Abstract
A serologically distinct avian metapneumovirus (aMPV) was isolated in the United States after an outbreak of turkey rhinotracheitis (TRT) in February 1997. The newly recognized U.S. virus was subsequently demonstrated to be genetically distinct from European subtypes and was designated aMPV serotype C (aMPV/C). We have determined the nucleotide sequence of the gene encoding the cell attachment glycoprotein (G) of aMPV/C (Colorado strain and three Minnesota isolates) and predicted amino acid sequence by sequencing cloned cDNAs synthesized from intracellular RNA of aMPV/C-infected cells. The nucleotide sequence comprised 1,321 nucleotides with only one predicted open reading frame encoding a protein of 435 amino acids, with a predicted Mr of 48,840. The structural characteristics of the predicted G protein of aMPV/C were similar to those of the human respiratory syncytial virus (hRSV) attachment G protein, including two mucin-like regions (heparin-binding domains) flanking both sides of a CX3C chemokine motif present in a conserved hydrophobic pocket. Comparison of the deduced G-protein amino acid sequence of aMPV/C with those of aMPV serotypes A, B, and D, as well as hRSV revealed overall predicted amino acid sequence identities ranging from 4 to 16.5%, suggesting a distant relationship. However, G-protein sequence identities ranged from 72 to 97% when aMPV/C was compared to other members within the aMPV/C subtype or 21% for the recently identified human MPV (hMPV) G protein. Ratios of nonsynonymous to synonymous nucleotide changes were greater than one in the G gene when comparing the more recent Minnesota isolates to the original Colorado isolate. Epidemiologically, this indicates positive selection among U.S. isolates since the first outbreak of TRT in the United States.
Avian metapneumovirus (aMPV) causes turkey rhinotracheitis (TRT) and is associated with swollen head syndrome (SHS) in chickens, which is usually accompanied by secondary bacterial infections that increase mortality. TRT was first reported in South Africa during the early 1970s, and viruses were subsequently isolated in Europe, Israel, and Asia (1, 12, 23). In February 1997, the National Veterinary Services Laboratory (NVSL) (Animal and Plant Health Inspection Service, U.S. Department of Agriculture [USDA]) officially isolated aMPV from commercial turkeys in Colorado (aMPV/CO) after an outbreak of TRT the previous year. During the first 10 months of this outbreak of TRT in the United States, it was not possible to detect virus serologically due to the absence of cross-reactivity of the U.S. aMPV isolates with reagents produced in Europe. An enzyme-linked immunosorbent assay (ELISA) was developed by the NVSL using inactivated aMPV/CO as an antigen, and serological evidence of aMPV infection was subsequently demonstrated in north-central U.S. turkey flocks (D. A. Senne, R. K. Edson, J. C. Pederson, and B. Panigrahy, unpublished data). In the United States, mortality due to aMPV infections in birds with both aMPV and bacterial infections has ranged from 0 to 30%, with condemnations due to air sacculitis. The absence of serological reactivity by aMPV/CO-infected birds with aMPV serotype A (aMPV/A) or serotype B (aMPV/B) isolates and genomic sequence diversity of the U.S. isolates clearly demonstrated emergence of new avian pneumovirus strains previously considered exotic to North America (39-41, 43).
Pneumoviruses are members of the family Paramyxoviridae which contain a nonsegmented, single-strand, negative-sense RNA genome that is approximately 15 kb long. Related viruses include human, bovine, ovine, and caprine respiratory syncytial viruses (RSVs) and pneumonia virus of mice (PnVM) (11), as well as the recently identified human MPV (hMPV) (48). Human RSV (hRSV) encodes 10 genes, compared to 6 or 7 in other paramyxoviruses. The 10 genes encode the nonstructural proteins (NS1 and NS2), nucleoprotein (N), phosphoprotein (P), matrix protein (M), small hydrophobic protein (SH), surface glycoprotein (G), fusion protein (F), second matrix protein (M2), and a viral RNA-dependent RNA polymerase (L). The pneumoviruses have an F protein that promotes cell fusion, but these viruses do not hemagglutinate and the G attachment proteins of these viruses do not have neuraminidase activity. These are important characteristics distinguishing the pneumoviruses from the other paramyxoviruses (11).
Classification of European aMPV isolates was initially based on physical characterization of the virion (10), electrophoretic mobilities of viral proteins (30), and number of mRNA species detected in aMPV-infected cells (8). The putative gene order of aMPV (3′N-P-M-F-M2-SH-G-L5′) is different from its hRSV counterpart (3′NS1-NS2-N-P-M-SH-G-F-M2-L5′), wherein the SH and G genes are located 5′ to the M2 gene (29). The extreme 3′ and 5′ ends of one European aMPV genome was determined and established that the NS1 and NS2 genes are absent in the avian viruses (37). This is different from their RSV counterparts and along with a smaller L gene results in aMPV having a genome of only 13.3 kb (38). Since aMPV has no NS1 or NS2 gene but has a M2 gene with structural characteristics similar to those of other pneumoviruses, it has become the type virus of a new genus within the Metapneumovirus (36). The recently identified hMPV has a genome structure essentially equivalent to aMPV with a reported genome length of 13,378 nucleotides (47).
The G gene of aMPV encodes the surface glycoprotein responsible for cell attachment and serves as one of the major antigens of pneumoviruses. The G protein is known to be the most variable protein in hRSV (7, 17, 21) and other MPVs, including aMPV/A, aMPV/B, and aMPV/D (4, 24). To further understand the molecular structure and epidemiology of aMPV, we completely sequenced and examined phylogenetic relationships of the cell surface glycoprotein (G) of the aMPV/C serotype circulating within the United States. Furthermore, the predicted amino acid sequence of the G protein of aMPV/C was compared with the sequences of the G proteins of other members of the Pneumovirinae subfamily, including the recently identified hMPV, isolated from patients with acute respiratory disease.
Viral propagation and cDNA synthesis of the G attachment protein gene.
aMPV strains Colorado (aMPV/CO/96/C), Minnesota 1a (aMPV/MN1a/97/C), Minnesota 2a (aMPV/MN2a/97/C), and Minnesota 7 (aMPV/MN7/99/C) were propagated in Vero cells (40, 41, 43). All viral stocks are maintained at passage level four. Briefly, Vero cells were cultured in growth medium (minimal essential medium [MEM], 5% fetal bovine serum, 1% 100× antibiotic-antimycotic solution [100× solution is 10,000 μg of streptomycin, 10,000 U of penicillin, and 25 μg of amphotericin B per ml]) to 95% confluence. Cells were infected at a multiplicity of infection (MOI) of 0.1, adsorbed for 1 h at 37°C, and overlaid with maintenance medium (MEM, 2% fetal bovine serum, 1% 100× antibiotic-antimycotic solution). Infected cells were incubated at 37°C with 5% CO2 for 48 to 72 h or until >90% cytopathic effect (CPE) was observed by light microscopy. Infected cells were harvested by scraping and centrifugation. Total RNA from aMPV/C-infected cells was purified by using RNAeasy Mini total RNA isolation kit (Qiagen, Valencia, Calif.) according to the manufacturer's protocol. Briefly, total aMPV/C-infected cell RNA was reverse transcribed (32) using aMPV/C G1 primer (5′ AACATGGAGCCCCTGAAAGTCTCT-3′) and aMPV G1321c primer (5′-TTTTTGGTTGTTGCCTGTCTCTT-3′) at 60°C utilizing Thermoscript (Invitrogen, Carlsbad, Calif.). The full-length gene was subsequently amplified by PCR (3) for 35 cycles utilizing primers G1 and G1321c with an annealing temperature of 60°C. The DNA fragment was isolated from an agarose gel using Qiaquick agarose gel purification kit (Qiagen) and cloned (31) into pCR-XL Topo cloning vector (Invitrogen) according to the manufacturer's protocol. Editing nucleotide sequences, prediction of amino acid sequences, and computer predictions of protein structures were done using the DNASTAR (Madison, Wis.) and GeneWorks 2.3 programs (Intelligenetics, Mountain View, Calif.). Sequence alignments were performed by the CLUSTAL W method (45). Nucleotide sequence analysis, including determination of synonymous and nonsynonymous substitutions (33), was conducted using the Molecular Evolutionary Genetics Analysis system (MEGA) (25). To determine relationships among aMPV isolates and to determine how protein sequence information is related to the current virus designations, analysis was performed by Phylogenetic Analysis Using Parsimony (PAUP*4.0b10) (44) after 2,000 bootstrap replications (18).
Nucleotide sequence of the aMPV/C G gene and analysis of the predicted protein.
The attachment G-protein gene of aMPV/CO/C was 1,321 bases long, which is 61 nucleotides longer than the G genes of aMPV/A and aMPV/B (24). The aMPV/C G gene is 136 nucleotides longer than the aMPV/D G gene (4) and 710 nucleotides longer than the hMPV G gene (47). The transcription start sequence of the aMPV/C G gene was identical to the conserved sequence (GGGACAAGU) of other aMPV genes (15, 28, 29, 41), followed by the AUG start codon at nucleotide position 14. There was one major open reading frame from residues 14 to 1321, encoding a predicted protein of 435 amino acids with an Mr of 48,843 and a net charge of 23.15 at neutral pH. A second putative leader sequence (GGGACAAGU) was identified at positions 715 to 723, followed by a second putative open reading frame spanning positions 728 to 1321, encoding a predicted protein of 197 amino acid residues with an Mr of 22,024 and a net charge of 11.20 at neutral pH.
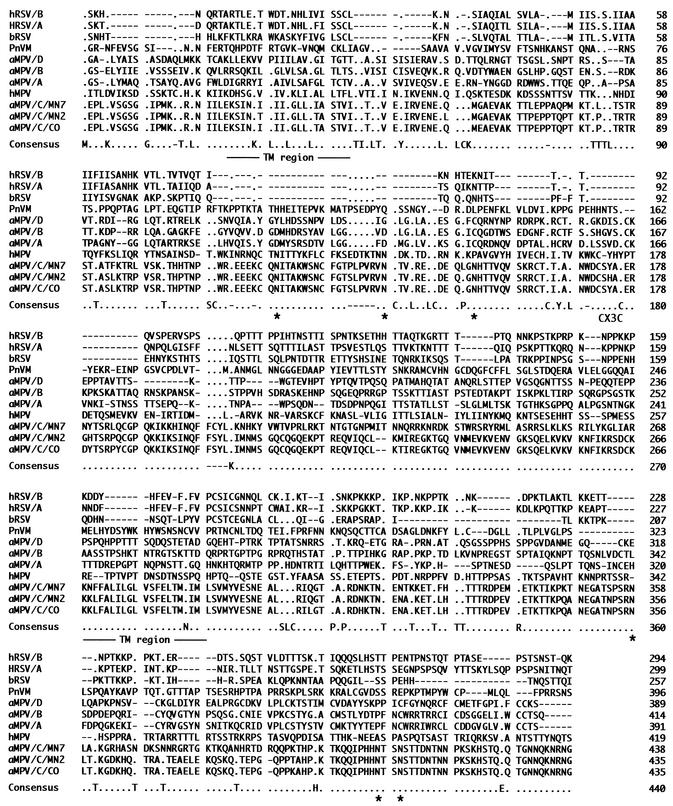
Analysis of the predicted aMPV/C G attachment protein by BLAST (2) revealed two mucin-like motifs encompassing amino acid positions 21 to 163 and 190 to 433 with potential transmembrane regions within these areas from residues 24 to 45 and 266 to 287 (Fig. 1). These highly basic regions are potentially important for heparin binding on the membrane surface of infected cells and play a key role in pneumovirus entry into cells (16). Between the two mucin-like regions is a conserved hydrophobic domain that includes the fractalkine-like CX3C chemokine sequence CSYAC from positions 172 through 176 (Fig. 1). The CX3C region of hRSV binds to the CX3CRI receptor, the specific receptor of the chemokine fractalkine. This is a chemokine motif found in the hRSV G protein that may be important in trafficking of pulmonary polymorphic neutrophils (PMNs) and natural killer cells during infection. It may also contribute to altered chemokine mRNA expression of macrophage inflammatory protein 1α (MIP-1α), MIP-1β, MIP-2, monocyte chemoattractant protein 1, and interferon-inducible protein 10 (IP-10) by bronchoalveolar leukocytes (46).
FIG. 1.
Predicted amino acid sequence alignment of recognized pneumovirus G attachment glycoproteins. Those amino acid residues that are strictly conserved in the Pneumovirinae subfamily are present in the consensus sequence. The predicted transmembrane (TM) region within the mucin-like portions of the protein, along with the putative CX3C motif and potential glycosylation sites (asterisks) conserved among the aMPV/C isolates, are indicated. A dot indicates absence of consensus, while a dash denotes absence of an amino acid relative to the majority of sequences.
Comparison of the nucleotide sequence of the aMPV/C G gene and predicted protein sequence with those of other members of the Pneumovirinae subfamily.
The G gene nucleotide sequences of aMPV/CO/C, aMPV/MN2a/C, aMPV/MN7/C, aMPV/A (L34032), aMPV/B (L34031), aMPV/D (AJ251085), PnVM (D11130), hRSV (AF065255 and AF065253), and bovine RSV (bRSV) (AF295543) were aligned with the combined SH and G genes of hMPV (AF371337) (GenBank accession numbers given in parentheses). The combined SH and G genes of hMPV were included due to similarities in the SH protein with the G protein of other members of the Pneumovirinae. Sequence identities of aMPV/CO/C gene to G genes of other pneumoviruses ranged from 33.3 to 98.6% (Table 1). The nucleotide sequence identity of the aMPV/CO/C and aMPV/MN2a/C G gene was 98.6% with 12 nonsynonymous and 6 synonymous differences resulting in a nonsynonymous to synonymous substitution ratio of 2. The greater number of nonsynonymous nucleotide substitutions suggests that positive selection has occurred (20) since aMPV/C was isolated in Colorado in 1996. The nucleotide sequence identity of the aMPV/CO/C and aMPV/MN7/C G genes was 86.3%, which was similar to the values reported for other genes in these two isolates (43), while the nucleotide sequence identity between the G gene of aMPV/CO/C and the combined SH-G genes of hMPV was 42.5% (Table 1). The sequence identities of aMPV/CO/C G gene with the aMPV/A, aMPV/B, aMPV/D, hRSV, and bRSV sequences was 33.3, 33.9, 34.2, 34.0, and 36.3%, respectively.
TABLE 1.
Nucleotide and predicted amino acid identities among the cell attachment (G) glycoproteins of pneumoviruses
| Pneumovirus | % Identity between G proteins of pneumovirusesa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| aMPV/CO/C | aMPV/MN2a/C | aMPV/MN7/C | aMPV/A | aMPV/B | aMPV/D | hRSV | bRSV | hMPV | |
| aMPV/CO/C | 98.6 | 86.3 | 33.3 | 33.9 | 34.2 | 34.0 | 36.3 | 42.5 | |
| aMPV/MN2a/C | 97.2 | 86.2 | 33.6 | 35.6 | 32.9 | 34.3 | 36.2 | 42.0 | |
| aMPV/MN7/C | 72.9 | 72.7 | 34.2 | 34.3 | 34.2 | 36.8 | 37.6 | 43.7 | |
| aMPV/A | 13.0 | 13.3 | 12.8 | 48.2 | 51.4 | 38.6 | 42.1 | 38.5 | |
| aMPV/B | 7.2 | 4.8 | 8.6 | 10.1 | 44.1 | 35.9 | 33.4 | 42.4 | |
| aMPV/D | 12.6 | 12.6 | 14.8 | 28.7 | 5.6 | 44.4 | 43.8 | 43.3 | |
| hRSV | 18.5 | 17.8 | 16.2 | 13.8 | 3.4 | 13.5 | 58.8 | 44.8 | |
| bRSV | 13.6 | 14.8 | 15.6 | 12.8 | 8.6 | 14.8 | 28.8 | 45.7 | |
| hMPV | 22.4 | 22.4 | 18.2 | 13.8 | 11.4 | 9.7 | 11.4 | 12.1 | |
Amino acid identities are shown in the lower left portion of the table, and nucleotide sequence identities are shown in the upper right portion of the table. Percent identity for combined SH and G genes is shown for hMPV. The specific isolates and strains used for aMPV subtypes and RSVs follow: for aMPV/A, isolate CVL 14/1; for aMPV/B, isolate 872S; for aMPV/D, isolate Fr/85/2; for hRSV, wild-type strain B1; for bRSV, isolate ATUE51908; and for hMPV, isolate 00-1.
The predicted amino acid sequence of the aMPV/CO/C G protein was aligned with the G-protein amino acid sequences of other members of the Pneumovirinae (Fig. 1), and percent identities for the aMPV/CO/C G protein and the G proteins of aMPV/A, aMPV/B, aMPV/D, hRSV, bRSV, and PnVM ranged from 4 to 16.5%. The percent sequence identity values increased to 97, 72, and 21 for aMPV/CO/C and aMPV/MN2a/C, aMPV/MN7/C, and hMPV (combined SH-G proteins), respectively. Several motifs were identified among either the MPVs or between the aMPV/C viruses and hRSV G proteins, but little or no consensus was apparent in the predicted amino acid G-protein sequences for all viruses examined. In the MPVs, there was a conserved CX2LX2LCX3P sequence within the amino-terminal mucin-like region, which may be important for heparin binding on cell surfaces (16). There are several common motifs in the G-protein sequences of aMPV/C and hRSV or bRSV; these motifs include KX4IXLXLL at the amino terminus, followed downstream by LXLXK. In the central portion of the G protein is a conserved motif, CX2LX2LCX3P. When the conserved hydrophobic domains containing the CX3C motif were examined separately, a similar pattern was detected. Specifically, the CX2CX5CX3C motif for hRSV and CXCX7CX3C motif for aMPV/C were present, with the four conserved cysteine residues previously shown to be involved in forming two disulfide bridges (5, 27).
Among the aMPV/C isolates (Fig. 1), the G-protein region from residues 161 through 178 containing the CX3C motif was highly conserved [C(T/I)CIYALNWDCS(Y/H)ACER]. There was only an Y-to-H substitution in the aMPV/MN2a/C protein at position 174 and an I-to-T substitution at position 165 of the aMPV/MN7/C isolate compared to the aMPV/CO/C protein. There were no amino acid substitutions in the predicted G proteins of aMPV/CO/C and aMPV/MN1a/C isolates, with 100% sequence identity. However, several amino acid substitutions occurred in the predicted G proteins of the aMPV/MN2a/C isolate and the aMPV/CO/C virus; these substitutions occurred throughout the predicted G protein and were reflected by the greater number of nonsynonymous nucleotide substitutions in the aMPV/MN2a/C gene relative to synonymous changes. In the aMPVs from the United States isolated early, this was reported for the fusion (F) protein gene (41) and was further substantiated for other aMPV/C structural genes (43). More striking differences were found in the G predicted amino acid sequence of the aMPV/MN7/C virus relative to either aMPV/CO/C or aMPV/MN2a/C, wherein a substantial number of substitutions occurred from residues 203 through 270 and from residues 363 through 380. Consequently, certain portions of the aMPV G protein may be more hypervariable, as occurs with other viral agents (6, 42). Finally, the G-protein variation among aMPV/C isolates potentially parallels similar evolution of hRSV (21) and hMPV (19, 34, 35) into two phylogenetically distinct groups.
Phylogenetic analysis of pneumovirus G genes.
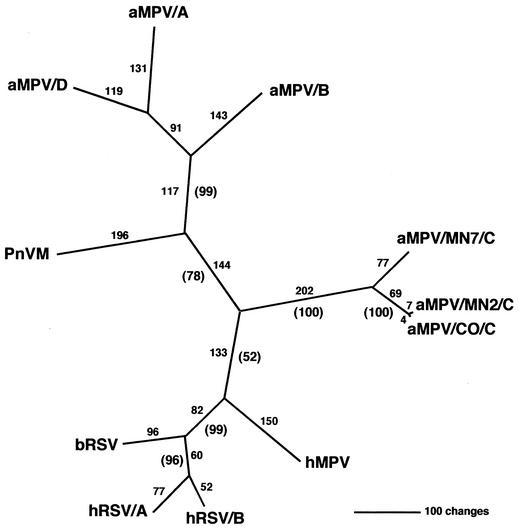
The G attachment glycoprotein gene is the most variable of the pneumovirus proteins, and this diversity has been utilized for phylogenetic classification and development of diagnostic methods (7, 17, 22, 26). As demonstrated for the N, P, M, F, and M2 genes (43), the aMPV/C isolates were phylogenetically distinct from aMPV subtypes A, B, and D (Fig. 2). This is the first phylogenetic analysis of all MPVs utilizing G-protein sequences. Most notably, aMPV/C isolates segregated as a distinct cluster closer to the human pneumoviruses, in particular hMPV. Antigenic differences occur between aMPV subtype A and B isolates, although they behave as one serotype in neutralization tests (9, 14). The U.S. aMPV/C isolate was serologically less related to aMPV/A or aMPV/B isolates, even though biochemical characteristics of all the MPVs are very similar (13). Genomic sequence analysis again confirmed the antigenic uniqueness of the U.S. virus (13) and supports classification of the U.S. isolate of aMPV as a subtype (subtype C) distinct from its European aMPV counterparts (40, 41, 43).
FIG. 2.
Phylogenetic relationships of the pneumoviruses based on the attachment glycoprotein sequences. After alignment of predicted amino acid sequences for the G attachment glycoprotein, an unrooted phylogram was generated by maximum parsimony. Absolute distances are presented above each branch, with bootstrap confidence levels presented in parentheses.
Nucleotide sequence accession numbers.
The nucleotide sequences for the gene sequences reported in this study have been deposited in GenBank, and the accession numbers are AF513020 for aMPV/CO/C, AY198393 for aMPV/MN2a/C, and AY198394 for aMPV/MN7/C.
Acknowledgments
We thank Joyce Bennett for automated nucleotide sequencing and Melissa Scott and Cassandra Smith for technical assistance.
This work was supported in part by USDA, ARS, CRIS project number 6612-32000-015-00D-085.
REFERENCES
- 1.Alexander, D. J. 1997. Newcastle disease and other Paramyxoviridae infections, p. 541-569. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. McDougald, and Y. M. Saif, Jr. (ed.), Diseases of poultry, 10th ed. Iowa State University, Ames, Iowa.
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, W. M. 1994. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc. Natl. Acad. Sci. USA 91:2216-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayon-Auboyer, M. H., C. Arnauld, D. Toquin, and N. Eterradossi. 2000. Nucleotide sequences of the F, L and G protein genes of two non-A/non-B avian pneumoviruses (APV) reveal a novel APV subgroup. J. Gen. Virol. 81:2723-2733. [DOI] [PubMed] [Google Scholar]
- 5.Beck, A., N. Zorn, M. C. Bussat, J. F. Haeuw, N. Corvaia, T. N. Nguyen, A. Van Dorsselaer, and J. Y. Bonnefoy. 2000. Synthesis and characterization of respiratory syncytial virus protein G related peptides containing two disulphide bridges. Dev. Biol. 103:231-236. [PubMed] [Google Scholar]
- 6.Bush, R. M., W. M. Fitch, C. A. Bender, and N. J. Cox. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 16:1457-1465. [DOI] [PubMed] [Google Scholar]
- 7.Cane, P. A., and C. R. Pringle. 1995. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J. Virol. 69:2918-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavanagh, D., and T. Barrett. 1988. Pneumovirus-like characteristics of the mRNA and proteins of turkey rhinotracheitis virus. Virus Res. 11:241-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, M. S., R. E. Gough, and D. J. Alexander. 1993. Antigenic differentiation of avian pneumovirus isolates using polyclonal antisera and mouse monoclonal antibodies. Avian Pathol. 22:469-479. [DOI] [PubMed] [Google Scholar]
- 10.Collins, M. S., R. E. Gough, S. A. Lister, N. Chettle, and R. Eddy. 1986. Further characterisation of a virus associated with turkey rhinotracheitis. Vet. Rec. 119:606. [PubMed] [Google Scholar]
- 11.Collins, P. L., K. McIntosh, and R. M. Chanock. 1996. Respiratory syncytial virus, p. 1313-1351. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 12.Cook, J. K. 2000. Avian pneumovirus infections of turkeys and chickens. Vet. J. 160:118-125. [DOI] [PubMed] [Google Scholar]
- 13.Cook, J. K., M. B. Huggins, S. J. Orbell, and D. A. Senne. 1999. Preliminary antigenic characterization of an avian pneumovirus isolated from commercial turkeys in Colorado, USA. Avian Pathol. 28:607-617. [DOI] [PubMed] [Google Scholar]
- 14.Cook, J. K., B. V. Jones, M. M. Ellis, L. Ling, and D. Cavanagh. 1993. Antigenic differentiation of strains of turkey rhinotracheitis virus using monoclonal antibodies. Avian Pathol. 22:257-273. [DOI] [PubMed] [Google Scholar]
- 15.Dar, A. M., S. Munir, S. M. Goyal, M. S. Abrahamsen, and V. Kapur. 2001. Sequence analysis of the nucleocapsid and phosphoprotein genes of avian pneumoviruses circulating in the US. Virus Res. 79:15-25. [DOI] [PubMed] [Google Scholar]
- 16.Feldman, S. A., R. M. Hendry, and J. A. Beeler. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 73:6610-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia, O., M. Martin, J. Dopazo, J. Arbiza, S. Frabasile, J. Russi, M. Hortal, P. Perez-Brena, I. Martinez, B. Garcia-Barreno, et al. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J. Virol. 68:5448-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedges, S. B. 1992. The number of replications needed for accurate estimation of the bootstrap P value in phylogenetic studies. Mol. Biol. Evol. 9:366-369. [DOI] [PubMed] [Google Scholar]
- 19.Howe, M. 2002. Australian find suggests worldwide reach for metapneumovirus. Lancet Infect. Dis. 2:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ina, Y. 1996. Pattern of synonymous and nonsynonymous substitutions: an indicator of mechanisms of molecular evolution. J. Genet. 75:91-115. [Google Scholar]
- 21.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, L. P., H. Q. Zheng, R. A. Karron, T. C. Peret, C. Tsou, and L. J. Anderson. 2002. Multiplex assay for detection of strain-specific antibodies against the two variable regions of the G protein of respiratory syncytial virus. Clin. Diagn. Lab. Immunol. 9:633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, R. C. 1996. Avian pneumovirus infection: questions still unanswered. Avian Pathol. 25:639-648. [DOI] [PubMed] [Google Scholar]
- 24.Juhasz, K., and A. J. Easton. 1994. Extensive sequence variation in the attachment (G) protein gene of avian pneumovirus: evidence for two distinct subgroups. J. Gen. Virol. 75:2873-2880. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: molecular evolutionary genetics analysis, version 1.01. The Pennsylvania State University, University Park, Pa.
- 26.Langedijk, J. P., A. H. Brandenburg, W. G. Middel, A. Osterhaus, R. H. Meloen, and J. T. van Oirschot. 1997. A subtype-specific peptide-based enzyme immunoassay for detection of antibodies to the G protein of human respiratory syncytial virus is more sensitive than routine serological tests. J. Clin. Microbiol. 35:1656-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langedijk, J. P., W. M. Schaaper, R. H. Meloen, and J. T. van Oirschot. 1996. Proposed three-dimensional model for the attachment protein G of respiratory syncytial virus. J. Gen. Virol. 77:1249-1257. [DOI] [PubMed] [Google Scholar]
- 28.Li, J., R. Ling, J. S. Randhawa, K. Shaw, P. J. Davis, K. Juhasz, C. R. Pringle, A. J. Easton, and D. Cavanagh. 1996. Sequence of the nucleocapsid protein gene of subgroup A and B avian pneumoviruses. Virus Res. 41:185-191. [DOI] [PubMed] [Google Scholar]
- 29.Ling, R., A. J. Easton, and C. R. Pringle. 1992. Sequence analysis of the 22K, SH and G genes of turkey rhinotracheitis virus and their intergenic regions reveals a gene order different from that of other pneumoviruses. J. Gen. Virol. 73:1709-1715. [DOI] [PubMed] [Google Scholar]
- 30.Ling, R., and C. R. Pringle. 1988. Turkey rhinotracheitis virus: in vivo and in vitro polypeptide synthesis. J. Gen. Virol. 69:917-923. [DOI] [PubMed] [Google Scholar]
- 31.Mead, D. A., N. K. Pey, C. Herrnstadt, R. A. Marcil, and L. M. Smith. 1991. A universal method for the direct cloning of PCR amplified nucleic acid. Bio/Technology 9:657-663. [DOI] [PubMed] [Google Scholar]
- 32.Myers, T. W., and D. H. Gelfand. 1991. Reverse transcription and DNA amplification by a Thermus thermophilus DNA polymerase. Biochemistry 30:7661-7666. [DOI] [PubMed] [Google Scholar]
- 33.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 34.Nissen, M. D., D. J. Siebert, I. M. Mackay, T. P. Sloots, and S. J. Withers. 2002. Evidence of human metapneumovirus in Australian children. Med. J. Aust. 176:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peret, T. C., G. Boivin, Y. Li, M. Couillard, C. Humphrey, A. D. Osterhaus, D. D. Erdman, and L. J. Anderson. 2002. Characterization of human metapneumoviruses isolated from patients in North America. J. Infect. Dis. 185:1660-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pringle, C. R. 1998. Virus taxonomy-San Diego 1998. Arch. Virol. 143:1449-1459. [DOI] [PubMed] [Google Scholar]
- 37.Randhawa, J. S., A. C. Marriott, C. R. Pringle, and A. J. Easton. 1997. Rescue of synthetic minireplicons establishes the absence of the NS1 and NS2 genes from avian pneumovirus. J. Virol. 71:9849-9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randhawa, J. S., S. D. Wilson, K. P. Tolley, D. Cavanagh, C. R. Pringle, and A. J. Easton. 1996. Nucleotide sequence of the gene encoding the viral polymerase of avian pneumovirus. J. Gen. Virol. 77:3047-3051. [DOI] [PubMed] [Google Scholar]
- 39.Seal, B. S. 2000. Avian pneumoviruses and emergence of a new type in the United States of America. Anim. Health Res. Rev. 1:67-72. [DOI] [PubMed] [Google Scholar]
- 40.Seal, B. S. 1998. Matrix protein gene nucleotide and predicted amino acid sequence demonstrate that the first US avian pneumovirus isolate is distinct from European strains. Virus Res. 58:45-52. [DOI] [PubMed] [Google Scholar]
- 41.Seal, B. S., H. S. Sellers, and R. J. Meinersmann. 2000. Fusion protein predicted amino acid sequence of the first US avian pneumovirus isolate and lack of heterogeneity among other US isolates. Virus Res. 66:139-147. [DOI] [PubMed] [Google Scholar]
- 42.Seibert, S. A., C. Y. Howell, M. K. Hughes, and A. L. Hughes. 1995. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol. Biol. Evol. 12:803-813. [DOI] [PubMed] [Google Scholar]
- 43.Shin, H. J., K. T. Cameron, J. A. Jacobs, E. A. Turpin, D. A. Halvorson, S. M. Goyal, K. V. Nagaraja, M. C. Kumar, D. C. Lauer, B. S. Seal, and M. K. Njenga. 2002. Molecular epidemiology of subgroup C avian pneumoviruses isolated in the United States and comparison with subgroup A and B viruses. J. Clin. Microbiol. 40:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swofford, D. 1998. PAUP*4.0: phylogenetic analysis using parsimony. Smithsonian Institution. Sinauer Associates, Inc., Sunderland, Mass.
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tripp, R. A., L. P. Jones, L. M. Haynes, H. Zheng, P. M. Murphy, and L. J. Anderson. 2001. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat. Immunol. 2:732-738. [DOI] [PubMed] [Google Scholar]
- 47.van den Hoogen, B. G., T. M. Bestebroer, A. D. Osterhaus, and R. A. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119-132. [DOI] [PubMed] [Google Scholar]
- 48.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]