Abstract
Twenty-eight emm12-type Streptococcus pyogenes isolates from patients with invasive and noninvasive infections or from asymptomatic carriers were genetically typed. Sequencing of drs (distantly related sic [streptococcal inhibitor of complement]) genes identified two novel alleles and revealed a polymorphism for drs similar to that of sic. No association was observed between the five different drs alleles and the five restriction patterns of the vir regulon for the isolates studied. These data suggest that drs sequencing may be useful for further differentiation of S. pyogenes isolates with emm12 and identical vir regulon restriction patterns.
The worldwide emergence of invasive Streptococcus pyogenes infections since the first description of streptococcal toxic shock syndrome almost 20 years ago (4) and the persistence of these infections are still unexplained (22). Particularly, isolates with prevalent emm types, largely emm1, followed by emm3, emm28, and emm12, have the ability to persist in the human environment for long periods, despite the fact that the world's population has likely been exposed to these strains on numerous occasions. Results of established molecular typing methods, including restriction fragment length polymorphism pattern analysis, random amplified polymorphic DNA (RAPD) analysis, and multilocus sequence typing, have revealed only very few individual clones of invasive emm1-type isolates over the past 2 decades (6, 17). Moreover, identical strains have also accounted for less serious infections, such as pharyngitis, cellulitis, and even asymptomatic carriage. Whether or not the high prevalence and persistence of such strains are due to an absence of protective immunity in a significant portion of the human population or to changes in the protective epitopes of the organism remains unclear (5, 7). Previous studies have shown that subtle differences, e.g., in the surface structure of the M1 protein, render some strains resistant to immune sera. Moreover, it has been postulated that natural selection on human mucosal surfaces for variants of the sic gene encoding the streptococcal inhibitor of complement (SIC) contributes to the emergence or reemergence of emm1-type S. pyogenes (13). Sequencing of the highly polymorphic sic genes of 1,132 of emm1-type S. pyogenes isolates derived from global sources has previously shown a high level of allelic diversity, with 220 distinct genes coding for 215 SIC protein variants. This polymorphism of sic has been applied to genetic subtyping of S. pyogenes isolates with emm1 and thus unambiguously differentiated isolates from temporally clustered invasive disease episodes (11). While the sic gene has previously been detected only within the vir(mga) regulon of all S. pyogenes strains harboring emm1 and outside the vir(mga) regulon of S. pyogenes strains with emm57 (1, 10), in a most recent study on S. pyogenes isolates from Japan, the presence of the sic gene was also reported with different frequencies for isolates harboring either emm2, emm4, emm12, emm28, emm75, emm89, emm94, or emm112 (15). drs (distantly related sic) genes have been detected subsequently within the vir(mga) regulon of S. pyogenes isolates harboring emm12 or emm55 (10). The deduced DRS protein sequences have a leader sequence very similar to that of SIC and a high degree of similarity to one C-proximal stretch of SIC.
The purpose of this study was to determine the genetic polymorphism of drs genes among invasive and noninvasive S. pyogenes isolates with emm12 and to evaluate drs as an epidemiological marker for the investigation of nosocomial infections and outbreaks.
(This work was presented in part at the 11th European Congress of Clinical Microbiology and Infectious Diseases, Istanbul, Turkey, 1 to 4 April 2001 [abstr. P1357], at the 53 Tagung der Deutschen Gesellschaft für Hygiene und Mikrobiologie, Aachen, Germany, 30 September to 4 October 2001 [abstr. P102], and at the 15th Lancefield International Symposium on Streptococci and Streptococcal Diseases, Goa, India, 6 to 11 October 2002 [abstr. P120]).
S. pyogenes isolates.
Twenty-eight epidemiologically unrelated emm12-type S. pyogenes isolates recovered from patients with invasive and noninvasive infections or from asymptomatic carriers that were referred to the National Reference Center for Streptococci in Aachen, Germany, between January 1996 and December 2000 were included in this study for further genetic typing.
emm gene sequencing.
emm typing of all 28 S. pyogenes isolates was performed according to the method of Podbielski et al. (19).
RAPD analysis.
PCR amplification was carried out according to previously published protocols by using either (i) the 9-mer arbitrary primer H2 (5′-CCT CCC GCC ACC-3′) (20) or (ii) the 10-mer arbitrary primers p14 (5′-GAT CAA GTC C-3′) and p17 (5′-GAT CTG ACA C-3′) (9).
vir(mga) regulon typing.
Typing of the vir(mga) regulon was performed by a modification of the method described by Gardiner et al. (9). To amplify the majority of the vir(mga) regulon, primer 5′-AAA CCG TAT CCT TTG ACG CAC TAG AGG ACA ATT TGC GAG ATT AG-3′ was used in combination with primer 5′-GAG CGC AAT GGGC AAG TTT ATC AAA TGG-3′. Cycling conditions included a preheat time of 2 min at 94°C, followed by 30 cycles of 94°C for 30 s, 60°C for 1 min, and 68°C for 7 min. Resulting PCR products were digested in three separate reactions for 2 h by using the restriction enzymes HaeIII, RsaI, and BsiYI (Roche Diagnostics, Mannheim, Germany). Fragments were separated by electrophoresis and visualized with ethidium bromide-stained agarose gel.
sic gene detection.
The presence of the sic gene was assessed by PCR with primers sic.I (5′-TAA GGA GAG GTC ACA AAC TA-3′) and sic.II (5′-TTA CGT TGC TGA TGG TGT AT-3′) as described previously (16).
drs gene sequencing.
For all 28 isolates, drs genes were sequenced on an ABI 310 automated DNA sequencer by using purified PCR products of the amplified vir(mga) regulon as described above and primers sicFdrs (5′-CAG CAG ATG AAG CAA GTA ATA GC-3′; positions 98 to 120 of drs12.01; accession number AJ 300679), sicRdrs (5′-CTT GTT TGT CAA TTT TGC TTT ACG ACC-3′; positions 861 to 835 of drs12.01) (10), id.for (5′-TTA AAG GAA TGG GGA ACA GCA G-3′; positions 317 to 338 of drs12.01), iu.rev (5′-TAT TAC TGC TGT TCC CCA TTC C-3′; positions 343 to 322 of drs12.01), hd.for (5′-CCT TCT GGT AAA AAC CCT C-3′; positions 535 to 553 of drs12.01), hu.rev (5′-GAG GGT TTT TAC CAG AAG G-3′; positions 553 to 535 of drs12.01), and e.for (5′-AGT AGT ATA CCA TCG CCA AG-3′; positions 745 to 764 of drs12.01) (2). The sequence editor DCSE, version 2.6, was used for editing, concatenation, and multiple alignments. The nucleotide sequence of drs12.01 was used as a reference.
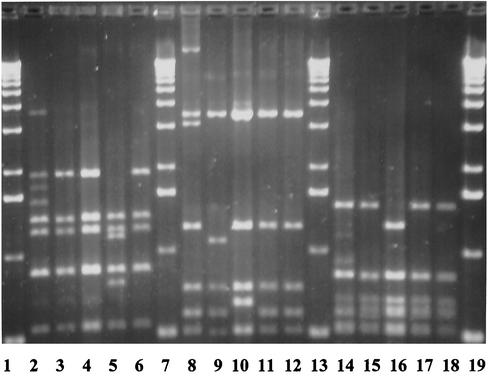
Results of molecular typing of the 28 S. pyogenes isolates are shown in Table 1. RAPD patterns were almost identical for all isolates with emm12. Subtyping of isolates identified five distinct restriction patterns of the vir(mga) regulon after digestion of the long PCR products with HaeIII, BsiYI, and RsaI. The different restriction patterns of the vir(mga) regulon are shown in Fig. 1. No PCR product of the expected size of the presently known sic alleles (between 800 and 1,300 bp) was detected by PCR in any of the strains studied. Sequencing of drs revealed five distinct alleles. Comparison of drs sequences with the previously published sequences of alleles of strains NS488 and DRV1 (10) and of drs12.01 and drs12.02 (2) identified two novel alleles: drs12.03 (accession no. AJ315146) and drs12.04 (accession no. AJ315147). The drs12.03 and drs12.04 alleles differed from drs12.01 by only a single nucleotide change and by three nucleotide changes, respectively, and resulted in variations in the deduced protein sequences. Figure 2 shows a partial alignment of the sequences of the new variants with those of the known drs alleles. No association was observed between individual drs alleles and the different restriction patterns of the vir(mga) regulon of the isolates studied. Except for drs12.04, all drs alleles found among invasive isolates were also found among isolates from noninvasive infections or carriers.
TABLE 1.
Results of molecular typing of S. pyogenes isolates harboring emm12
| Isolate | Clinical sign(s) and/or symptom(s)a | Site(s) of isolation | RAPD patternb
|
vir regulon pattern | drs allele | |
|---|---|---|---|---|---|---|
| A | B | |||||
| AC-1460 | Cellulitis, STSS | Blood, wound | I | I | 1 | drs12.04 |
| AC-1574 | STSS, necrotizing fasciitis | Blood | I | I | 1 | drs12.01 |
| AC-1575 | Erysipelas, STSS | Blood | I | I | 1 | drs12.01 |
| AC-1780 | Nasal carriage | Nasopharynx | I | I | 1 | drs12.03 |
| AC-1781 | Nasal carriage | Nasopharynx | I | I | 1 | drs12.01 |
| AC-1985 | Osteomyelitis | Joint fluid | D* | I | 2 | drs12.01 |
| AC-2080 | SIDS | Lung tissue | I | D** | 1 | drs12.03 |
| AC-2152 | Pharyngitis, rheumatic fever | Nasopharynx | I | I | 1 | drs12.01 |
| AC-2272 | STSS | Blood | I | I | 1 | drs12.03 |
| AC-2276 | SIDS | Trachea | I | I | 1 | drs12.02 |
| AC-2352 | Pharyngitis | Nasopharynx | I | I | 5 | drs12.02 |
| AC-2353 | Pharyngitis | Nasopharynx | I | I | 5 | drs12.01 |
| AC-2416 | Bacteremia | Blood | I | I | 1 | drs12.01 |
| AC-2417 | Bacteremia | Blood | I | I | 1 | drs12.01 |
| AC-2432 | Pharyngitis | Nasopharynx | I | I | 5 | drs12.02 |
| AC-2445 | Pharyngitis | Nasopharynx | I | I | 4 | drs12.02 |
| AC-2874 | Bacteremia | Blood | I | I | 1 | drs12.01 |
| AC-3024 | Bacteremia | Blood | I | I | 1 | drs12.01 |
| AC-3292 | Surgical wound infection, abscess | Wound | I | I | 1 | drs12.02 |
| AC-3297 | Abscess, cellulitis, STSS | Wound | I | I | 1 | drs12.02 |
| AC-3681 | SIDS | Meninges, pleura | I | I | 1 | drs12.01 |
| AC-3735 | Necrotizing fasciitis, myositis | Blood, wound | I | I | 1 | drs12.01 |
| AC-3854 | Septic arthritis, necrotizing fasciitis | Joint fluid | I | I | 1 | drs12.03 |
| AC-3954 | Sepsis | Blood | I | D** | 1 | drs12.01 |
| AC-3959 | Necrotizing fasciitis | Pleural fluid | I | I | 2 | drs12.01 |
| AC-3967 | Sepsis | Blood | I | D** | 1 | drs12.01 |
| AC-4075 | Necrotizing fasciitis | Blood | I | I | 1 | drs12.01 |
| AC-4098 | Recurrent tonsillitis | Nasopharynx | I | I | 3 | drs.NS488 |
STSS, streptococcal toxic shock syndrome; SIDS, sudden infant death syndrome.
RAPD patterns were obtained by using either primer H2 (A) or primers p14 and p17 (B). I, pattern indistinguishable; D*, pattern lacking three distinct bands; D**, pattern with one aberrant band out of 15 distinct bands.
FIG. 1.
Restriction fragments of the vir(mga) regulon after digestion of the PCR product with HaeIII (lanes 2 to 6), BsiYI (lanes 8 to 12), and RsaI (lanes 14 to 18). Lanes 1, 7, 13, and 19, DNA molecular weight marker; lanes 2, 8, and 14, isolate AC-2353 (type 5); lanes 3, 9, and 15, isolate AC-2445 (type 4); lanes 4, 10, and 16, isolate AC-4098 (type 3); lanes 5, 11, and 17, isolate AC-1985 (type 2); lanes 6, 12, and 18, isolate AC-1460 (type 1).
FIG. 2.
Partial alignment of the nucleotide sequences of drs alleles drs12.01, drs12.02, drs12.03, drs12.04, drs.NS488, and drs.DRV1 and of the deduced protein sequences DRS12.01, DRS12.02, DRS12.03, DRS12.04, DRS.NS488, and DRS.DRV1. The allele drs12.01 serves as a reference.
In contrast to findings of previous studies that have shown RAPD analysis to have additional discriminatory power over that of streptococcal M protein serotyping (20), the finding of identical RAPD patterns for almost all isolates in this study suggests that most current isolates with emm12 are closely genetically related and supports the need for reliable and reproducible genetic subtyping methods for conducting epidemiological investigations with S. pyogenes isolates harboring the same emm type.
The fact that none of the isolates studied harbored any of the known sic alleles within or outside the vir(mga) regulon is in agreement with previous results of studies from Australia and Europe that suggest that the presence of sic is confined to S. pyogenes strains with certain emm types (1, 10) and contrasts with the finding of a recent study that detected sic in 13 of 18 emm12-type S. pyogenes isolates from Japan (15). Whether or not the high prevalence of sic in emm12-type S. pyogenes isolates in Japan is due to epidemiological differences or to recent recombination events needs to be studied further. Even though drs genes have been detected only recently in S. pyogenes isolates harboring emm12 or emm55 (10), the finding of a substantial level of genetic diversity, including two novel distinct drs alleles, suggests that drs genes in S. pyogenes strains with emm12 may also exhibit a genetic polymorphism paralleling that of sic in S. pyogenes strains with emm1. All drs variants can be theoretically linked to each other by only a few molecular events, and all variants result in differences in the deduced protein sequences. This fact suggests a preference for nonsynonymous drs mutations over synonymous mutations, analogous to the molecular features of sic (11).
Previous hybridization studies and nucleotide sequence analysis from our laboratory have shown that the variations in sizes of restriction fragments of the vir(mga) regulon between invasive S. pyogenes isolates with emm1 were largely associated with variations within the sic gene (3). In contrast, other studies have demonstrated that even though the variability of sic was revealed by polymorphic fragments in the virR-emm1-restriction fragment length polymorphism analysis, the lack of a correlation between sizes of the polymorphic restriction fragment and the sic allele rather suggested a size variation in an insertion segment further downstream (18). Comparison of the vir regulon restriction patterns in this study revealed five different patterns. The fact that no association between the drs polymorphism and the vir(mga) regulon polymorphism was observed in this study suggests that drs may be a valuable target for epidemiological investigations of nosocomial infections and outbreak situations caused by S. pyogenes with emm12 and identical vir(mga) regulon patterns.
The finding that most drs alleles and vir(mga) regulon restriction patterns found among invasive isolates were also found among isolates from patients with pharyngitis or from asymptomatic carriers suggests that there is no association between the severity of disease and individual drs alleles. This observation provides insights into the reservoir for invasive S. pyogenes isolates and supports the hypothesis that the upper respiratory tract is the principal reservoir from which organisms causing invasive diseases are disseminated (8).
It has previously been shown that SIC is a secreted protein produced by M1 strains of S. pyogenes and that it inhibits the formation of the membrane attack complex of complement in vitro by binding to the C5b-9 complex (1). Intensive intranasal infection studies in a mouse model have shown a significantly impaired ability to colonize and persist in the upper respiratory tract for a sic-negative mutant compared to that for its parental strain (14). Moreover, recent studies have shown that SIC can act as an antiphagocytic effector molecule in the interior of host cells, thus providing important information on the interactions between SIC and host cells (12). Previous studies on emm1-type S. pyogenes isolates from patients with pharyngitis and treatment failure have shown that the selection of sic variants most likely occurs on human mucosal surfaces (2). Since epidemic waves are composed of highly heterogenous subclones based on distinct variants of sic (13), this diversification may contribute to an increased fitness of the isolates in the human-pathogen interactions and M1 epidemics (13). Even though previous studies have shown that DRS proteins like SIC are immunogenic in natural infections (21), it still needs to be determined in further studies whether or not SIC and DRS share common biological properties.
Nucleotide sequence accession number.
Sequences of alleles drs12.03 and drs12.04 have been deposited under accession no. AJ315146 and AJ315147, respectively.
Acknowledgments
This report is based in part on a project supported by grant RKI-415/1369 235 of the Federal Ministry of Health of Germany (Bundesministerium für Gesundheit).
We take full responsibility for the content of this publication.
We appreciate the excellent technical assistance of Marlies Breuer-Werle and Bruno Weidenhaupt.
REFERENCES
- 1.Akesson, P., A. G. Sjöholm, and L. Björck. 1996. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J. Biol. Chem. 271:1081-1088. [DOI] [PubMed] [Google Scholar]
- 2.Brandt, C. M., F. Allerberger, B. Spellerberg, R. Holland, R. Lütticken, and G. Haase. 2001. Characterization of consecutive Streptococcus pyogenes isolates from patients with pharyngitis and bacteriological treatment failure: special reference to prtF1 and sic/drs. J. Infect. Dis. 183:670-674. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, C. M., B. Spellerberg, R. Holland, and R. Lütticken. 2000. Correspondence between vir(mga)-regulon typing and sic gene polymorphism in Streptococcus pyogenes isolates with emm1-sequences, p. 163-166. In D. R. Martin and J. R. Tagg (ed.), Streptococci and streptococcal diseases: entering the new millennium. Proceedings of the 14th Lancefield International Symposium on Streptococci and Streptococcal Diseases. Securacopy, Porirua, New Zealand.
- 4.Cone, L. A., D. R. Woodard, P. M. Schlievert, and G. S. Tomory. 1987. Clinical and bacteriologic observations of a toxic shock-like syndrome due to Streptococcus pyogenes. N. Engl. J. Med. 317:146-149. [DOI] [PubMed] [Google Scholar]
- 5.de Malmanche, S. A., and D. R. Martin. 1994. Protective immunity to the group A streptococcus may be only strain specific. Med. Microbiol. Immunol. 183:299-306. [DOI] [PubMed] [Google Scholar]
- 6.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eriksson, B. K., A. Villasenor-Sierra, M. Norgren, and D. L. Stevens. 2001. Opsonization of T1M1 group A streptococcus: dynamics of antibody production and strain specificity. Clin. Infect. Dis. 32:E24-E30. [DOI] [PubMed]
- 8.Fiorentino, T. R., B. Beall, P. Mshar, and D. E. Bessen. 1997. A genetic-based evaluation of the principal tissue reservoir for group A streptococci isolated from normally sterile sites. J. Infect. Dis. 176:177-182. [DOI] [PubMed] [Google Scholar]
- 9.Gardiner, D., J. Hartas, B. Currie, J. D. Mathews, D. J. Kemp, and K. S. Sriprakash. 1995. Vir typing: a long-PCR typing method for group A streptococci. PCR Methods Appl. 4:288-293. [DOI] [PubMed] [Google Scholar]
- 10.Hartas, J., and K. S. Sriprakash. 1999. Streptococcus pyogenes strains containing emm12 and emm55 possess a novel gene coding for distantly related SIC protein. Microb. Pathog. 26:25-33. [DOI] [PubMed] [Google Scholar]
- 11.Hoe, N., K. Nakashima, D. Grigsby, X. Pan, S. J. Dou, S. Naidich, M. Garcia, E. Kahn, D. Bergmire-Sweat, and J. M. Musser. 1999. Rapid molecular genetic subtyping of serotype M1 group A streptococcus strains. Emerg. Infect. Dis. 5:254-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoe, N. P., R. M. Ireland, F. R. DeLeo, B. B. Gowen, D. W. Dorward, J. M. Voyich, M. Liu, E. H. Burns, Jr., D. M. Culman, A. Bretscher, and J. M. Musser. 2002. Insight into the molecular basis of pathogen abundance: group A streptococcus inhibitor of complement inhibits bacterial adherence and internalization into human cells. Proc. Natl. Acad. Sci. USA 99:7646-7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoe, N. P., K. Nakashima, S. Lukomski, D. Grigsby, M. Liu, P. Kordari, S. J. Dou, X. Pan, J. Voupio-Varkila, S. Salmelinna, A. McGeer, D. E. Low, B. Schwartz, A. Schuchat, S. Naidich, D. De Lorenzo, Y. X. Fu, and J. M. Musser. 1999. Rapid selection of complement-inhibiting protein variants in group A streptococcus epidemic waves. Nat. Med. 5:924-929. [DOI] [PubMed] [Google Scholar]
- 14.Lukomski, S., N. P. Hoe, I. Abdi, J. Rurangirwa, P. Kordari, M. Liu, S.-J. Dou, G. G. Adams, and J. M. Musser. 2000. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect. Immun. 68:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma, X., H. Kikuta, N. Ishiguro, M. Yoshioka, T. Ebihara, T. Murai, I. Kobayashi, and K. Kobayashi. 2002. Association of the prtF1 gene (encoding fibronectin-binding protein F1) and the sic gene (encoding the streptococcal inhibitor of complement) with emm types of group A streptococci isolated from Japanese children with pharyngitis. J. Clin. Microbiol. 40:3835-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mejia, L. M. P., K. E. Stockbauer, X. Pan, A. Cravioto, and J. M. Musser. 1997. Characterization of group A Streptococcus strains recovered from Mexican children with pharyngitis by automated DNA sequencing of virulence-related genes: unexpectedly large variation in the gene (sic) encoding a complement-inhibiting protein. J. Clin. Microbiol. 35:3220-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musser, J. M., V. Kapur, S. Kanjilal, U. Shah, D. M. Musher, N. L. Barg, K. H. Johnston, P. M. Schlievert, J. Henrichsen, D. Gerlach, R. M. Rakita, A. Tanna, B. D. Cookson, and J. C. Huang. 1993. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (Scarlet fever toxin). J. Infect. Dis. 167:337-346. [DOI] [PubMed] [Google Scholar]
- 18.Mylvaganam, H., B. Bjorvatn, and A. Osland. 2001. Polymorphism of the virulence regulon and allelic variations of the sic gene among the emm1 isolates of group A Streptococcus from western Norway. Microb. Pathog. 30:71-79. [DOI] [PubMed] [Google Scholar]
- 19.Podbielski, A., B. Melzer, and R. Lütticken. 1991. Application of the polymerase chain reaction to study the M protein(-like) gene family in beta-hemolytic streptococci. Med. Microbiol. Immunol. 180:213-227. [DOI] [PubMed] [Google Scholar]
- 20.Seppälä, H., Q. He, M. Osterblad, and P. Huovinen. 1994. Typing of group A streptococci by random amplified polymorphic DNA analysis. J. Clin. Microbiol. 32:1945-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sriprakash, K. S., J. Hartas, and A. White. 2002. Antibodies to streptococcal inhibitor of complement function and M peptides in a poststreptococcal glomerulonephritis endemic region of Australia. J. Med. Microbiol. 51:589-594. [DOI] [PubMed] [Google Scholar]
- 22.Stevens, D. L. 2001. Invasive streptococcal infections. J. Infect. Chemother. 7:69-80. [DOI] [PMC free article] [PubMed] [Google Scholar]