine/tyrosine kinase activity’. The score is simply the product of the GO level multiplied by the fraction of agreeing servers. In this manner the scoring function rewards the prediction of more specific (high level) terms. Note that ‘unknown function’, ‘unknown process’ and ‘unknown cellular compartment’ are special cases of a level-1 ontology term.
Figure 1.
A partial screen shot of JAFA results. The query sequence was the Steroidogenic Acute Regulatory Protein, or StAR, a cholesterol binding and transporting enzyme (Swissprot: STAR_HUMAN). Columns, from left to right, Ontology, ontology type. One of molecular function, cellular location or biological process; ACC, GO accession number, linked to AmiGo, a GeneOntology browser; GO level, distance in vertices from the GO root node; Name, the GO term, in English. Mouse-over for elaboration; Score, the product of the GO level by the ratio of agreeing servers; Servers agreed, servers agreeing upon this GO term. Colored squares represent different servers. Squares are linked to original server results; QuickGO tree, link to a GO graph visualizer, from EBI; Comment, additional comments, mainly for obsolete GO terms.
JAFA also provides all the queried server results in XML and MS-Excel® format, which makes JAFA itself embeddable in other programs. Finally, JAFA also queries the NCBI BLAST server, and provides raw BLAST (2) results for inspection.
CONCLUSIONS AND FUTURE WORK
JAFA provides a compilation of several function prediction resources aimed at serving the life sciences community. Experimental biologists will find JAFA useful for querying unannotated sequences. Computational biologists can use the XML output produced by JAFA to investigate the attributes of different annotation programs. We would like to ask computational biologists who are developing function prediction programs to consider making them available via JAFA as well. Please contact the corresponding author for details.
In the future, we aim to add a structure based prediction methods to JAFA. We are also exploring ways of developing a better consensus scoring method that will reflect the capabilities for the various queried programs. Finally, we are also developing a stand-alone version of JAFA, to be used for more massive data mining efforts.
TECHNICAL ASPECTS
The JAFA user interface was written in Zope (http://zope.org), an open source application server for building content management systems. Biopython (http://biopython.org), an open source toolkit for computational biology was used in many parts of JAFA. PyXLWriter (http://sourceforge.net/projects/pyxlwriter) was used to generate MS-Excel files.
Acknowledgments
We would like to thank Dr Martin Jambon for stimulating discussions. JAFA is funded by NIH grant P01-GM63208. Funding to pay the Open Access publication charges for this article was provided by NIH grant P01-GM63208 and the Burnham Institute for Medical Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg I. Automated function prediction: the genomic challenge. Brief. Bioinform. 2006. accepted for publication. [DOI] [PubMed]
- 2.Altschul S., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian W., Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J. Mol. Biol. 2003;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 4.Enault F., Suhre K., Claverie J.-M. Phydbac ‘Gene Function Predictor’: a gene annotation tool based on genomic context analysis. BMC Bioinformatics. 2005;6:247. doi: 10.1186/1471-2105-6-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whisstock J.C., Lesk A.M. Prediction of protein function from protein sequence and structure. Q Rev. Biophys. 2003;36:307–340. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 6.Rost B., Liu J., Nair R., Wrzeszczynski K.O., Ofran Y. Automatic prediction of protein function. Cell Mol. Life Sci. 2003;60:2637–2650. doi: 10.1007/s00018-003-3114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ofran Y., Punta M., Schneider R., Rost B. Beyond annotation transfer by homology: novel protein-function prediction methods to assist drug discovery. Drug Discov. Today. 2005;10:1475–1482. doi: 10.1016/S1359-6446(05)03621-4. [DOI] [PubMed] [Google Scholar]
- 8.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan S., Situ G., Decker K., Schmidt C.J. GoFigure: automated Gene Ontology annotation. Bioinformatics. 2003;19:2484–2485. doi: 10.1093/bioinformatics/btg338. [DOI] [PubMed] [Google Scholar]
- 10.Martin D.M., Berriman M., Barton G.J. GOtcha: a new method for prediction of protein function assessed by the annotation of seven genomes. BMC Bioinformatics. 2004;5:178. doi: 10.1186/1471-2105-5-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groth D., Lehrach H., Hennig S. GOblet: a platform for Gene Ontology annotation of anonymous sequence data. Nucleic Acids Res. 2004;32:W313–W317. doi: 10.1093/nar/gkh406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zdobnov E.M., Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]