Abstract
Pulsed-fieldgel electrophoresis (PFGE) is the most common genotypic method used in reference and clinical laboratories for typing methicillin-resistant Staphylococcus aureus (MRSA). Many different protocols have been developed in laboratories that have extensive experience with the technique and have established national databases. However, the comparabilities of the different European PFGE protocols for MRSA and of the various national MRSA clones themselves had not been addressed until now. This multinational European Union (EU) project has established for the first time a European database of representative epidemic MRSA (EMRSA) strains and has compared them by using a new “harmonized” PFGE protocol developed by a consensus approach that has demonstrated sufficient reproducibility to allow the successful comparison of pulsed-field gels between laboratories and the tracking of strains around the EU. In-house protocols from 10 laboratories in eight European countries were compared by each center with a “gold standard” or initial harmonized protocol in which many of the parameters had been standardized. The group found that it was not important to standardize some elements of the protocol, such as the type of agarose, DNA block preparation, and plug digestion. Other elements were shown to be critical, namely, a standard gel volume and concentration of agarose, the DNA concentration in the plug, the ionic strength and volume of running buffer used, the running temperature, the voltage, and the switching times of electrophoresis. A new harmonized protocol was agreed on, further modified in a pilot study in two laboratories, and finally tested by all others. Seven laboratories' gels were found to be of sufficiently good quality to allow comparison of the strains by using a computer software program, while two gels could not be analyzed because of inadequate destaining and DNA overloading. Good-quality gels and inclusion of an internal quality control strain are essential before attempting intercenter PFGE comparisons. A number of clonally related strains have been shown to be present in multiple countries throughout Europe. The well-known Iberian clone has been demonstrated in Belgium, Finland, France, Germany, and Spain (and from the wider HARMONY collection in Portugal, Slovenia, and Sweden). Strains from the United Kingdom (EMRSA-15 and -16) have been identified in several othercountries, and other clonally related strains have also been identified. This highlights the need for closer international collaboration to monitor the spread of current epidemic strains as well as the emergence of new ones.
Methicillin-resistant Staphylococcus aureus (MRSA) continues to be a major cause of nosocomial infection throughout Europe, with the exception of certain countries such as those of Scandinavia and The Netherlands, where the incidence remains relatively low (15, 33). Typing plays an important role in understanding the epidemiology of MRSA and evaluating the effectiveness of infection control and antimicrobial prescribing measures. The many different methods employed for MRSA typing include antibiograms and chemical resistograms, phage typing, ribotyping, pulsed-field gel electrophoresis (PFGE) (26), and PCR-based methods (14, 31, 32).
The requirements for any typing system include typeability, reproducibility, discriminatory power, stability, and ease of interpretation and use (17, 24). One of the first efforts towards standardization of a typing technique was seen with phage typing of S. aureus by the International Union of Microbiological Societies (IUMS) subcommittee for staphylococcal typing, which combined the experimental phages from different laboratories in order to define a set that would type most of the then-current epidemic strains of MRSA (21). However, in recent years, phage typing has fallen out of favor due to the increasing number of nontypeable isolates encountered in some countries, the costs of quality assurance, and a perceived lack of reproducibility in some laboratories (4, 21).
PFGE, by far the most widespread molecular typing tool in developed countries, is considered to be the method of choice for DNA fingerprinting of MRSA and other bacterial pathogens (16). However, despite the recent explosive increase in PFGE use, there have been a large variety of published PFGE protocols in the literature and few successful attempts to standardize or harmonize them, making intercenter reproducibility seem a rather distant goal. In 1996, a study by Cookson et al. (11), involving three laboratories in Belgium, the United Kingdom, and the United States, examined 12 different MRSA strains with two different types of electrophoretic apparatus and different protocols as optimized in each center. Although there were few disagreements in recognizing similar strains, there was a lack of interlaboratory reproducibility of the electrophoretic patterns themselves. Two of these laboratories then joined with 10 others in 10 countries in 1998 (30) to examine a well-designed collection of MRSA strains by using their own established in-house PFGE protocols, as well as a recently standardized commercial kit. A number of laboratories had difficulties with the protocol for the commercial kit, while comparison of the different in-house protocols again showed poor reproducibility.
HARMONY is a European Union (EU)-funded project, one of whose main goals is the harmonization of typing methods with respect to nosocomial pathogens. The HARMONY typing group originally comprised 12 laboratories in 10 countries (Belgium, Denmark, Finland, France, Germany, Greece, The Netherlands, Spain [two laboratories], Sweden [two laboratories], and the United Kingdom). The primary aims of the group were to establish a collection of European epidemic and other important MRSA strains and to develop, if possible, a standardized protocol for PFGE of MRSA to facilitate international comparisons and tracking of isolates. Laboratories in other countries, including Ireland, Poland, Portugal, Scotland, and Slovenia, have also contributed to the collection and database.
The participating laboratories had already established their own MRSA PFGE databases and were concerned that a standardized protocol might result in inferior discrimination for their national strains. DNA preparation, for example, was perceived to be optimal in each of the laboratories, with different protocols that had been developed over many years. In addition, methods described in the literature often lacked the detail to be reproduced accurately, and the group was not convinced that a standardized protocol could produce optimal discrimination for differing MRSA strains around the EU or was necessary. Thus, it was not possible to impose a centralized standardized protocol as has been the case for food-borne pathogens in the United States (25).
We describe here the two-phase process that was negotiated within the group and resulted in a new, harmonized PFGE typing protocol, which identified those parts of the protocol requiring standardization. The outcome was higher intercenter reproducibility, local acceptability, and the establishment of a web-based database of harmonized MRSA SmaI restriction patterns that has now become the basis for the IUMS global MRSA database. We will describe in a future paper other techniques that have been used to further validate European strain types.
MATERIALS AND METHODS
Strains.
The participants sent to the coordinating laboratory (Laboratory of Hospital Infection, Central Public Health Laboratory) their prototypic epidemic MRSA (EMRSA) strains (defined as spreading within and between several hospitals) or important MRSA strains (defined as causing significant infections in one hospital), as well as some of the more important variants of these, along with their phenotypic and genotypic characteristics and the strain designations as determined in the originating laboratories. The initial collection of 36 EMRSA strains, selected to represent the spectrum of PFGE types submitted from seven countries, along with 13 subtypes of some of these and the reference standard, NCTC 8325, was sent to all participants with instructions to run 21 of the most prevalent types identified (Table 1) in a specified order with their own in-house and the initial harmonized protocols.
TABLE 1.
Characteristics of the 21 prototype strains of EMRSAa
| Strain | Isolate no. | Yr (country) first isolated | City or region | Outbreaks | PFGE profile | Phage typeb | Antibiogramc | Resistogramc | Toxin gene(s) detected | Urease production |
|---|---|---|---|---|---|---|---|---|---|---|
| Belgium E1 | 97S96 | 1990 | Brussels | >79 hospitals, 1990-1997 | A1 | 77, 83A, 84 | CEGKMNRST | Cd/EB/PI/MC/PMA | sea | + |
| Belgium E2 | 97S99 | 1992 | Brussels | >60 hospitals, 1992-1997 | L1 | 84, 85 | CEMS | sec, seg, sei | + | |
| Belgium E3 | 97S101 | 1995 | Antwerp | 17 hospitals, 1992-1997 | C1 | 29 | CKMRST | Cd/EB/P1 | seg, sei | + |
| Finland E1 | 61974 | 1992 | Helsinki | >10 hospitals | C2 | 75, 77, 83A, 83C | CEGKMS | seg, sei | + | |
| Finland E5 | 75916 | 1995 (Turkey) | Helsinki | 8 hospitals | B2 | 29, 52, 75, 77, 83A, 83C | CEGKMMuNS | Cd/PI | sea, seg, sei, tst | + |
| Finland E24 | 98541 | 1998 | Lohja | 6 hospitals | G1 | 100: NTd | CEGKMNST | Cd/EB/PI/MC/PMA | − | − |
| Finland E10 | 62176 | 1992 | Kotka | 8 hospitals | A4 | 29, 47, 54, 75, 77, 84, 85 | CEGKMNRST | Cd/EB/PI/MC/PMA | sea | + |
| Finland E7 | 54518 | 1990 (Italy) | Turku | 4 hospitals | A2 | 100:NT | CEGKMNRSTTei | Cd/EB/PI/MC/PMA | sea | + |
| France A | 162 | 1993 (Belgium) | Ghent | Yes | A5 | 85, 90, 932 | CEKMNS | sea | + | |
| France B | 97121 | 1996 | Paris | Yes | H1 | 100: NT | CKMN | sea (+sed, sej, or −) | + | |
| France C | BM10828 | 1993 | Bordeaux | Yes | A6 | 47, 53, 54, 75, 77, 83A, 84, 85, D16 | CEFGKMNRSTTei | Cd/EB/PI/MC/PMA | sea | + |
| Greece 1 | 3680 | 1994 | Athens | >3 hospitals | F1 | 84, 90, 932 | CEFGKMNRST | Cd/MC/PMA | sea | + |
| Spain E1 | 5 | 1989 | Seville | 11 regions | A1 | 29, 54, 77, 84, 85 | CEFGKMNRST | Cd/EB/PI/MC/PMA | sea | + |
| North German I | 134/93 | 1993 | Gottingen | 28 hospitals, 1998 | A3 | 77, 932 | CEFGKMRST | Cd/ASA/EB/PI | −(sea, seb) | + |
| South German II | 1155-2/98 | 1998 | Oberhausen | 35 hospitals, 1998 | D1 | 29 | CEGKMNS | Cd/EB/PI/MC/PMA | seg, sei | + |
| Hannover III | 1000/93 | 1993 | Hannover | 29 hospitals, 1998 | 11 | 6, 47, 54, 75, 77 | CEGKMNST | Cd/ASA/MC/PMA | seb | + |
| Berlin IV | 825/96 | 1996 | Berlin | 39 hospitals, 1998 | K1 | 100: NT | CEKMNS | seg, sei | + | |
| United Kingdom EMRSA-1 | NCTC 11939 | Early 1980s | >100 hospitals | 11 | 6, 83A, 85, 88A, 932 | EGKMST | Cd/EB/PI/MC/PMA | sea | + | |
| United Kingdom EMRSA-3 | M307 | 1980s | >100 hospitals | E1 | 75, 83A, 932 | EGKMNST | Cd/EB/PI/MC/PMA | seg, sei | + | |
| United Kingdom EMRSA-15 | 90/10685 | 1991 | Southeast England | >100 hospitals | M1 | 75+− | CEMe | sec, seg, sei | − | |
| United Kingdom EMRSA-16 | 96/32010 | 1993 | Kettering | >100 hospitals | B1 | 29, 52, 75, 77, 83A, 83C | CEKMN | Cd/EB/P1 | sea, seg, sei, tst | + |
Subtypes may vary to a certain extent.
Underlining indicates results at 100 times the routine test dilution.
Cd, cadmium acetate; ASA, sodium arsenate; EB, ethidium bromide; PI, propamide isothionate; MC, mercuric chloride; PMA, phenylmercuric acetate; C, ciprofloxacin; E, erythromycin; F, fusidic acid; G, gentamicin; K, kanamycin; M, methicillin; Mu, mupirocin; N, neomycin; R, rifampin; S, streptomycin; T, tetracycline; Tei, teicoplanin. All strains were sensitive to pristinamycin and vancomycin. Underlining indicates raised teicoplanin MICs.
NT, nontypeable.
Most common pattern associated with this strain.
Study phase I.
PFGE was performed in the coordinating laboratory on all isolates submitted, using a protocol determined by the group to give the optimum separation of the DNA fragments based on a previous study (31). This initial harmonized protocol had a long run time (30 h) with a pulsing switch time of 1 to 80 s in a 1.2% gel with 0.5× Tris-borate-EDTA (TBE) circulated at 12°C (17). Lambda or NCTC 8325 was permitted as a reference standard in this first phase, depending on what each center was already using. Gels were analyzed by visual interpretation of the banding patterns according to the criteria of Tenover et al. (27). They were also analyzed with GelCompar software (Applied Maths, Kortrijk, Belgium), using the Dice coefficient, and represented by unweighted pair grouping by mathematical averaging (UPGMA) with 1% band tolerance and 0.5% optimization settings. A cutoff of 80% was applied to all dendrograms generated, above which strains were considered to be closely related; this was previously found to correlate with one to six band differences on visual examination (23).
In the initial phase, the HARMONY group decided that it was not necessary to standardize the DNA block preparation or digestion, the concentration and size (dimension to volume ratio) of the gel, or the volume of the buffer used. Table 2 lists the details of the protocols used in the 10 laboratories in eight countries. Each center thus sent to the coordinating laboratory the gel images produced by both in-house and initial harmonized protocols either as TIFF files or Polaroid photographs, along with the results of any analyses performed using their in-house procedures. Clusters of similar strains as identified by each participant, comments, problems, and any other observations were collated and reanalyzed at the coordinating laboratory. These observations were reviewed at a subsequent meeting (see Results).
TABLE 2.
Details of in-house protocols for PFGE of MRSA strainsa
| Study center | Pulse switches | Total run time (h) | Agarose (%) | Buffer (TBE) | Temp (°C) | Reference standard |
|---|---|---|---|---|---|---|
| 1b | 1-80 s | 30 | 1.2 | 0.5× | 12 | Lambda |
| 2 | 5-15 s for 10 h, 15-45 s for 15 h | 25 | 1.0 | 0.5× | 14 | NCTC 8325 |
| 3 | 5-60 s | 23 | 1.0 | 0.5× | 14 | Lambda and NCTC 8325 |
| 4 | 5-15 s for 7 h, 15-60 s for 19 h | 26 | 1.0 | 1× | 14 | NCTC 8325 |
| 5 | 1-45 s | 20 | 1.2 | 0.5× | 10 | NCTC 8325 |
| 6 | 5-60 s | 23 | 1.0 | 0.5× | 14 | Lambda and NCTC 8325 |
| 7 | 5-60 s | 23 | 1.0 | 0.5× | 14 | Lambda and NCTC 8325 |
| 8 | 0.5-5.5 s for 2 h, 7.5-27.5 s for 18 h, 50-65 s for 1 h | 21 | 1.0 | 0.5× | 14 | Lambda |
| 9 | 5-50 s | 24 | 1.0 | 0.5× | 12 | Lambda |
| 10 | 5-15 s for 10 h, 15-45 s for 12 h | 22 | 1.0 | 0.5× | 14 | Lambda |
Center 8 used the CHEF DR-III apparatus; all others used the CHEF DR-II. Laboratories 1 and 2 additionally used the CHEF Mapper, and laboratory 3 also used the CHEF DR-III. In phase I, the images were received as TIFF files, with one exception, which was received as a Polaroid photograph. All images for phase II were received as TIFF files.
Initial harmonized protocol as determined by the group.
The entire collection of MRSA strains was characterized by phage typing at the routine test dilution and 100 times the routine test dilution (5, 20), antimicrobial susceptibility testing against a panel of 14 antibiotics according to the British Society of Antimicrobial Chemotherapy guidelines (7, 8), toxin gene carriage by use of a multiplex PCR assay (18), and urease production by use of Christensen's urea slopes incubated for 5 days at 37°C (3). The collection was additionally analyzed by resistogram typing against a panel of six chemicals and dyes (28, 29). See the footnotes to Table 1 for further details.
Study phase II.
Following phase I, the majority of the group were found to favor the Belgian protocol over all of the others. However, the coordinating laboratory and Belgian laboratory modified the latter's protocol to improve the resolution of the higher-molecular-weight bands observed in the longer initial harmonized protocol. This reduced the overall run time to less than 24 h, which was considered critical by several laboratories. The new protocol was tested once again on the same set of 21 prototypic MRSA strains as for phase I by each participant.
A number of other parameters were standardized at this stage (see Results), but other aspects, such as DNA block preparation and plug digestion, were not considered to have an impact on the final result. However, a standard protocol, including all parameters, is included in this paper (and on the HARMONY website at http://www.phls.co.uk/International/Harmony/microtyping.htm) for new workers to the field.
HARMONY PFGE protocol.
DNA purification was performed as for each laboratory's in-house protocol. Otherwise, the group recommended the following protocol, which was adapted from the original protocols used in laboratories 1 and 2.
The MRSA suspension incorporated into the agarose block was standardized to a density equivalent to approximately 9 × 108 cells/ml for standard blocks (1 mm thick) or 3 × 109 cells/ml for ultrathin blocks (0.5 mm thick; used by the Belgian center but not available commercially) mixed with an equal volume of a 2% low-melting-point agarose and poured into the block molds. Plugs (2- to 4-mm portions of a block ideally freshly prepared, but they can be kept for up to 6 months by storing in Tris-EDTA buffer at 4°C) for each isolate were equilibrated in SmaI buffer at 4°C for 15 to 30 min prior to digestion and then covered with 100 μl of SmaI reaction buffer containing 20 U of SmaI restriction enzyme (10 U for plugs from ultrathin molds). The reaction tubes were mixed gently and incubated at the temperature recommended by the manufacturer (usually 25 to 30°C) for at least 4 h or overnight.
A 1% gel was prepared by using molecular grade agarose in 150 ml of 0.5× TBE and poured into a freshly cleaned gel-casting frame (20.5 by 14 cm) once the agarose had cooled to 60°C. The gel tray and comb were wiped with alcohol and a lint-free tissue immediately prior to pouring the agarose in order to avoid dust particles, which can cause speckling in the gel. Once set, the gel was positioned centrally in the contour-clamped homogeneous electric field (CHEF) (Bio-Rad, Hercules, Calif.) tank and covered with 1,700 ml of 0.5× TBE. The running temperature was set at 14°C. The recommended reference standard, NCTC 8325, was positioned in every fifth or sixth lane to allow later normalization of electrophoretic patterns across the gel. The total run time was 23 h, the first-block switch time was 5 to 15 s for 10 h, and the second-block switch time was 15 to 60 s for 13 h. The voltage for the run was 6 V/cm or 200 V. For the CHEF Mapper system, the included angle was 120° and the ramp factor was linear.
The gels were stained for 30 to 45 min in ethidium bromide (1 μg/ml) and destained for 45 min or longer in distilled water. Gels were viewed under UV transillumination and photographed by using each laboratory's gel documentation system. Digital images were stored electronically as TIFF files and analyzed visually and with GelCompar (Applied Maths), using the Dice coefficient, and represented by UPGMA with 1% tolerance and 0.5% optimization settings. As previously, a similarity cutoff of 80% (23) and the criterion of a difference of ≤6 bands as described by Tenover et al. (27) were both used to define a cluster.
RESULTS
Phase I PFGE intercenter comparison.
All of the gel photographs and analyses from phase I were examined by visual comparison of patterns for the number of band differences by each of the participating laboratories. Figure 1 shows the results obtained at the coordinating laboratory for the 21 strains analyzed by the initial harmonized protocol with NCTC 8325 as the reference standard. Three clusters were identified by PFGE: cluster A included Belgium E1, Finland E7 and E10, France A and C, North German I, and Spain E1; cluster B included Finland E5 and United Kingdom EMRSA-16; and cluster C included Belgium E3 and Finland E1. Centralized analysis with GelCompar was not possible due to problems with resizing of gel images and often-poor resolution of bands as a result. Table 3 shows the identification of these clusters according to each center's own analysis, where submitted to the coordinating laboratory. PFGE clusters A and B were recognized in all laboratories by using both in-house and initial harmonized protocols, while cluster C was identified in three of the five laboratories.
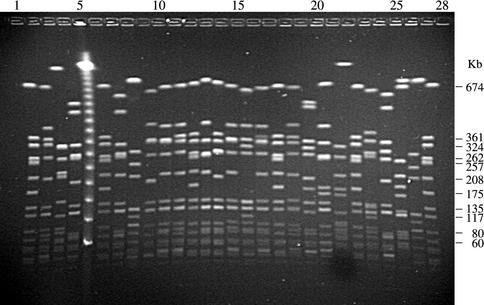
FIG. 1.
Gel image obtained in center 1 following PFGE of representative type strains of European EMRSA by using the initial harmonized protocol and NCTC 8325 as the reference standard. Molecular sizes in kilobases are shown for the reference standard. Lanes 1, 6, 12, 18, 23, and 28, NCTC 8325; lane 2, Belgium E1; lane 3, Belgium E2; lane 4, Belgium E3; lane 5, lambda; lane 7, Finland E1; lane 8, Finland E5; lane 9, Finland E24; lane 10, Finland E10; lane 11, Finland E7; lane 13, France A; lane 14, France B; lane 15, France C; lane 16, Greece 1; lane 17, Spain E1; Lane 19, North German I; lane 20, South German II; lane 21, Hannover III; lane 22, Berlin IV; lane 24, United Kingdom EMRSA-1; lane 25, United Kingdom EMRSA-3; lane 26, United Kingdom EMRSA-15; lane 27, United Kingdom EMRSA-16.
TABLE 3.
Comparison of UPGMA clusteringsa
| PFGE cluster | No. of laboratories
|
|||
|---|---|---|---|---|
| Phase I, protocol
|
Phase II, analysis
|
|||
| In-house gel (n = 5) | Initial har- monized gel (n = 5) | In-house analysis (n = 4) | Coordinating laboratory analysisb (n = 7) | |
| Ac | 5 | 5 | 4 | 7 |
| B | 5 | 5 | 4 | 7 |
| C | 3 | 3 | 3d | 6e |
Analysis was done by Dice coefficient for both gels run by using the in-house and initial harmonized protocols in each center for phase I and analyses performed locally (in four laboratories) and centrally in the coordinating laboratory (for seven laboratories) on gels run by using the final harmonized protocol in each center for phase II. PFGE clusters A to C (see text) were identified in the Laboratory of Hospital Infection, Central Public Health Laboratory, by using the initial harmonized protocol. PFGE cluster A includes Belgium E1; Finland E7 and E10; France A, B, and C; North German I; and Spain E1. Cluster B includes Finland E5 and United Kingdom EMRSA-16. Cluster C includes Belgium E3 and Finland E1.
Analyzed singly (i.e., not compared with the other gels).
There was a difference of up to one of seven strains between laboratories reporting.
One of these includes two additional strains not otherwise considered to be related.
One of these includes United Kingdom EMRSA-3.
The initial harmonized protocol created problems for two laboratories, with bands poorly resolving due to overloading of DNA or their running off the gel. According to each center's own evaluation, the results obtained with the initial harmonized protocol were worse than their in-house methods in three laboratories, similar in two laboratories, and improved in another two laboratories. The group agreed at meetings and via e-mail exchanges that the initial harmonized protocol produced the best resolution of the higher-molecular-weight bands, while the very similar German and Belgian in-house protocols were best at resolving the more important mid- to low-molecular-weight bands of the gel. The group also found the 30-h run of the initial harmonized protocol method to be impractical in cases where there were large numbers of gels run each week. Several different methods for block preparation and DNA digestion, developed over many years in the participating laboratories, were used and found to have worked well, making standardization unnecessary. However, the group felt that guidance should be given in the methods for phase II for those with little experience or where problems had been encountered.
The disappointing lack of comparability of the initial harmonized protocol between most of the laboratories suggested that some additional parameters needed standardization. It was also noted that while most laboratories used experienced personnel to perform the PFGE, two were unable to do so. In one there was the consequence that even their in-house gel was of a lower quality than those where skilled operators had been involved, and in the other there was inadequate digestion of DNA and significant overloading, producing diffuse bands. This added further to the study's analytical problems that persisted into phase II, where the improved protocol was evaluated.
Experiments were conducted in the coordinating laboratory to explore the importance of some additional parameters. These found that standardization of the volumes of agarose and buffer improved reproducibility. In addition, three agaroses from two different manufacturers (molecular biology certified agarose [Bio-Rad] and SeaKem and SeaKem Gold [FMC Bioproducts, Rockland, Maine]) produced similar electrophoretic results, suggesting that this was not a critical parameter when these brands were used. One center found additionally that electrophoresis-grade agarose from Life Technologies also resulted in equivalent separation of bands. The group favored the use of S. aureus strain NCTC 8325 in phase II, since lambda molecular-weight-standards from different suppliers were found to migrate differently through the gel (see Fig. 3). In addition, lambda from one supplier required pretreatment at 52°C for 8 min prior to electrophoresis; otherwise, the phage concatemers failed to separate. Thus, for phase II it was agreed to standardize the running buffer volume, gel volume/gel tray size ratio (thickness), and temperature (ensuring that it was the same as measured in the tank) and to develop a shorter run time.
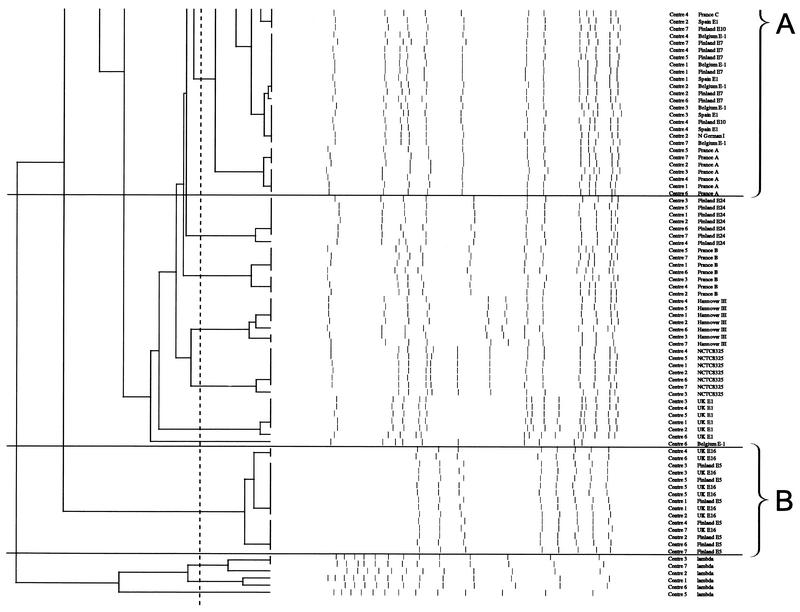
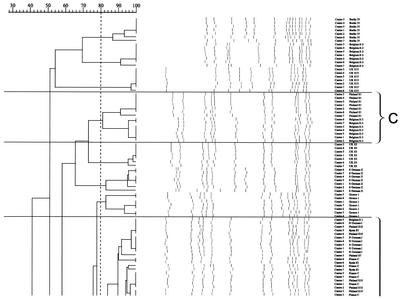
FIG. 3.
Dendrogram of all 21 type strains from gels from seven laboratories analyzed in the Laboratory of Hospital Infection, Central Public Health Laboratory, using GelCompar with percent similarity calculated by Dice coefficient (tolerance of 1% and optimization of 0.5%) and represented by UPGMA. Bands larger than 674 kb (as bands above the highest band of the standard will not be normalized) and smaller than 36 kb (the third-lowest band of the standard), as resolution of bands below this is often poor, were not defined for the purpose of further analysis of the gel. Clones A, B, and C are shown (see text for details).
Phase II results.
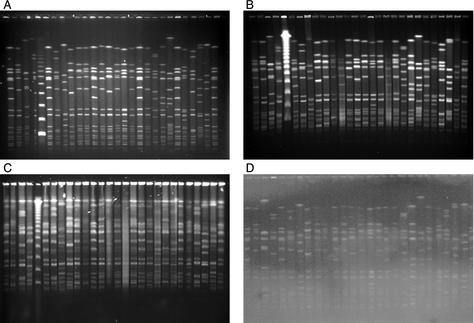
Nine laboratories ran the new protocol. Seven gels were of sufficient quality for analysis with GelCompar in the coordinating laboratory (Fig. 2A shows an example). Interestingly, their quality was now so improved that they were readily comparable by the program and gave an almost identical clustering of strains (Fig. 3). Comparison of the gels run in laboratories 1 and 2 showed almost 100% correlation between corresponding strains, except for two strains, France C and Spain E1, which differed by one band each (data not shown), perhaps due to the use of ultrathin molds for preparing blocks for PFGE in center 2. For the gels from laboratories 1, 2, 4, and 5, the correlation between corresponding isolates from the five gels was 100% for 13 of the 21 strains and ≥94% for the remaining 8 strains. For laboratories 3, 6, and 7 there were some discrepant results due to full normalization not being possible where some bands at the lower- or higher-molecular-weight ends of the NCTC 8325 reference markers were degraded and thus not visible. In one laboratory, the gel did not run as far as expected (Fig. 2B). This had been observed previously in phase I. The reason for this was later identified during an exchange of workers between laboratories. After interactions with Bio-Rad, we discovered that some machines were using the default temperature measurement setting from the cooler rather than activating the probe that measures the tank temperature. This resulted in suboptimal migration down the gel. In addition, in laboratory 3, the South German II isolate differed on repeated testing from those of the other laboratories by four bands, suggesting it had undergone several genetic rearrangements or that a contaminant had been introduced at some point. It was not possible to analyze the gels from two laboratories. In one this was due to overloading of the gel as indicated by the large amounts of undigested DNA in the wells and poor resolution of bands (Fig. 2C), and in the other it was due to inadequate destaining (Fig. 2D).
FIG. 2.
Four examples of gel images of the 21 representative type strains of European EMRSA strains digested with SmaI and with the final harmonized protocol for PFGE as performed in 9 of the 10 laboratories. NCTC 8325 is the reference standard used to normalize the gels. Bands larger than 674 kb (as bands above the highest band of the standard will not be normalized) and smaller than 36 kb (the third-lowest band of the standard) (as resolution of bands below this is often poor) were not defined for the purpose of further analysis of the gel. (A) Example of a good gel. (B) An underrunning gel due to a lower running temperature. (C) An overloaded gel. (D) An inadequately destained gel. See the text for further details and the legend to Fig. 1 for the running order of the 21 strains.
Comparison of PFGE types of important European MRSA.
The results of the various analyses of the strains are shown in Table 3. Computer-assisted analysis of the gel run in the coordinating laboratory by the final harmonized standard protocol and using an 80% cutoff when looking at the dendrogram of similarity revealed three clusters of international MRSA strains. This was in agreement with the criterion of Tenover et al. of one to six band differences for interpreting strain relatedness. PFGE cluster A now comprised the eight strains Belgium E1, North German I, Spain E1, Finland E7 and E10, and France A, B, and C; cluster B included Finland E5 and United Kingdom E16; and cluster C contained Belgium E3 and Finland E1. Looking at the dendrogram comparing all seven gels, clustering similar to that described above was observed. Comparing corresponding isolates across gels showed that all clustered at ≥82% (due to the problems with gels from laboratories 3, 6, and 7 discussed above).
Table 1 includes other typing characteristics for the 21 prototypic strains that should be useful in a clinical laboratory, as well as information on where and when the strains were first identified. Data on the full collection can be found on the HARMONY website at http://www.phls.co.uk/International/Harmony/microtyping.htm. Table 4 outlines the general characteristics of the three clusters. It is important to note that these results represent the predominant strains and that some of the subtypes encountered may have slightly different characteristics, requiring further studies to confirm their epidemiological significance. Phage, antibiogram, and resistogram typing may be useful tools to examine isolates locally and even nationally, but the results here show that they are not as definitive for long-term international comparisons. Some general comments can be made regarding susceptibility. For example, PFGE cluster A is more multiply resistant to antibiotics and chemicals. It has been found that for some strains belonging to this cluster, teicoplanin MICs that are elevated and are at the breakpoint according to British Society of Antimicrobial Chemotherapy criteria (7). Although toxin typing results agree with the clusterings produced, analysis of the whole collection (see the HARMONY website) showed that subtypes within PFGE clusters can vary in the toxin genes they carry; for example, no toxins were detected in the prototypic North German I strain, but sea was detected in three of its subtypes. In addition, enterotoxin genes sea and seb were found in one other subtype, while the Hannover III strain was found to carry seb.
TABLE 4.
General characteristics of the three PFGE clusters identified
| PFGE cluster (no. strains) | Phage type | Antibiograma,b | Resistograma | Toxin profileb |
|---|---|---|---|---|
| A (8) | Various | CEGKMNRST | Cd/EB/PI/MC/PMA | sea |
| B (2)c | 29/52/75/77/83A/83C | CEGKMS | Cd/EB/PI | sea, seg, sei, tst |
| C (2) | Various | CEGKMRST | Cd/EB/PI or fully sensitive | seg, sei |
See Table 1 for explanation of antibiotic, chemical, and toxin codes.
Underlining indicates that the characteristic may or may not be present.
Belgium E1, North German I, Spain E1, Finland E7 and E10, and France A, B, and C.
Finland E5 and United Kingdom EMRSA-16.
Belgium E3 and Finland E1.
DISCUSSION
An international group of reference and university hospital laboratories with considerable experience with MRSA and PFGE collaborated to develop a European MRSA strain collection and a PFGE protocol that resulted in a high level of intercenter gel comparability. Strains were contributed from all participating countries, and an internet-accessible European MRSA database was established. Except for the DNA concentration in the plug, standardization of the DNA preparation was not necessary, as each center had already optimized these steps for its own laboratory. However, the group agreed that a standardized protocol for new or inexperienced workers should include details of the desirable cell suspension based on the protocols used in laboratories 1 and 2. One used a standard mold (1 mm thick) and the other used an ultrathin mold (0.5 mm thick), but they produced comparable results.
One problem that was highlighted in our study was the disparity of the temperature measurement in different CHEF apparatuses. The companies are going to reinforce their advice to customers This detail would not have been identified but for an exchange of workers between laboratories, which further illustrates the value of such projects and the detail that is required in methodological descriptions to attain the goal of intercenter standardization and comparability of strains.
The ultimate goal of standardization is to facilitate intercenter comparisons of isolates and strains, aiming at determining their degree of relatedness. In the second phase of the study, four of the seven gels analyzed resulted in almost complete concordance, while three achieved an acceptable level of reproducibility. This highlights the need for laboratories to reevaluate their protocols if consistently suboptimal results are obtained, for example, due to plugs overloaded with DNA or blocks that have not been stored appropriately.
The second phase of the study also showed that many, but clearly not all, of the methodological problems seen in phase I were eventually overcome by close cooperation between laboratories and standardization of certain parameters. The most important variable appeared to be the skill of the operator, as overloading of the gel and inadequate destaining were evident. We therefore recommend including an internal quality control to assess the reproducibility and quality of the gel before inclusion in the database for intra- or intercenter comparison. The quality control strain should ideally demonstrate >95% similarity between gels to ensure acceptability. When the gels from laboratories 1 and 2 were compared by using GelCompar, almost 100% correlation was observed between corresponding isolates from each. There were a few minor differences observed, particularly with respect to the strains that make up the major clonal group called cluster A in this study. This group, otherwise known as the Iberian clone, was confirmed by comparison with the strain identified as being the prototype submitted to the HARMONY collection by Herminia de Lencastre (22). It emerged in the early 1990s and had been previously characterized by other techniques (1).
A recent multicenter study by Chung et al. (9) compared gels run in different laboratories by using a fully standardized protocol. It was unclear how much experience with the PFGE technique these laboratories had before participating in this study. They showed that, even when the full process was standardized, some of the problems (amount of loaded DNA; overstaining or under-destaining of the gel; too much DNA in the blocks, indicating insufficient lysis; blocks not stored properly; and problems related to the electrophoresis) experienced in both phases of our study also occurred in that study. Again, this highlights the importance of using skilled operators for performing the technique if we are to aspire to national and international comparison and tracking of strains.
The Nordic countries (Denmark, Finland, Norway, and Sweden) have produced a standardized protocol that is used for interlaboratory comparison (C. S. Elsberg, S. Salmenlinna, J. V. Fussing, J. Vuopio-Varkila, V. T. Rosdahl, and the Nordic MRSA Study Group. Abstr. 9th Int. Symp. Staphylococci Staphylococcal Infect., abstr. 90, 2000). The protocol was based on the most common parameters from the in-house PFGE protocols in 8 of the 12 laboratories involved. Overall, there was very good correlation between the laboratories, confirmed by visual and BioNumerics (Applied Maths) analysis. They found that interlaboratory comparison of MRSA could be achieved, as the minor differences observed could be attributed to the poor resolution of TIFF files exchanged. In particular, they found that attention should be paid to the variable photographic quality in the lower part of the gels (between 48.5 and 97kb, when lambda is used as a reference standard). They also highlighted the need for a reduction in the workload and identification time.
Standardization of the image capture process and optimal settings for analysis are also required. In this study, it was necessary to standardize the size of TIFF files to allow comparison by using GelCompar or BioNumerics analysis software. For intercenter comparisons, it is important to specify the part of the gel that is to be included in the analysis. Bands above the highest-molecular-size band of the NCTC 8325 marker (674 kb) should not be included, as this part of the gel will not have been normalized. Similarly, bands below the third-lowest band of the marker (36 kb) should also be excluded due to the often-poor resolution of the lower-molecular-size bands.
The exchange of isolates and central analysis revealed the international spread of MRSA clones in the EU. This proved to be far more extensive than had been realized previously and justified the establishment of the HARMONY network, which the IUMS subcommittee now wishes to extend globally. Already several other countries have joined the network, and additional techniques have been performed to further explore the clonality of the EMRSA strains (unpublished data).
Following from a previous study by Deplano et al. (13), an extremely important outcome of this study is that a number of clonally related strains have been shown to be present in multiple countries throughout Europe. The well-known Iberian clone (PFGE cluster A) has been demonstrated in Belgium, Finland, France, Germany, and Spain (and from the wider HARMONY collection in Portugal, Slovenia, and Sweden). Other strains in the collection (from Greece, the United Kingdom, and Ireland) may also be related, although more distantly, which supports the clonal nature of MRSA (2). Other clonally related strains have also been identified. For example, the South German II strain is closely related to the most prevalent strain in Slovenia. The United Kingdom strains EMRSA-15 and EMRSA-16 (which belongs to PFGE cluster B) have recently been identified in a number of other European countries as a result of this study. EMRSA-15 has been found in Finland (S. Salmenlinna, personal communication), Germany (W. Witte, personal communication), and Sweden, while EMRSA-16 has been identified in Finland via Turkey, in Sweden via Cyprus, and Denmark via Sweden (C. S. Elsberg, J. Mondrup, L. Larsson, S. Murchan, and C. Welinder-Olsson, Abstr. 9th Int. Symp. Staphylococci Staphylococcal Infect., abstr. 232, 2000). Studies have also shown that this strain is present in Switzerland (6), and this highlights the need for closer international collaboration to monitor the spread of current epidemic strains as well as the emergence of new ones. Furthermore both EMRSA-15 and EMRSA-16 have been identified in Belgium since at least 2001 in six and two hospitals, respectively (M. Struelens and coworkers, personal communication). PFGE cluster C, representing strains in Belgium and Finland, may also be distantly related to the South German II and United Kingdom EMRSA-3 strains.
Reliable and reproducible intercenter comparisons are possible by using the standardization approach outlined here (an approach we have termed harmonization). However, as with many other techniques, it is important to remember that the largest impediment to getting reproducible results is the skill of the operator. At least 5 of the 10 laboratories have adopted this new protocol for routine use in epidemiological studies, and such an approach will be required if laboratories are to externally query the IUMS HARMONY database that is soon to be established by using the BioNumerics software package in the coordinating laboratory. Other laboratories have also agreed to use this new protocol for the purpose of comparing strains with the central database. This internet-based database with the reference profiles (type strains and predominant subtypes) will be maintained by the coordinating laboratory as the IUMS reference center and administered via the IUMS subcommittee. Other laboratories performing PFGE will be able to compare their isolates of interest with the reference collection. These isolates should then be submitted to the central laboratory for validation once the agreed national center has confirmed that it is a new strain by repeating the PFGE with the relevant reference strain by both computer-assisted and visual comparison. It will then be added to the collection and reference panel library.
The database includes antimicrobial susceptibility and toxin typing characteristics, and more will be added, including other genotypic data, in the near future. Some of these characteristics are helpful in interpreting the relationship and evolution of strains within a country (12). However, interpreting them can be quite a challenge: some toxins, for example, are known to be phage related, and perhaps the variations we have observed may be related to this process (10).
The group was aware of the PulseNet food-borne enteric disease surveillance network that had been established in four laboratories in 1995 (25). PulseNet had shown that when a protocol was strictly followed, near-perfect intercenter reproducibility was possible for most of the participating laboratories. There is now a rigorous quality assurance and accreditation system in place. Interestingly, when a laboratory introduced an unspecified variation in the protocol, a problem with interlaboratory comparison occurred. It was not deemed appropriate to adopt this approach ab initio with the HARMONY laboratories. Instead, we wanted to learn from each other's expertise and experience. The final protocol is thus “owned” by the group and seen to be “fit for the purpose.” It has been adopted by all of the laboratories for interlaboratory comparisons and by several for all their routine work. It will be interesting to see if, as experience and further quality assurance work progress, all laboratories are willing to use the protocol for their routine work and build new databases for their own countries.
A similar Canadian PFGE comparison and standardization study was published (19) just as the work of HARMONY was completed. Their results look very promising, although they are working further to improve their intercenter reproducibility, using, at this juncture, five quality assurance strains. It will be important for the IUMS to continue progression of the HARMONY initiative globally, discuss the various published and other initiatives, and build on the whole group's experiences and opinions.
We are also examining other typing methods, such as use of methicillin resistance genes (mecRIA and SCCmec), multilocus sequence typing, binary typing, and ribotyping, to establish which of these will be recommended and when, to confirm the relationships between known PFGE patterns, and to confirm the relationships of new strains to those already in the collection. There is still a need to address the international nomenclature of strains and the relative roles and utility of the various typing systems in this process. It is the intention of the IUMS subcommittee to work on this. The results of epidemiological typing data on all MRSA isolates from bloodstream infections (in collaboration with the European Antimicrobial Resistance Surveillance System) and other clinical and carriage isolates could give further information on the spread of EMRSA clones throughout Europe.
In conclusion, the HARMONY typing group has established a European collection and database of EMRSA strains and developed a harmonized protocol for PFGE that has been widely adopted for international comparisons, thus providing important information on spread of epidemic clones of MRSA.
Acknowledgments
We thank all the laboratories that have contributed to the EMRSA collection and database, Marina Warner from the Antimicrobial Resistance Monitoring and Reference Laboratory for invaluable technical assistance with susceptibility testing, and Mark Ganner and Ana New from the Staphylococcus aureus Reference Service at Colindale for phage typing. We also extend our gratitude to all other laboratories that have contributed strains, data, and expertise to the EMRSA collection and database for their enthusiasm and support for the project.
This project was supported by HARMONY, DGXII contract no. BMH4-CT96. B.C. was the project leader.
REFERENCES
- 1.Aparicio, P., J. Richardson, S. Martin, A. Vindel, R. R. Marples, and B. D. Cookson. 1992. An epidemic methicillin-resistant strain of Staphylococcus aureus in Spain. Epidemiol. Infect. 108:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayliffe, G. A. J. 1997. The progressive inter-continental spread of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 24(Suppl. 1):S74-S79. [DOI] [PubMed]
- 3.Ayliffe, G. A. J., A. Buckles, M. S. Casewell, B. D. Cookson, R. A. Cox, G. J. Duckworth, G. L. French, A. Griffiths-Jones, R. Heathcock, H. Humphreys, C. T. Keane, R. R. Marples, D. C. Shanson, R. Slack, and E. Tebbs. 1998. Revised guidelines for the control of methicillin-resistant Staphylococcus aureus infections in hospitals. Report of a combined working party of the British Society of Antimicrobial Chemotherapy, the Hospital Infection Society, and the Infection Control Nurses' Association. J. Hosp. Infect. 39:253-290.9749399 [Google Scholar]
- 4.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair, J. E., and R. E. O. Wilson. 1961. Phage typing of staphylococci. Bull. W. H. O. 24:771-784. [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc, D., A. N. Banuls, P. M. Hauser, P. Moreillon, P. Francioli, M. Tibayrenc, and the Swiss MRSA Group. 2001. Methicillin-resistant Staphylococcus aureus: phylogenetic relatedness between European epidemic clones and Swiss sporadic clones. Microbial Drug Resistance 6:231-238. [DOI] [PubMed] [Google Scholar]
- 7.British Society for Antimicrobial Chemotherapy. 1998. BSAC standardised disc sensitivity testing method. Summer newsletter of the British Society for Antimicrobial Chemotherapy. British Society for Antimicrobial Chemotherapy, Birmingham, United Kingdom.
- 8.British Society for Antimicrobial Chemotherapy. 1991. A guide to sensitivity testing. Working party report of the British Society for Antimicrobial Chemotherapy on antibiotic sensitivity testing. J. Antimicrob. Chemother. 27(Suppl. D):1-50. [PubMed] [Google Scholar]
- 9.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, and the Multi-Laboratory Project Collaborators. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multi-laboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 10.Coleman, D. C., D. J. Sullivan, R. J. Russell, J. P. Arbuthnott, B. F. Carey, and H. M. Pomeroy. 1989. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of B-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J. Gen. Microbiol. 135:1679-1697. [DOI] [PubMed] [Google Scholar]
- 11.Cookson, B. D., P. Aparicio, A. Deplano, M. Struelens, R. Goering, and R. Marples. 1996. Inter-centre comparison of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 44:179-184. [DOI] [PubMed] [Google Scholar]
- 12.Cuny, C., H. Claus, and W. Witte. 1996. Discrimination of S. aureus strains by PCR for r-RNA gene spacer size polymorphism and comparison to SmaI-macrorestriction patterns. Zentralbl. Bakteriol. 283:466-476. [DOI] [PubMed] [Google Scholar]
- 13.Deplano, A., W. Witte, W. J. van Leeuwen, Y. Brun, and M. J. Struelens. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighbouring countries. Clin. Microbiol. Infect. 6:239-245. [DOI] [PubMed] [Google Scholar]
- 14.Deplano, A., A. Schuermans, J. Van Eldere, W. Witte, H. Meugnier, J. Etienne, H. Grundmann, D. Jonas, G. T. Noordhoek, J. Dijkstra, A. van Belkum, W. van Leeuwen, P. T. Tassios, N. J. Legakis, A. van der Zee, A. Bergmans, D. S. Blanc, F. C. Tenover, B. C. Cookson, G. O'Neill, M. J. Struelens, et al. 2000. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis. J. Clin. Microbiol. 38:3527-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Antimicrobial Resistance Surveillance System. 2002. Newsletter no. 4. National Institute of Public Health and the Environment, Bilthoven, The Netherlands.
- 16.Kaufmann, M. E. 1998. Pulsed-field gel electrophoresis, p. 33-50. In N. W. Woodford and A. Johnson (ed.), Methods in molecular medicine, vol. 15. Molecular bacteriology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 17.Maslow, J. N., M. E. Mulligan, and R. D. Arbeit. 1993. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin. Infect. Dis. 17:153-162. [DOI] [PubMed] [Google Scholar]
- 18.McLauchlin, J., G. L. Narayanan, V. Mithani, and G. O'Neill. 2000. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Protect. 63:479-488. [DOI] [PubMed] [Google Scholar]
- 19.Mulvey, M. R., L. Chui, J. Ismail, L. Louie, C. Murphy, N. Chang, M. Alfa, and the Canadian Committee for the Standardisation of Molecular Methods. 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:3481-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker, M. T. 1983. The significance of phage typing patterns in Staphylococcus aureus, p. 33-62. In C. S. F. Easmon and C. Adlam (ed.), Staphylococci and staphylococcal infections. Academic Press Inc., London, England.
- 21.Richardson, J. F., V. T. Rosdahl, W. J. van Leeuwen, A. M. Vicekery, A. Vindel, and W. Witte. 1999. Phages for methicillin-resistant Staphylococcus aureus: an international trial. Epidemiol. Infect. 122:227-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos Sanches, I., M. Ramirez, H. Troni, M. Abecassis, M. Padua, A. Tomasz, and H. de Lencastre. 1995. Evidence for the spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J. Clin. Microbiol. 33:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Struelens, M. J., A. Deplano, C. Godard, N. Maes, and E. Serruys. 1992. Epidemiological typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macro-restriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Struelens, M. J., and the Members of the European Study Group on Epidemiological Markers of the European Society for Clinical Microbiology and Infectious Diseases. 1996. Consensus guidelines for evaluation, appropriate use and interpretation of molecular epidemiological typing systems for bacterial pathogens. Clin. Microbiol. Infect. 2:2-11.11866804 [Google Scholar]
- 25.Swaminathan, R., T. J. Barrett, S. B. Hunter, R. V. Tauxe, and the CDC Pulsenet Task Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover, F. C. 1994. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Townsend, D. E., W. B. Grubb, and N. Ashdown. 1983. Gentamicin resistance in methicillin-resistant Staphylococcus aureus. Pathology 15:169-174. [DOI] [PubMed] [Google Scholar]
- 29.Townsend, D. E., W. B. Grubb, and N. Ashdown. 1983. Genetics of drug resistance in methicillin-resistant Staphylococcus aureus from Australian hospitals. J. Hosp. Infect. 4:331-337. [DOI] [PubMed] [Google Scholar]
- 30.van Belkum, A., J. Kluytmans, W. van Leeuwen, R. Bax, W. Quint, E. Peters, A. Fluit, C. Vandenbroucke-Grauls, A. van den Brule, H. Koeleman, W. Melchers, J. Meis, A. Elaichouni, M. Vaneechoutte, F. Moonens, N. Maes, M. Struelens, F. Tenover, and H. Verbrugh. 1995. Mulicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 31:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Foery, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Belkum, A. 2000. Molecular epidemiology of methicillin-resistant Staphylococcus aureus strains: state of affairs and tomorrow's possibilities. Microb. Drug Resist. 6:173-188. [DOI] [PubMed] [Google Scholar]
- 33.Voss, A., D. Milatovic, C. Wallrauch-Schwarz, V. T. Rosdahl, and I. Braveny. 1994. Methicillin-resistant Staphylococcus aureus in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 13:50-55. [DOI] [PubMed] [Google Scholar]