Abstract
A new real-time PCR assay for quantitation of Encephalitozoon intestinalis DNA was developed which used a TaqMan fluorescent probe for specific detection. Serial dilutions of E. intestinalis spore suspensions obtained from tissue culture were used as external standards. The detection limit of the technique was 20 spores per ml, with a good interassay reproducibility (coefficient of variation of 7.1% for the suspension containing 20 spores/ml, 5.0% for the suspension containing 75 spores/ml and below 3.5% for higher concentrations). Quantitative detection of E. intestinalis DNA was similar whether the serial dilutions of spores were made in distilled water or in a stool suspension, allowing the use of the assay for stool specimens. The assay was then applied to 14 clinical specimens from 8 immunocompromised patients with proven E. intestinalis infection. The quantitation of the parasitic burden was achieved in stools, blood, urine, tissue biopsies, and bronchopulmonary specimens. The highest parasitic burdens were noted in stools, urine, and bronchopulmonary specimens, reaching 105 to 106 spores/g or ml. Dissemination of the infection was also evidenced in some patients by demonstration of E. intestinalis DNA in blood and serum. We conclude that real-time PCR is a valuable tool for quantitation of E. intestinalis burden in clinical specimens.
Microsporidia are primitive unicellular eukaryotes that are obligate intracellular parasites of eukaryotic cells from a large number of invertebrate and vertebrate hosts, including primates (15). They lack eukaryotic cytoplasmic organelles such as mitochondria and peroxisomes (19). Two species, Enterocytozoon bieneusi and Encephalitozoon intestinalis, are responsible for most infections in humans, mainly occurring in severely immunocompromised human immunodeficiency virus (HIV)-infected patients. Although less frequent than E. bieneusi, E. intestinalis can be responsible for chronic diarrhea and for systemic infections with dissemination to the kidneys, lungs, and biliary tract (1, 7, 10, 13, 19).
The diagnosis of microsporidiosis relies on direct visualization of spores in clinical specimens by light or fluorescent microscopy (16, 18). Electron microscopy enables Encephalitozoon genus identification (2, 20). Specific PCR is used for E. intestinalis identification (3, 4, 6, 8, 9, 12). Species identification is important as, unlike for E. bieneusi infections, treatment with albendazole is effective for E. intestinalis infections (11).
In this study, we developed a new quantitative PCR assay as a complementary tool for the assessment of E. intestinalis parasitic burden in various clinical specimens.
MATERIALS AND METHODS
Experimental study. (i) Parasites and cultures.
The strain of E. intestinalis used in this study, kindly provided by T. Van Gool (Amsterdam, The Netherlands), was isolated from the stools of an HIV-infected patient (17). It was identified by electron microscopy and specific PCR (5) and maintained in tissue culture with the human glioblastoma cell line U-373-MG (ATCC HTB17) (14). Fresh spores were collected from culture supernatants and centrifuged at 1,500 × g for 5 min. The pellet was resuspended in distilled water and then stored at 4°C until use.
(ii) Dilutions of spores.
Serial dilutions of spores, containing 20 to 5 × 106 spores per ml were prepared in distilled water. Similarly, serial spore dilutions were prepared by using a stool suspension as a diluent to examine the possible effect of stools on the performance of PCR. For this purpose, we used 10 g of stool from a control patient without intestinal parasitic infection, diluted the specimen in 40 ml of distilled water, and used this stool suspension to prepare spore dilutions.
(iii) DNA extraction.
Two hundred microliters of each spore dilution, either in distilled water or in stool suspension, were incubated with 24 μg of lyticase (Roche Molecular Biochemicals, Meylan, France)/ml at 37°C, and then DNA was extracted by using the High Pure PCR template preparation kit (Roche Molecular Biochemicals) according to the manufacturer's instructions. Extracted DNA was resuspended in 200 μl of 10 mM Tris buffer (pH 8.5).
Clinical study. (i) Clinical specimens.
Stored clinical specimens from 8 immunocompromised patients with proven E. intestinalis infection were used. Seven patients were HIV-infected and had already experienced AIDS-defining opportunistic diseases. They were all male homosexuals with CD4 cell counts below 50 per mm3. All had a history of chronic intermittent diarrhea. Fever was also documented in two patients, and cholangitis was documented in one patient. Another patient was HIV-seronegative but received chemotherapy for Hodgkin's lymphoma. His CD4 cell count was 50 per mm3. He presented with fever and bronchopulmonary symptoms but also had abdominal pain. In addition, 10 stool specimens from patients without microsporidiosis and 4 stool specimens from E. bieneusi-infected patients were tested.
(ii) Diagnosis of E. intestinalis infection.
The diagnosis of E. intestinalis infection in these 8 patients relied upon demonstration of typical spores by light and Uvitex 2B microscopy (16, 18) followed by specific PCR (9). A semiquantitative assessment of parasitic burden (microscopy score) was performed by the same trained investigator (C. Sarfati) using Uvitex 2B-stained slides. A score of 0 indicated no spore seen after the observation of 200 microscopic fields at a magnification of ×1,000, a score of 1 was given for rare spores (less than 1 spore per 10 microscopic fields at a magnification of ×1,000), a score of 2 was given for some spores (from 1 spore per 10 fields to 2 spores per field), and a score of 3 was given for numerous spores (more than 2 spores per field).
(iii) DNA extraction from clinical specimens.
For stool specimens, approximately 1 g of formed stool or 1 ml of diarrheic stool was suspended in 4 ml of distilled water and stored at −20°C until use. Two hundred microliters of each stool specimen suspension was thawed and then centrifuged at 1,500 × g for 5 min. The pellet was washed with 200 μl of phosphate-buffered saline (pH 7.2), then centrifuged at 1,500 × g for 5 min, and resuspended in 200 μl of phosphate-buffered saline. Lung biopsy samples were first incubated at 37°C for 4 h with 200 μl of tissue lysis buffer and 40 μl of proteinase K from the Roche Molecular Biochemicals High Pure PCR template preparation kit. For other types of samples (urine, sputum, bronchoalveolar lavage fluid, blood, and serum), 200 μl of the clinical specimens was directly used. Each sample was lysed for 30 min with 24 μg of lyticase/ml at 37°C, and then DNA extraction was performed by using the High Pure PCR template preparation kit according to the manufacturer's instructions. The extracted DNA was resuspended in 200 μl of 10 mM Tris buffer (pH 8.5).
Real-time quantitative PCR.
Several primers and one TaqMan fluorescent probe were designed within the consensus sequence of the E. intestinalis small-subunit rRNA gene (GenBank accession no. U09929) with Primer Express software (Applied Biosystems, Foster City, Calif.). In a preliminary experiment, each was tested for E. intestinalis performance in a real-time PCR. The primer set FEI1 (5′-GCAAGGGAGGAATGGAACAGAACAG-3′) and REI1 (5′-CACGTTCAGAAGCCCATTACACAGC-3′) was selected, amplifying a 127-bp fragment between positions 988 and 1114. TheTaqMan fluorescent probe (5′-CGGGCGGCACGCGCACTACGATA-3′ at positions 1036 to 1058) was labeled at the 5′ end with 6-carboxyfluorescein and at the 3′ end with 6-carboxytetramethylrhodamine. Real-time quantitative PCR was performed on an ABI Prism 7700 sequence detection system in a 50-μl volume containing 5 mM MgCl2, 1× Applied Biosystems TaqMan buffer A, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 400 μM dUTP, 0.2 μM FEI1 primer, 0.2 μM REI1 primer, 0.4 μM TaqMan fluorescent probe, 0.5 U of Applied Biosystems AmpErase uracil DNA glycosylase, 1.25 U of Applied Biosystems AmpliTaq Gold, and 5 μl of spore-extracted standard DNA or patient sample DNA. After 2 min at 50°C and 10 min at 95°C, amplification consisted of 45 cycles of 15 s of denaturation at 95°C followed by 1 min of annealing and extension at 65°C. Results were expressed as threshold cycle (Ct) values corresponding to the cycle at which PCR enters the exponential phase, i.e., when the 6-carboxyfluorescein fluorescence exceeds 10 times the standard deviation (SD) of the mean baseline emission for cycles 3 to 15.
Each set of PCR assays comprised serial dilutions of E. intestinalis spores as external standards. Each dilution was tested in triplicate. A standard curve was established between the Ct values and the spore numbers by using the Applied Biosystems sequence detection system software, version 1.6.3. For each clinical specimen, also tested in triplicate, this standard curve was used to estimate the spore number by interpolation of the Ct value obtained by real-time PCR. Negative controls (distilled water) were included in each set of experiments.
Statistical analysis.
All statistical analyses were performed with GraphPad Prism, version 3.02 for Windows (GraphPad Software, San Diego, Calif.). The means ± SDs of Ct values from dilutions of the spore suspension were calculated to assess interassay reproducibility of real-time PCR by using 5 independent experiments. A Student test was performed for comparison of Ct values for the suspension containing 20 spores per ml versus negative controls. The relation between the decimal logarithm of spore number and Ct values was assessed performing standard ordinary least-square linear regression. Slope equality for real-time PCR experiments performed with spores diluted in distilled water or in a stool suspension was assessed with an F test.
RESULTS
Sensitivity and reproducibility of real-time PCR.
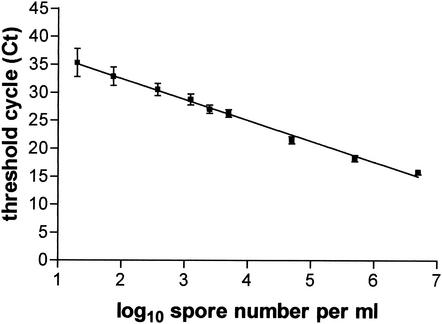
The sensitivity and reproducibility of real-time PCR was assessed by repeated testing of serial dilutions of an E. intestinalis spore suspension from 20 to 5 × 106 spores per ml in 5 independent real-time PCR experiments. The mean standard curve is represented in Fig. 1. A significant amplification was obtained with the suspension containing 20 spores/ml (Ct = 35.32 ± 2.35), compared to a Ct of >45 for negative controls (P < 0.0001). The parameters of the regression curve of Ct values versus the decimal logarithm of spore numbers were as follows: r2 = 0.984 ± 0.009; slope, P = −3.692 ± 0.204; y intercept, Ct = 37.11 ± 1.31. The interassay coefficient of variation of the Ct values was calculated for each spore dilution: it was 7.1% for the suspension containing 20 spores/ml, 5.0% for the suspension containing 75 spores/ml, and below 3.5% for suspensions containing higher concentrations.
FIG. 1.
Correlation between Ct values (means ± SD) and number of spores per milliliter from 5 independent real-time PCR experiments on serial dilutions of E. intestinalis spores.
Similarly, real-time PCRs were performed in parallel on DNA extracted from serial dilutions of spores prepared with a stool suspension or with distilled water (data not shown). The slope of the resulting standard curve was similar whether the spore suspension was diluted in distilled water or in stool suspension, according to an F test (F = 2.9, P = 0.10).
Clinical specimens.
The results of real-time PCR performed on samples from patients with microsporidiosis and in uninfected controls are presented in Table 1. No amplification was observed for samples obtained from E. bieneusi-infected patients or from patients without microsporidiosis. In patients infected with E. intestinalis, parasitic burden could be determined in various types of samples taken at the same time, including tissue biopsy samples and blood. The highest parasitic burdens were noted in stools, urine, and bronchopulmonary specimens, reaching 105 to 106 spores/g or ml. In two patients, dissemination of infection was assessed by the positivity of PCR in serum or whole blood.
TABLE 1.
E. intestinalis DNA quantitation by real-time PCR in various clinical specimens from 8 E. intestinalis-infected patients, 4 E. bieneusi-infected patients, and 10 patients without microsporidiosis
| Patient no.f | Cause of immunodeficiency | Specimen source(s) | Microscopya result | MSb | PCRc result | Real-time PCR result (mean ± SD) |
|---|---|---|---|---|---|---|
| 1 | HIV infection | Stool | Presence | 3 | Positive | 4.6 × 105 ± 0.6 × 105/g |
| 2 | HIV infection | Urine | Presence | 3 | Positive | 8.0 × 105 ± 0.0 × 105/ml |
| 3 | HIV infection | Urine | Presence | 3 | Positive | 1.4 × 106 ± 0.0 × 106/ml |
| Serum | NDd | ND | Positive | 1.0 × 102 ± 0.1 × 102/ml | ||
| 4 | HIV infection | Urine | Presence | 2 | Positive | 1.1 × 103 ± 0.1 × 103/ml |
| 5 | HIV infection | Stool | Presence | 2 | Positive | 1.1 × 104 ± 0.1 × 104/g |
| 6 | HIV infection | Whole blood | Presence | 1 | Positive | 4.6 × 102 ± 0.5 × 102/ml |
| 7 | Hodgkin's lymphoma | Pleural fluid | Absence | 0 | Negative | Negative |
| Lung biopsy | Absence | 0 | Positive | 1.4 × 104 ± 0.1 × 104/g | ||
| 8 | HIV infection | Induced sputum | Presence | 2 | Positive | 1.6 × 105 ± 0.1 × 105/ml |
| BALe fluid | Presence | 2 | Positive | 1.3 × 105 ± 0.1 × 105/ml | ||
| Stool | Presence | 2 | Positive | 1.4 × 105 ± 0.1 × 105/ml | ||
| Urine | Presence | 2 | Positive | 8.3 × 104 ± 0.7 × 104/ml | ||
| Whole blood | ND | ND | Negative | Negative | ||
| 9 | HIV infection | Stool | Presence | 2 | Negative | Negative |
| 10 | HIV infection | Stool | Presence | 2 | Negative | Negative |
| 11 | HIV infection | Stool | Presence | 3 | Negative | Negative |
| 12 | HIV infection | Stool | Presence | 3 | Negative | Negative |
| 13-22 | None | Stools | Absence | 0 | Negative | Negative |
Presence or absence of spores on microscopic examination with Uvitex 2B and Weber's modified trichrome stains (16, 18).
MS, microscopy score (semiquantitative assessment of parasitic burden made by using Uvitex 2B-stained slides observed at a magnification of ×1,000: 0 indicated no spore, 1 indicated less than 1 spore per 10 microscopic fields, 2 indicated between 1 spore per 10 fields and 2 spores per field, and 3 indicated more than 2 spores per field).
E. intestinalis conventional PCR is described in reference 9.
ND, not done.
BAL, bronchoalveolar lavage.
Patients 1 to 8 (n = 8) were infected with E. intestinalis. Patients 9 to 12 (n = 4) were infected with E. bieneusi. Patients 13 to 22 (n = 10) did not have microsporidiosis.
DISCUSSION
Specific PCR is considered to be a sensitive and reliable method for the diagnosis of microsporidiosis (8). However, this technique is exposed to a significant risk of contamination and does not allow quantitation of parasitic burden. Real-time PCR offers the advantage of no post-PCR handling, thus reducing the risk of contamination with previously amplified products. Additionally, the use of a TaqMan fluorescent probe rather than SybrGreen detection better guaranties the specificity of the measured signal.
In this study, we described a new real-time PCR technique for quantitative detection of E. intestinalis in biological samples. Using spores obtained from tissue culture, we first showed that this technique was reliable for parasite quantitation over a wide range of spore concentrations diluted either in distilled water or in a stool suspension. In both cases, the lower threshold of detection was 20 spores/ml.
When applied to clinical samples, we showed that none of the specimens obtained from patients without microsporidiosis (n = 10) or from E. bieneusi-infected patients (n = 4) was positive by E. intestinalis real-time PCR. For patients infected with E. intestinalis, however, real-time PCR was successfully applied to several types of clinical samples, including stools, urine, sputum, tissue biopsy, blood, and serum, and allowed to estimate the level of infection in body fluids and tissues. Although only a limited number of clinical specimens was studied, our results confirmed that E. intestinalis infection predominantly involves the digestive tract, the urinary tract, and the lungs, since we found high parasite burdens in stools, urine, and sputum. Dissemination was also evidenced by the demonstration of E. intestinalis DNA in the blood of two patients, although at a lower level, as previously described (4).
In terms of routine laboratory practice, E. intestinalis real-time PCR can be performed by using any type of clinical specimen. Stool should be preferentially examined, since intestinal localization is predominant, but other samples can be considered to assess extraintestinal dissemination of infection. Real-time PCR offers the advantage of not requiring any post-PCR processing for PCR product detection, which decreases turnaround time and the risk of amplimer contamination. Additionally, the use of a simple commercially available DNA extraction kit enhances the practicability of the assay for clinical laboratories, and the entire assay, including DNA extraction, requires less than 5 h.
In conclusion, this method is a useful tool for quantitation of E. intestinalis burden in vitro and in clinical specimens. Quantitative determination of parasite burden in clinical specimens from E. intestinalis-infected patients can provide valuable information about the natural history of the infection and could also be used to assess the effect of different treatments in vivo.
Acknowledgments
This study was supported in part by the Centre d'Etude et de Recherche en Infectiologie.
We thank A. M. Deluol, from the Hôpital Saint-Antoine (Assistance Publique-Hôpitaux de Paris), who provided us with blood from an HIV-infected patient with disseminated E. intestinalis infection, L. Kitanovski from the University Children's Hospital of Ljubljana (Ljubljana, Slovenia), who provided us with pulmonary tissue from a patient with Hodgkin's lymphoma and disseminated E. intestinalis infection, and Y. J. F. Garin for helpful discussions and assistance with statistical analysis.
REFERENCES
- 1.Cali, A., D. P. Kotler, and J. M. Orenstein. 1993. Septata intestinalis n.g., n.sp., an intestinal microsporidian associated with chronic diarrhea and dissemination in AIDS patients. J. Protozool. 40:101-112. [DOI] [PubMed] [Google Scholar]
- 2.Canning, E. U. 1993. Microsporidia, p. 299-385. In J. P. Kreier and J. R. Baker (ed.), Parasitic protozoa, 2nd ed., vol 6. Academic Press, New York, N.Y.
- 3.David, F., A. R. J. Schuitema, C. Sarfati, O. Liguory, R. A. Hartskeerl, F. Derouin, and J. M. Molina. 1996. Detection and species identification of intestinal microsporidia by polymerase chain reaction in duodenal biopsies from human immunodeficiency virus-infected patients. J. Infect. Dis. 174:874-877. [DOI] [PubMed] [Google Scholar]
- 4.Franzen, C., A. Muller, P. Hartmann, M. Kochanek, V. Diehl, and G. Fatkenheuer. 1996. Disseminated Encephalitozoon (Septata)intestinalis infection in a patient with AIDS. N. Engl. J. Med. 335:1610-1611. [DOI] [PubMed] [Google Scholar]
- 5.Hartskeerl, R. A., T. van Gool, A. R. J. Schuitema, E. S. Didier, and W. J. Terpstra. 1995. Genetic and immunological characterization of the microsporidian Septata intestinalis Cali, Kotler and Orenstein, 1993: reclassification to Encephalitozoon intestinalis. Parasitology 110:277-285. [DOI] [PubMed] [Google Scholar]
- 6.Kock, N. P., H. Petersen, T. Fenner, I. Sobottka, C. Schmetz, P. Deplazes, N. J. Pieniazek, H. Albrecht, and J. Schottelius. 1997. Species-specific identification of microsporidia in stool and intestinal biopsy specimens by the polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 16:369-376. [DOI] [PubMed] [Google Scholar]
- 7.Kotler, D. P., and J. M. Orenstein. 1999. Clinical syndromes associated with microsporidiosis, p. 258-292. In M. Wittner and L. M. Weiss (ed.), The microsporidia and microsporidiosis. ASM Press, Washington, D.C.
- 8.Liguory, O., F. David, C. Sarfati, A. R. J. Schuitema, R. A. Hartskeerl, F. Derouin, J. Modaï, and J. M. Molina. 1997. Diagnosis of infections caused by Enterocytozoon bieneusi and Encephalitozoon intestinalis using polymerase chain reaction in stool specimens. AIDS 11:723-726. [DOI] [PubMed] [Google Scholar]
- 9.Liguory, O., S. Fournier, C. Sarfati, F. Derouin, and J. M. Molina. 2000. Genetic homology among thirteen Encephalitozoon intestinalis isolates obtained from human immunodeficiency virus-infected patients with intestinal microsporidiosis. J. Clin. Microbiol. 38:2389-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molina, J. M., E. Oksenhendler, B. Beauvais, C. Sarfati, A. Jaccard, F. Derouin, and J. Modaï. 1995. Disseminated microsporidiosis due to Septata intestinalis in patients with AIDS: clinical features and response to albendazole therapy. J. Infect. Dis. 171:245-249. [DOI] [PubMed] [Google Scholar]
- 11.Molina, J. M., C. Chastang, J. Goguel, J. F. Michiels, C. Sarfati, I. Desportes-Livage, J. Horton, F. Derouin, and J. Modaï. 1998. Albendazole for treatment and prophylaxis of microsporidiosis due to Encephalitozoon intestinalis in patients with AIDS: a randomized double-blind controlled trial. J. Infect. Dis. 177:1373-1377. [DOI] [PubMed] [Google Scholar]
- 12.Ombrouck, C., L. Ciceron, S. Biligui, S. Brown, P. Marechal, T. van Gool, A. Datry, M. Danis, and I. Desportes-Livage. 1997. Specific PCR assay for direct detection of intestinal microsporidia Enterocytozoon bieneusi and Encephalitozoon intestinalis in fecal specimens from human immunodeficiency virus-infected patients. J. Clin. Microbiol. 35:652-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orenstein, J. M., D. T. Dieterich, and D. P. Kotler. 1992. Systemic dissemination by a newly recognized intestinal microsporidia species in AIDS. AIDS 6:1143-1150. [DOI] [PubMed] [Google Scholar]
- 14.Santillana-Hayat, M., C. Sarfati, S. Fournier, F. Chau, J. M. Molina, and F. Derouin. 2002. Effects of chemical and physical agents on viability and infectivity of Encephalitozoon intestinalis using cell culture and flow cytometry. Antimicrob. Agents Chemother. 46:2049-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprague, V., J. J. Becnel, and E. I. Hazard. 1992. Taxonomy of the phylum Microspora. Crit. Rev. Microbiol. 18:285-395. [DOI] [PubMed] [Google Scholar]
- 16.Van Gool, T., F. Snijders, P. Reiss, J. K. Eeftinck Schattenkerk, M. A. van den Bergh Weerman, J. F. Bartelsman, J. J. Bruins, E. U. Canning, and J. Dankert. 1993. Diagnosis of intestinal and disseminated microsporidial infections in patients with HIV by a new rapid fluorescence technique. J. Clin. Pathol. 46:694-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Gool, T., E. U. Canning, H. Gilis, M. A. van den Bergh Weerman, J. K. Eeftinck Schattenkerk, and J. Dankert. 1994. Septata intestinalis frequently isolated from stool of AIDS patients with a new cultivation method. Parasitology 109:281-289. [DOI] [PubMed] [Google Scholar]
- 18.Weber, R., R. T. Bryan, R. L. Owen, C. M. Wilcox, L. Gorelkin, and G. S. Visvesvara. 1992. Improved light-microscopical detection of microsporidia spores in stool and duodenal aspirates. N. Engl. J. Med. 326:161-166. [DOI] [PubMed] [Google Scholar]
- 19.Weber, R., R. T. Bryan, D. A. Schwartz, and R. Owen. 1994. Human microsporidial infections. Clin. Microbiol. Rev. 7:426-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber, R., D. A. Schwartz, and P. Deplazes. 1999. Laboratory diagnosis of microsporidiosis, p. 315-362. In M. Wittner and L. M. Weiss (ed.), The microsporidia and microsporidiosis. ASM Press, Washington, D.C.