Abstract
A method based on solid-phase cytometry for the detection and enumeration of single cells of Cryptococcus neoformans in serum and cerebrospinal fluid is described. Both viable and nonviable cells are detected by using fluorescence viability labeling and immunofluorescence. This 30-min procedure has a detection limit of 3 to 6 cells per ml.
Rapid methods for the diagnosis of cryptococcosis include India ink staining (IIS) (3), detection of a capsular antigen (3, 5), and recently PCR (2). IIS can detect 103 to 104 cells of Cryptococcus neoformans per ml within 1 min (3). Antigen detection is fast and sensitive but subject to occasional false positives (3, 5). We report the use of solid-phase cytometry (SPC) for the detection and enumeration of single cells of C. neoformans in serum and cerebrospinal fluid (CSF). SPC compares favorably with IIS in terms of speed but has a substantially lower detection limit.
Laboratory strains of different species, i.e., C. neoformans var. neoformans (n = 17), C. neoformans var. gattii (n = 2), Candida albicans (n = 4), Candida krusei (n = 3), Candida glabrata (n = 3), Candida tropicalis (n = 4), and a Trichosporon sp. (n = 4), were obtained from the Belgian Coordinated Collections of Microorganisms.
CSF samples (n = 3) and sera (n = 25) from patients with cryptococcosis previously proven by latex agglutination (IMMY-Immuno-mycologics, Norman, Okla.) were tested. In addition, C. neoformans was detected on three tracheoesophageal voice prostheses (TVPs) following detachment of the cells by vigorous mixing in 10 ml of buffered peptone.
Primary rabbit polyclonal antibodies (Novocastra, Newcastle upon Tyne, United Kingdom) against capsular polysaccharides of C. neoformans and secondary fluorescein-labeled goat anti-rabbit immunoglobulin G (Molecular Probes, Eugene, Oreg.) were diluted (1:100) in phosphate-buffered saline, pH 7.2.
Two 50-μl aliquots of a sample (CSF, serum, or an extract from a TVP) were filtered over Cycloblack polyester membranes (diameter, 25 mm; pore size, 2 μm) (Chemunex, Ivry-sur-Seine, France), and the retained cells were fluorescently labeled. To this end, the first membrane was incubated for 30 min at 30°C on a 25-mm-diameter cellulose pad (Millipore, Bedford, Mass.) impregnated with 600 μl of the viability stain ChemChromeV3 (Chemunex) (diluted 1:100 in Chemsol B2 [Chemunex]).
The second membrane was incubated for 30 min at 10°C on a glass fiber pad (Gelman Sciences, Ann Arbor, Mich.) impregnated with a mixture of primary and secondary antibodies (each diluted 1:100). The membranes were washed three times with 1 ml of phosphate-buffered saline and scanned with the ChemScan (Chemunex) (4).
The identity of presumed C. neoformans cells was confirmed by inspecting their typical morphology with an epifluorescence microscope (BX40; Olympus, Tokyo, Japan) connected to the ChemScan (4).
To isolate C. neoformans from serum and CSF, bird seed agar (BSA) (Becton Dickinson, Lexington, Ky.) was inoculated with 50 μl of the sample and incubated for 24 h to 2 weeks at 37°C. Positive results were observable at 24 to 48 h. The identity of colonies was confirmed with IIS and a urease test (2 to 4 h at 30°C).
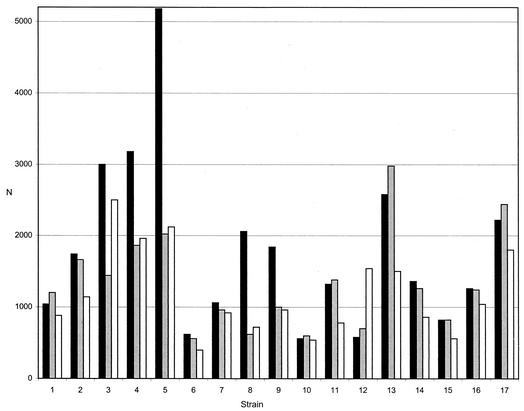
Numbers obtained in SPC for pure cultures of C. neoformans (n = 17) were compared with plate counts on BSA. Viability labeling yielded substantially higher numbers than the plate method except for one strain (Fig. 1). This difference was rationalized by the known ability of SPC to detect viable but nonculturable cells (6). Conversely, plate counts exceeded the SPC numbers after immunofluorescence labeling for 5 of the 17 test strains (Fig. 1). For 11 strains, viability counts were equal to or higher than those obtained with immunofluorescence (Fig. 1). A possible explanation for this unexpected result may be the occasionally poor accessibility of the antigen for the antibodies, leading to incomplete labeling and/or fluorescence intensity below the critical threshold value. Cross-reactivity of antibodies was observed with Candida glabrata, Candida tropicalis, and the Trichosporon sp.
FIG. 1.
Agreement between SPC (viability [black bars] and immunofluorescence labeling [gray bars]) and plate methods [white bars] for laboratory strains (n = 17). N, number of counts per milliliter in SPC or CFU per milliliter in the plate method.
The recovery at high numbers of cells was determined by mixing 500 μl of a suspension (102 cells) with 500 μl of CSF (4 strains) or 500 μl of serum (10 strains). The entire volume (1 ml) was filtered, resulting in approximately 100 cells on the membrane. Recoveries were 111.2% ± 9.7% (viability labeling) and 124.4% ± 19.0% (immunofluorescence labeling) for CSF (n = 4) and 111.9% ± 15.7% (viability labeling) and 119.9% ± 22.8% (immunofluorescence labeling) for serum (n = 10). To determine the recovery at low cell numbers, CSF (0.05, 0.5, 1.0, and 2.5 ml) or serum (50 and 100 μl) was supplemented with 100 μl of a suspension of C. neoformans in peptone to yield a final concentration of approximately 1 to 10 cells per total sample volume filtered. The experimental SPC counts were 5, 4, 3, and 7 (viability labeling) and 12, 5, 6, and 6 (immunofluorescence labeling). For spiked sera (50 or 100 μl) the corresponding values were 5 and 5 (viability) and 9 and 11 (immunofluorescence). The plate yielded two (0.05 ml), three (0.5 ml), three (1.0 ml), and four (2.5 ml) colonies for CSF and four (50 μl) and four (100 μl) colonies for serum.
From only 3 of the 28 samples could C. neoformans be cultured on BSA, yielding 1, 3, and 4 colonies per 50 μl. SPC detected 20 to 2,260 (viability labeling) and 80 to 2,160 (immunofluorescence) cells per ml in samples which were positive for the cryptococcal antigen at the time of sampling and which had subsequently been stored for up to 20 years. The numbers detected after immunological labeling were equal or higher than those obtained after viability labeling. The sera had been partly prepared from whole blood by gravity sedimentation rather than centrifugation, so that it was not unexpected that they still contained cryptococci. Fluorescent cells had the globose shape and capsule characteristic of C. neoformans var. neoformans. Furthermore, given the known history of the samples, i.e., their positivity in the latex agglutination test, these cells were most likely cryptococci.
SPC counts for the TVPs colonized by C. neoformans (1) containing culturable cells were 33 ± 16 (viability labeling) and 44 ± 8 (immunofluorescence), with a corresponding plate count of 28 ± 2 (n = 3) (counts given above include standard deviations).
The total analysis time for the SPC method is only 30 min. This allows the extremely rapid diagnosis of life-threatening cryptococcosis, particularly in those cases where numbers of C. neoformans cells are below the detection limits of other rapid methods.
Acknowledgments
This study was supported in part by an Award for Young Investigators from the British Society for Medical Mycology.
REFERENCES
- 1.Bauters, T. G. M., M. Moerman, G. Pini, H. Vermeersch, and H. J. Nelis. 2001. Colonization of a voice prosthesis by Cryptococcus neoformans. Med. Mycol. 39:379-381. [DOI] [PubMed] [Google Scholar]
- 2.Bialek, R., M. Weiss, K. Bekure-Nemariam, L. K. Najvar, M. B. Alberdi, J. R. Graybill, and U. Reischl. 2002. Detection of Cryptococcus neoformans DNA in tissue samples by nested and real-time PCR assays. Clin. Diagn. Lab. Immunol. 9:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall, A., and J. R. Perfect. 1998. Diagnosis and laboratory techniques, p. 381-405. In A. Casadevall and J. R. Perfect (ed.), Cryptococcus neoformans. ASM Press, Washington, D.C.
- 4.D'Haese, E., and H. J. Nelis. 2002. Rapid detection of single cell bacteria as a novel approach in food microbiology. J. AOAC (Assoc. Off. Anal. Chem.) Int. 85:979-983. [PubMed] [Google Scholar]
- 5.Dromer, F., E. Gueho, O. Ronin, and B. Dupont. 1993. Serotyping of Cryptococcus neoformans by using a monoclonal antibody specific for capsular polysaccharide. J. Clin. Microbiol. 31:359-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mignon-Godefroy, K., J.-G. Guillet, and C. Butor. 1997. Solid phase cytometry for detection of rare events. Cytometry 27:336-344. [PubMed] [Google Scholar]