Abstract
Background
The abundance and the conservation of the repeated element (rep) genes in Ichnoviruses genomes suggest that this gene family plays an important role in viral cycles. In the Ichnovirus associated with the wasp Hyposoter didymator, named HdIV, 10 rep genes were identified to date. In this work, we report a relative quantitative transcription study of these HdIV rep genes in several tissues of the lepidopteran host Spodoptera frugiperda as well as in the H. didymator wasps.
Results
The data obtained in this work indicate that, in the early phases of infection (24 hours), HdIV rep genes each display different levels of transcripts in parasitized 2nd instar or HdIV-injected last instar S. frugiperda larvae. Only one, rep1, is significantly transcribed in female wasps. Transcript levels of the HdIV rep genes were found as not correlated to their copy number in HdIV genome. Our results also show that HdIV rep genes display different tissue specificity, and that they are primarily transcribed in S. frugiperda fat body and cuticular epithelium.
Conclusion
This work is the first quantitative analysis of transcription of the ichnovirus rep gene family, and the first investigation on a correlation between transcript levels and gene copy numbers in Ichnoviruses. Our data indicate that, despite similar gene copy numbers, not all the members of this gene family are significantly transcribed 24 hours after infection in lepidopteran larvae. Additionally, our data show that, as opposed to other described HdIV genes, rep genes are little transcribed in hemocytes, thus suggesting that they are not directly associated with the disruption of the immune response but rather involved in other physiological alterations of the infected lepidopteran larva.
Background
Polydnaviruses are obligatory endosymbionts of some endoparasitic Hymenoptera from Ichneumonid and Braconid families. They are integrated as provirus in wasp chromosomes. Viral replication occurs in calyx cells of the wasp ovary, and leads to the formation of multiple circular dsDNA encapsidated molecules. Viral particles accumulate in the oviducts and are injected through oviposition in the lepidopteran host larva.
Polydnaviruses do not replicate in the parasitized lepidopteran host, but infect several host tissues, what leads to viral gene expression in these tissues. Polydnaviruses induce major physiological alterations in parasitized host such as immune disruption, developmental arrest, hormonal alterations and a decrease in hemolymph storage proteins [1-5].
Recent sequencing programs of the polydispersed polydnavirus genomes reveal that a large proportion of the genes encoded by the circular DNA molecules are organized in gene families. This characteristic is common to the two polydnavirus families, the Ichnoviruses (IV), associated with ichneumonid wasps, and the Bracoviruses (BV), associated with braconid wasps [6,7]. We are studying the Ichnovirus associated with the endoparasitoid wasp Hyposoter didymator (HdIV), where several gene families have been identified so far [7,8]. Although only a fraction of HdIV genome is presently known, 10 members of a gene family named repeated element (rep) gene family have already been identified in the genome. Originally described by Theilmann & Summers [9] on the basis of multiple cross-hybridization between several Campoletis sonorensis IV (CsIV) genome segments, members of the rep family possess a conserved 540-bp repeated element motif, found singly or in multiple repeats [9-12]. The rep gene family is the largest conserved ichnovirus gene family identified to date [7,13]. Indeed, 30 members of the rep gene family have been reported in the fully sequenced CsIV genome, whereas 36 and ten rep genes are described in Hyposoter fugitivus IV (HfIV) and Tranosema rostrale IV (TrIV), respectively [7].
Although the function of the rep genes has not yet been elucidated, their conservation among ichnoviruses and their abundance in viral genomes both suggest that they play an important role in viral cycles. To date, transcription studies for ichnoviruses rep genes have been carried out by Northern blot analysis [12,14] or by RT-PCR [11] and have indicated that members of this gene family may be transcribed in both wasp and caterpillar hosts [11,14] and in different tissues of the parasitized lepidopteran host [12,14]. Variations in the number of transcripts during the first day after parasitism have also been suggested for members of this gene family by Northern-blot analysis [14]. Altogether, these results seem to indicate that rep genes show a wide range of expression patterns, making it difficult to identify any putative physiological function. Based on the abundance of rep genes in ichnoviruses genomes, one might expect that they have diverged in their expression pattern, acquiring specificity for given tissues, hosts or development stages.
In this work, we report the relative quantitative transcription study of the 10 rep genes identified to date in HdIV. The transcription studies were carried out on several tissues of the lepidopteran host Spodoptera frugiperda as well as in H. didymator adult wasps. Our data indicate that 24 hours after infection HdIV rep genes display different levels of transcription in parasitized or HdIV-injected S. frugiperda. Surprisingly, one of the rep genes, rep1, is significantly transcribed in female wasps. However, rep genes remain preferentially transcribed in the lepidopteran host compared to the wasp host. Our data show that transcription levels of the HdIV rep genes are not correlated to their copy number in HdIV genome. In addition, each HdIV transcribed rep gene displays tissue specificity, and the primary targets are the lepidopteran host fat body and cuticular epithelium.
Results and discussion
In HdIV, 10 rep genes are identified at present. Three have been previously described in HdIV segment SH-E (rep1, rep2 and rep3, [12]) and one in segment SH-G (named rep12 in this work, [15]). More recently, six additional sequences have been identified, which are available in the GenBank database (accession numbers in Table 4).
Table 4.
List of the gene-specific primers used in relative quantitative PCR analysis. Gene names and accession numbers are indicated. (*) Accession numbers for S. frugiperda correspond to Spodobase identifying numbers http://bioweb.ensam.inra.fr/spodobase/
| gene name | accession number | qPCR forward primer (5'-3') | qPCR reverse primer (5'-3') |
| HdIV | |||
| rep1 | AF364055 | AACGTGGAAACTTTGTCGCC | CGTTCCTGGAGGGACTACCC |
| rep2 | AF364055 | TCGGTGTGCTGATTGTGAGC | TCATGTCCCAAGTCACACGG |
| rep3 | AF364055 | GCCCCTGCCATTTGAAAAAT | TCGCGAATGCAGTAGCACTG |
| rep4 | AY499565 | CGGCGTGTCACAAACTGTTG | GCTTCAAGATGTTGCCCCATT |
| rep5 | AY499566 | GGAAGACCGCCTGCTTATCA | CCTCCGAATAAAGGCGTCAGT |
| rep6 | AY499567 | AAGGCCAGAAGAAGATCGCC | AGAGGCATGAGCCAGTCCC |
| rep7 | AY499568 | TCGTATCGTTCCACCGGGTA | CAGCCAGATGGTGGAAGCTC |
| rep8 | AY499569 | GTTTTGCCCCAATGGTGATG | TGCCACAGTTTTGCTCGAAC |
| rep11 | AY501383 | same as rep6 gene | same as rep6 gene |
| rep12 | AF479654 | GGGTCGCAATGAAGGTGCTA | CTGGCGAGTGTGTTTGCAAT |
| H. didymator | |||
| 18S RNA | AY433942 | CATCGTGGTGCTCTTCATTGA | CAAAGTAAACGTACCGGCCC |
| S. frugiperda | |||
| E2 ubiquitin ligase | SF9L03548 (*) | ACTTGTGGCCCGCATACACT | GGATCGGCACAATAAATGGG |
| RNA polymerase II | SF9L00930 (*) | TGCCATCGGGAAAATGAAAT | TTCTCTGCACCTTATTGGGTCTC |
Genome distribution and sequence analysis of HdIV rep genes
Characterisation of the segments containing the six new rep genes (rep4, rep5, rep6, rep7, rep8 and rep11) was achieved by Southern-blot analysis and PCR amplification of the corresponding circular molecules.
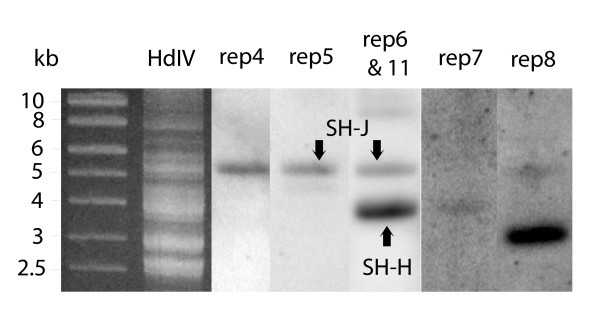
Southern-blot of HdIV segmented genome was performed with oligonucleotide probes specific to each of the newly identified rep genes, except for rep6. Indeed, rep6 and rep11 share 98 % nucleotide identity in their coding sequence and thus the rep6 probe was expected to cross-hybridize with rep11. As shown in Figure 1, the rep4 primer resulted in a hybridization band that co-localised with the hybridization band obtained with the rep5 primer (Figure 1, compare rep4 and rep5 lanes). The rep6 probe hybridized with two HdIV segments, firstly with a segment that co-localizes with the rep5 segment and secondly with another segment of lower size (Figure 1, rep6 lane). The faint hybridization band obtained with the rep7 probe had a size similar to that of the lower size segment to which the rep6 probe hybridized (Figure 1, compare rep7 and rep6 lanes). Lastly, the rep8 probe hybridized with a segment of smaller size compared to the other rep gene-containing HdIV segments. Thus, the Southern-blot analysis indicated that the 6 new rep genes are encoded by at least 3 different HdIV segments (Table 1).
Figure 1.
Characterization of the HdIV genomic segments encoding the novel 6 rep genes by Southern-blot analysis with gene-specific oligonucleotide probes. The molecular weight marker corresponds to linear DNA (kb). Purified HdIV DNA was separated on 1% agarose gel and stained with BET (HdIV), then transferred to Nylon membrane for hybridization with oligonucleotide probes specific to rep4 (rep4), rep5 (rep5), rep7 (rep7) and rep8 (rep8) genes. Due to high similarity between the rep6 and rep11 coding sequences, the rep6 probe (rep6&11) should allow detection of both genes. SH-J, containing rep5 and rep11 genes, and SH-H, containing the rep6 gene, are indicated by vertical arrows.
Table 1.
HdIV segments predicted to encode the rep genes analysed in this work. Segment names and putative sizes are indicated. Segment names were given alphabetically from the shortest to the longest; however only segments for which a real (completely sequenced; SH-E and SH-G) or estimated (PCR fragment; SH-J, SH-H and SH-A2 containing rep7 sequence) size could be given were named. SH-x and SH-y stand for segments of unknown size (since molecular weigh marker represents linear DNA).Rep7 is underlined because of discrepancy between PCR and Southern-blot results. For each segment, the rep gene(s) identified after sequencing of PCR amplification fragments or by Southern-blot analysis are reported
| segment | size (kbp) | sequencing/PCR | Southern-blot |
| SH-J | ~6 | rep5, rep11 | rep4 |
| SH-y | ~5 | -------- | rep7? |
| SH-H | ~5 | rep6 | |
| SH-G | 5.6 | rep12 | |
| SH-E | 4.6 | rep1, rep2, rep3 | |
| SH-x | ? | -------- | rep8 |
| SH-A2 | 3.1 | rep7 |
The HdIV rep-encoding segments were further analysed by PCR. Primers specific to the rep5 gene amplified a ~6 kbp fragment, whereas those designed within the rep6 gene amplified a ~5 kbp fragment. The HdIV super-helical (SH) segments are named alphabetically from the shortest to the longest. Thus, based on their size, the segment containing the rep5 gene was named SH-J and the one containing rep6 was named SH-H. Presence of the rep5 and rep6 genes was confirmed by partial sequencing of the two PCR fragments (GenBank accession numbers DQ295920 and DQ295919, for the segments SH-J and SH-H respectively). Sequencing revealed that SH-J also contained a sequence corresponding to the rep11 gene, thus confirming Southern-blot analysis where the rep6 probe hybridized with SH-H and cross-hybridized with rep11 present in SH-J (Figure 1, rep6 lane) whereas the rep5 probe hybridized solely with SH-J (Figure 1, compare rep5 and rep6 lanes). PCR using primers specific to the rep7 gene resulted in a 3.1 kbp fragment. Sequencing of this PCR fragment (GenBank accession number DQ295918) revealed a sequence identical to rep7. However, Southern-blot analysis suggested that a larger segment encodes this gene (Figure 1, rep7 lane). The discrepancy between the two results could be explained by the existence of two segments encoding this gene, similar to the rep1 gene, which is encoded by both SH-E and SH-Evar [12]. However, this result will need to be confirmed by identification and sequencing of the rep7 hybridizing segment.
Therefore, based on both Southern-blot and PCR results, we can conclude that the 10 HdIV rep genes are encoded by at least 5 different HdIV molecules (Table 1).
Members of the rep family are characterized by a conserved 540-bp repeated element motif, found singly or in multiple repeats [9,11]. All the 10 rep genes identified to date in HdIV encode proteins containing a single repeated element motif (Figure 2). However, only part of the HdIV genome is presently known and therefore we cannot exclude existence of multiple-repeat containing genes in this Ichnovirus.
Figure 2.
ClutalX alignment of deduced amino acid sequences of HdIV and selected ichnoviruses rep genes. The first 2 letters indicate ichnovirus species (Cs: Campoletis sonorensis; Hd: Hyposoter didymator; Hf: Hyposoter fugitivus; Tr: Tranosema rostrale), followed by the name of the segment containing the corresponding gene (except for Hd ichnovirus) then the rep gene number. Arrow-heads indicate beginning and ending of the conserved repeated element motif as defined by Theilmann & Summers [14]. Different shades of grey indicate conserved residues. Consensus sequence represents conserved residues: in capital letters: residues with >80% identity; p: polar residue; h: hydrophobic residue; l: aliphatic residue; +: positive residue; b: big residue; s: small residue.
All the rep genes described to date lack intron and encode proteins with no predicted signal peptide [11,12,14]. The HdIV deduced rep proteins analysed in this work follow this rule. Moreover, immunofluorescence studies in cell lines transfected with rep proteins coupled in their C-terminal part to GFP confirm that the GFP-rep1, rep3 and rep5 proteins are intracellular (Galibert et al., unpub.).
ClustalX alignment [16] of rep proteins from HdIV and from other Ichnoviruses reveals a high degree of conservation in the repeated element motif (Figure 2). In contrast to the repeat element motif, the N-terminal and C-terminal sequences greatly diverge among the different rep sequences. The close similarity between rep6 and rep11 (at both nucleotide and amino acid levels) suggests the genes have diverged recently. Surprisingly, rep11 lacks the C-terminal part of the repeated element motif, compared to the other rep proteins.
Whole rep protein sequences of several Ichnoviruses containing a single repeat and accessible on GenBank database (10 HdIV, 2 HfIV, 1 TrIV and 25 CsIV proteins) were aligned by ClustalX [16] to generate trees (data not shown). Results did not indicate a clustering by virus species, regardless of the method used, distance and parsimony (PHYLIP package [17]), but rather a dispersion of HdIV sequences among the other ichnovirus sequences (data not shown). This distribution was different from that seen in previous studies, comparing a lower number of sequences, where rep proteins clustered by virus species [12]. Phylogenetic analysis of this important and diversified gene family would require supplementary studies, in order to understand if rep genes are derived from a single ancestor gene or if several rep genes existed prior to the association between an ichneumonid wasp and a polydnavirus ancestor.
Transcription in the parasitized lepidopteran host
Transcription of the 10 HdIV rep genes was analysed by quantitative PCR during the first 24 hours following parasitism in S. frugiperda larvae parasitized at their 2nd instar. Larvae parasitized by H. didymator rapidly exhibit reduced food consumption and growth, and their development is arrested at the end of the fourth larval instar, after the 8 days needed for completion of parasitoid larval development.
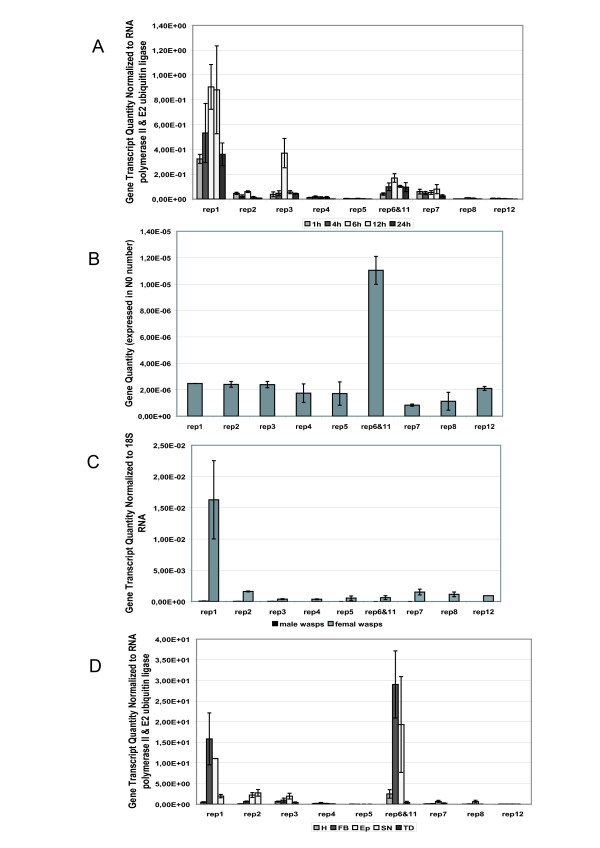
Our data reveal that in the initial phases of parasitism important differences are found between the transcript levels of the different HdIV rep genes when considering the overall expression of the genes (Figure 3A). The highest level of transcripts corresponds to the rep1 gene, followed by the rep7, rep3 and rep2 genes. For example, 1 hour after parasitism, the ratios of rep1 transcripts (N0 value) to rep3, rep2 and rep7 transcripts are 8.4 ± 0.5, 7.0 ± 0.2 and 5.4 ± 0.3, respectively; 24 hours after parasitism, the ratios of rep1 compared to rep3, rep2 and rep7 are 8.2 ± 0.3, 43.3 ± 0.3, and 14.2 ± 0.3, respectively. Because of the high degree of identity between the rep6 and rep11 sequences, we were not able to design pairs of primers specific for each of the genes. Therefore, the results obtained in quantitative PCR include both genes. Nonetheless, rep6 and rep11 transcript levels are generally similar to rep7 and significantly higher than other rep genes such as rep4, rep8, rep5 or rep12. The rep5 and rep12 transcripts are detected at very low levels in the parasitized larvae (557.6 ± 0.3 fold and 1789.0 ± 0.3 fold respectively less than rep1 at 24 h post-parasitism) suggesting that transcription of these two genes in the lepidopteran host may have no real biological significance.
Figure 3.
Expression profiles and gene copy number of the 10 rep genes identified in HdIV by relative quantitative PCR. A. Transcript levels in 2nd instar S. frugiperda parasitized larva, over 1-h to 24-h time course study. B. Relative gene copy numbers in HdIV genome. C. Transcript levels in H. didymator adult female and male wasps. D. Transcript levels in different tissues of last instar S. frugiperda larvae 24 hours after injection of HdIV (H: Hemocyte; FB: Fat Body; Ep: Cuticular Epithelium; SN: Nervous System (Head); TD: Digestive Track). Data are means ± SE of starting quantity of fluorescence (N0 value) for 6–9 measurements. For A, C and D, data are normalized to housekeeping genes RNA polymerase II and E2 ubiquitin ligase. For details, address to Methods chapter.
As indicated in Figure 3A, transcript levels remain relatively constant inside the whole parasitized S. frugiperda larvae over the first 24 hours of parasitism, for each of the HdIV rep genes with the exception of the rep3 gene, which appears to have transcript levels that are 6-fold higher at 6–9 hours post-parasitism compared to other points in the kinetic. These results are consistent with those obtained by Theilmann & Summers [14] in CsIV who observed through Northern blot experiments that some rep genes were slightly more transcribed 2 h and 6 h after parasitism than latter in parasitism (1d-8d). Nevertheless, the biological significance of this peak of transcription for the HdIV rep3 gene needs to be further investigated.
Transcript levels of the HdIV rep genes were also analysed during the time course of parasitism (data not shown). Preliminary results indicate that the differences in transcript levels between the HdIV genes are similar to those observed in early phase of parasitism. Moreover, transcript levels remain constant over the duration of parasitoid development. Therefore, the 10 HdIV rep genes studied here do not show variations in the course of parasitism as it has been described for some bracovirus genes [18].
In HdIV, the differences in transcript levels of the rep genes inside the whole parasitized S. frugiperda larvae are not related to their corresponding gene copy number in the HdIV segmented genome. Indeed, rep1 is more transcribed than both rep2 and rep3 although the 3 genes are found on the same viral segments SH-E and SH-Evar [12] and thus display the same gene copy numbers in the HdIV genome. Different patterns and levels of transcripts in the parasitized host for genes located on the same polydnavirus segment have also been previously described for the CsIV [19] and for the Bracovirus associated with Chelonus inanitus [18]. The absence of a correlation between transcript level and gene copy number was further assessed by estimating the relative copy numbers of each of the 10 rep genes from purified HdIV DNA (Figure 3B). As expected, our quantitative PCR assay revealed similar numbers of gene copies for the genes encoded by the same segments, rep1, rep2 and rep3. Furthermore, our results indicate that rep1, for which a high level of transcripts was detected, and rep12, a gene that is almost not transcribed, have similar numbers of copies within the HdIV genome (Figure 3B, compare rep1 and rep12). Overall, our data indicate that there are no significant differences within the HdIV genome between the copy number for rep1, rep4, rep5, rep7, rep8 and rep12. The rep6 and rep11 genes represent an exception (Figure 3B, rep6&11). Since rep6 and rep11 genes are both amplified by rep6 primers, the N0 value indicated in Figure 3B corresponds to the sum of rep6 (segment SH-H) and rep11 (segment SH-J) gene copies. The proportion of the N0 value due to rep11 can be estimated by the value obtained for rep5, since both genes are on the same segment SH-H. This indicates that rep11 gene (on SH-H segment) represents 15.5% of the total N0 value, whereas the rep6 gene (on SH-J segment) represents 84.5% of the N0 value. On the other side, quantification of the signal intensity obtained on Southern-blot (Figure 1, column rep6&11) indicates that SH-J (containing rep6 gene) and SH-H (containing rep11 gene) represent 78% and 15% hybridization signal, respectively. A third hybridization signal with a high molecular weight segment, representing 7% of the total signal intensity, was also detected in Southern-blot (Figure 1). Taken together, our results indicate that SH-J, containing the rep6 gene, is represented at least 5 times more than other rep-containing segments. Thus, although more abundant, rep6 is less transcribed than rep1 in parasitized larvae, result that confirms absence of correlation between copy numbers and transcription levels for the analysed HdIV rep genes.
To conclude, our results indicate that rep genes transcript levels are variable inside the parasitized caterpillars and are not linked to their relative copy numbers on HdIV genome thus suggesting that transcript levels of the HdIV rep genes are directly correlated to their promoter activities.
Transcription in the wasp host
Since some of the CsIV rep genes are transcribed in both lepidopteran and hymenopteran hosts [11,14], we investigated transcription of HdIV rep genes in 2–3 days old H. didymator female and male adult wasps. At this time, viral replication is taking place in the calyx cells [20]. In the female wasps, the rep genes are transcribed, but at a very low level, with the exception of rep1, which was significantly more transcribed compared to the other rep genes (Figure 3C). In the male wasps, transcript level is more than 200-fold lower than in females, suggesting that transcription of HdIV rep genes is residual in male wasps. This result differs from previous reports on C. inanitus bracovirus where 5 out of 6 analysed CiBV genes were transcribed at similar levels in male and female wasps [18]. The finding that transcription of rep1 gene is restricted to H. didymator females suggests an unexpected complex regulation of gene transcription, regardless transcripts are generated from the integrated or from the excised viral DNA. The remaining question is if rep1 transcription is restricted or not to the replicative calyx cells and thus if it may be related to HdIV viral particle production.
The HdIV rep1 gene is therefore the most transcribed rep gene in both parasitized S. frugiperda and whole adult female wasps. Whether transcription of rep1 gene is more important in parasitized S. frugiperda larvae than in the female wasp remains to be clearly established. By assuming that reverse transcription and PCR efficiencies were identical in the samples issued from both S. frugiperda larvae and female wasps, we were able to compare the N0 values obtained. In both samples, non-normalized N0 values are around 4E-07, which indicates that rep1 transcript levels are similar in both insect hosts. We can therefore assume that transcription of the rep1 gene in female wasps has a biological significance although it remains to be clarified whether the related protein has a function in the wasp and if this function is the same as that in the parasitized lepidopteran host.
Pattern of transcription in different tissues of HdIV-infected S. frugiperda larvae
In order to assess if HdIV rep genes have tissue specific patterns of transcription, quantitative analysis was performed in different tissues of S. frugiperda last instar larvae. Our results show that, 24 hours after HdIV injection, the HdIV rep genes are preferentially transcribed in the fat body and cuticular epithelium, and to a lower extent in the nervous system of the infected host (Figure 3D). Finding of a preferential transcription of the rep genes within these 3 tissues is consistent with previous results obtained by Northern-blot analysis for the HdIV rep1 gene [12].
In HdIV-injected last instar larvae, as in the parasitized 2nd instar larvae, rep4, rep5, rep8, rep12, but also rep7 show very low transcript levels in all tissues examined, whereas rep1, rep6, and to a lower extent, rep2 and rep3, are detected at higher levels (Figure 3D). In this assay, where tissues are analysed individually, rep1 transcripts are not any longer the most abundant. Indeed, rep6 transcripts level is similar to that of rep1 transcripts, in particular in the fat body and cuticular epithelium (despite a high variation between the biological samples for rep6 in cuticular epithelium, as indicated by the standard error, Figure 3D). This result has to be modulated by the fact that, in this assay, both rep6 and rep11 transcripts were measured and the proportion of each of the two genes is not known. However, rep6 preferential transcription in HdIV-injected last instar larvae fat body was corroborated by Northern blot analysis using rep genes specific oligonucleotide probes. Indeed, only one hybridization signal was detected, which corresponded to the rep6 specific probe in the fat body tissue (data not shown).
Our results indicate that the highest levels of rep gene transcripts are detected in the fat body and the cuticular epithelium (Figure 3D). Other ichnovirus genes of unknown function, such as TrIV1, also target primarily the fat body and the cuticular epithelium, with few transcripts detected in hemocytes [21]. In these two tissues, the HdIV rep6 and rep1 are the most represented transcripts, both at comparable levels. The rep2 and rep3 transcripts are also detected in fat body and cuticular epithelium, but at levels approximately 10 to 15-fold lower than those of rep1 and rep6 genes. In the nervous system, we detected mainly rep2 and rep1 transcripts, although at lower levels than in fat body and cuticular epithelium. For example, rep2, the highest transcribed gene in the nervous system, has 5-fold fewer transcripts than the rep1 gene in fat body. Compared to others tissues, transcripts in the digestive tract are almost undetectable for all the genes considered, suggesting that rep genes do not target this tissue. Whether this is due to promoter activity or virus penetration in this tissue remains to be determined.
Injection of purified viral HdIV particles inside S. frugiperda last instar larvae induces the inhibition of the cellular immune response and results in reduction of larval growth leading to abnormal or lack of pupation. Interestingly, the rep genes are expressed at low levels in hemocytes, as opposed to other HdIV genes [8,15,22] or genes from other polydnaviruses, which frequently preferentially target the blood cells [18,19,23,24]. The only rep gene that is transcribed significantly in the hemocytes is rep6 but transcript levels are still 6-fold less than in fat body and cuticular epithelium.
Based on the nature of the tissues where HdIV rep genes are preferentially transcribed and on the fact that rep proteins remain intracellular, we can hypothesize that members of this gene family play a small or an indirect role in cellular immune-suppression. The rep genes may thus mediate other physiological alterations of the parasitized caterpillar such as developmental/growth arrest.
Conclusion
This study by relative quantitative PCR allowed us to demonstrate that a number of HdIV rep genes are not transcribed at the same levels in the parasitized lepidopteran host. Even if transcript levels do not account for protein activity and needs, we can make hypotheses to explain the low transcript levels seen for some of the rep genes (rep4, rep5, rep8, rep12). Firstly, rep genes could be involved in host range for H. didymator wasp and those genes could be more transcribed inside other hosts. Another possibility is that these low transcribed rep genes have become pseudogenes, through genomic rearrangement in the wasp DNA. For example HdIV SH-G contains rep12 and HdGorf1, but the two open reading frames are on complementary strands [15]. Differences in transcript level between HdGorf1, which are similar to those of rep1 (data not shown), and rep12 could be related to their orientation on the viral segment. A third possibility would be that the rep genes that were not detected in fat body, cuticular epithelium or nervous system are expressed in other, less abundant tissues such as the endocrine glands.
HdIV rep genes seem to be specifically transcribed into the Lepidoptera host rather than in the Hymenoptera host, except maybe for the rep1 gene. In infected S. frugiperda larvae, the rep genes transcripts are detected mostly in fat body, cuticular epithelium and nervous system. Interestingly rep3 gene transcripts are found at the same level than rep1 transcripts in Sf9 cells infected with HdIV (data not shown), showing that viral gene regulation can differ in in vivo and in vitro systems.
The question whether rep genes have the same functions in different tissues has yet to be answered. Based on their transcription profiles, it is possible that rep genes do not have a direct role in the disruption of the immune response of the infected lepidopteran larva, but rather that they contribute to the manipulation of lepidopteran host larval growth and development.
Methods
Insect material
Rearing of Spodoptera frugiperda larvae and Hyposoter didymator wasps, as well as HdIV virus and DNA purifications, were conducted as described in [8].
For transcriptional studies in parasitized S. frugiperda larvae, second instar larvae were placed in presence of H. didymator female wasps for 3 hours. Negative controls corresponded to non-parasitized larvae.
To study the transcription of HdIV rep genes in HdIV-injected S. frugiperda larvae tissues, purified virions were injected into S. frugiperda last instar larvae (3 wasps equivalent/larva, representing 28 μl). For negative controls, last instar larvae were injected with an identical volume of saline buffer (PBS).
Southern blot analysis for identification of HdIV rep genes
Identification of HdIV segments containing the new rep genes was carried out by Southern blot analysis. 3 μg of purified HdIV DNA and linear DNA molecular weight marker (Eurogentec) were migrated on 1% agarose gel and transferred on positively charged nylon membranes (Boehringer). Gene specific oligonucleotide probes were selected in the coding sequence of each rep gene (sequences in Table 2). Specificity of the probe was ascertained with Blastn at the NCBI http://www.bioweb.ensam.inra.fr/spodobase/. Membranes were pre-hybridized for 3 hours at the same temperature than hybridization (see below) in a solution containing 5X Denhardt, 5X SSC, 0.1% SDS, and 100 μg/ml of salmon sperm DNA. Hybridization was carried out for 20 hours with oligonucleotide probe specific of each rep gene (hybridization temperature is indicated next to the primer in Table 2). The probes were labelled using γ-32P-ATP with T4 polynucleotide kinase (Promega). A DNA weight marker was hybridized with linear pUC-18 DNA labelled with α-32P-dCTP in a random priming reaction in the same conditions as described above (hybridization temperature 42°C). Membranes were rinsed at room temperature twice for 5 minutes in 2X SSC; 0.1% SDS solution, and once for 10 minutes in 0.2X SSC; 0.1% SDS solution. PhosphorImaging was performed on a STORM 840 apparatus (Amersham). Quantification of bands intensity was performed using ImageQuant 5.2 software from Amersham.
Table 2.
List of the gene-specific oligonucleotide probes used in Southern-blot. Hybridization temperature is indicated next to the primer
| Gene | oligonucleotide (5'-3') | Pre-hybridization & hybridization temperature |
| rep4 | GATGTTGCCCCATTTCTAGAACCGCAACAG | 48°C |
| rep5 | AGGGGCCCCACGCGGTAGACGAAACCCACG | 54°C |
| rep6 &11 | GCCCGCGGAACGTGAAGGTGTCCACCGGGT | 50°C |
| rep7 | CCTGCGAAATTTCTTGATACACCACAGCCT | 47°C |
| rep8 | TTCTCGTTGCAGCCCGTGACAGGCGCGAGC | 53°C |
PCR amplification of HdIV rep-containing segments
Characterization of HdIV segments containing the new rep genes was conducted by PCR with primers specific for each rep gene (Table 3). PCR was conducted with High Fidelity Taq DNA polymerase (Invitrogen) in standard conditions with 0.1 μg of DNA as a template and an annealing temperature of 60°C.
Table 3.
List of primers used to amplify the rep-containing HdIV segments by Polymerase Chain Reaction
| HdIV segment | rep gene | forward primer (5'-3') | reverse primer (5'-3') |
| SH-A2 | rep7 | ATCTTAAAGTGAGCACTATTGACGC | CCTGCGAAATTTCTTGATACACCACAGCCT |
| SH-H | rep6 | AAGTGTTGCTTGACTCGGCT | TCAAGTCCAGGTTTCGGATC |
| SH-J | rep5 | CTTGGTTACTCCAGCCCTTG | ACTCCTCCGAATAAAGGCGT |
Sequence analysis
Alignment of the deduced amino acid sequences encoded by the 10 HdIV rep genes, the HfIV rep genes (Hfc3rep (GenBank: AY597815) and Hfb15 rep (GenBank: AY570798)), the TrIV TrFrep gene (GenBank: AF421353) and the CsIV CsBrep gene (GenBank: AAA42923) was carried out with ClustalX [16] using default settings.
RNA isolation
To study the transcript levels of the rep genes in parasitized S. frugiperda larvae, total RNA was isolated from second instar larvae 1 h, 4 h, 6 h, 12 h and 24 h after parasitism. For each time point, 15 larvae were collected and homogenized in 1 mL TRIzol reagent (Invitrogen). For tissue specific transcription analysis, tissues were collected from 10 last instar HdIV-injected S. frugiperda larvae, 24 h after injection of HdIV or PBS. The tissues collected were hemocytes, digestive track, head (for nervous system), fat body and cuticular epithelium (including the muscles attached to the cuticle). With the exception of hemocytes, which were directly collected in TRIzol reagent, tissues samples were rinsed in PBS prior to collection. Tissues were then ground in 1 mL of TRIzol reagent. For the wasps' samples, 2 days old female and male wasps (20 of each) were ground in 1 mL TRIzol reagent. For each assay, RNA was collected from three independent sets of insects (biological replicates).
Total RNAs were extracted following the manufacturer's protocol. Total RNA samples were then incubated overnight at -20°C in 2 M of LiCl, centrifuged 30 min at 7500 g, rinsed 2 times with ethanol 75% and re-suspended in nuclease free water (Promega). RNA samples were quantified through spectrometry. The quality of the extracted RNA was confirmed on a 1% agarose gel.
To eliminate contaminating DNA, 8 μg of each RNA sample were treated with 8U of RQ1 DNAse (Promega) for 3 h at 37°C, following the manufacturer protocol. Samples were then ethanol precipitated with sodium acetate, rinsed twice in 75% ethanol and re-suspended in nuclease free water. The RNA samples treated with RQ1 DNAse were checked by PCR for the absence of contaminating DNA before being submitted to RT-PCR. For the S. frugiperda RNA samples, the absence of genomic contaminating DNA was controlled with primers amplifying the actin sequence (forward 5'-CAACTGGGACGACATGGAGAAGAT-3'; reverse 5'-CCACCGATCCATACGGAGTATTTC-3'). The absence of viral DNA contamination was controlled with primers amplifying the sequence of HdIV rep6 gene (forward 5'-ATGCCGTTCAACGAAGATCACGAC-3'; reverse 5'-GCTGCACCATGGCCGGAACTG-3'). For the H. didymator RNA samples, we used the primers amplifying the rep6 gene to control both absence of wasp and viral genomic DNA. The following protocol was used: 0.5 μg of each RNA sample served as matrix for RT-PCR using SuperScript™ III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen) and PCR using Platinium Taq DNA polymerase with the same buffer as the RT-PCR kit. The PCR program for both PCR and RT-PCR was 48°C 30 min; 94°C 5 min and 30 cycles 94°C 30 s; 55°C 30 sec; 1 min 68°C.
cDNA synthesis for relative quantitative PCR
Reverse transcription was carried out on 8 μg of total RNA using SuperscriptII reverse transcriptase (Invitrogen), following the manufacturer's protocol. 1U of RNAsin plus (Promega) was added in the reaction medium. Reverse transcription was carried out for 3 h.
Relative quantitative PCR
For Relative Quantitative PCR, primers were designed with the Primer Express software (version 2, Applied Biosystem). The gene specificity of the primers was verified using BLASTn (NCBI). The list of primers used is shown in Table 4. The primers for rep1, rep2 and rep3 were designed in such a way that the genes encoded by both SH-E and SH-Evar [12] were amplified.
For transcription studies, each of the 3 biological replicate samples was analysed in triplicate and non-template controls were included in duplicate or triplicate in each assay. Reactions were performed in 96-well PCR plates (ABgene). For PCR using HdIV DNA as template, 0.16 ng of DNA was used, and the experiment was conducted on two sets of independently collected DNA samples (biological replicates). For the PCR using cDNA as template, an amount of cDNA corresponding to 100 ng reverse transcribed total RNA was used. Each template was amplified in a volume of 25 μl containing 1X PCR buffer (Invitrogen), 3 mM of MgCl2, 200 μM dNTP mix (Invitrogen), 0.2 μl of 1/2000 dilution stock solution of SYBR green I (Invitrogen), 0.5 μM of ROX dye (Interchim), 0.4 μM of couples of primers and 0.1U of Platinium Taq DNA polymerase (Invitrogen).
Relative Quantitative PCR were performed on an ABI PRISM 7000 apparatus (Applied Biosystems) using the following thermal profile: 95°C 2 min and 40 cycles: 95°C 15 sec, 60°C 1 min. The specificity of the amplicons synthesised during the PCR was ascertained by performing a dissociation curve protocol from 60°C to 95°C.
Relative quantitative PCR results analysis
Analysis of Relative Quantitative PCR results was performed with the program LinReg PCR developed by Ramakers et al. [25], using the Rn values (SYBR green I fluorescence normalized to ROX passive dye fluorescence, given by the Sequence Detection Software of Applied Biosystem) as entries. This approach gives the initial number of molecules presents in each sample (N0 value). The mean of the 3 technical replicates N0 values was calculated.
Transcription results, obtained in S. frugiperda larvae, were first normalized, according to Vandesompele et al. [26], to the geometrical mean of 2 selected housekeeping genes: the RNA polymerase II and the E2 ubiquitin-conjugating enzyme. These two genes were chosen because their ratio was constant regardless of the tissue studied. Transcription results obtained in H. didymator wasps were first normalized to the 18S RNA gene.
For comparison between biological replicates, we introduced a second normalization step, aimed at reducing variability due to possible different quantities of virus inoculated or parasitism rates. Using the geNorm program [26], we first controlled that, for a same tissue or for a same time in the kinetic study, each rep gene behaves similarly in the 3 biological samples. Then using the geNorm program, a normalization factor was calculated for each tissue or time point, taking the most stable genes identified by the previous control, with the M value (internal gene-stability measure) set to 3. After normalisation, average values and standard errors were calculated for the 3 biological replicates.
Normalisation of the rep gene copy numbers on HdIV genome between the 2 biological samples was carried out using geNorm program as described above.
Abbreviations
HdIV: Hyposoter didymator IchnoVirus
PCR: Polymerase Chain Reaction.
rep: repeat element gene
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
L. Galibert conducted the experiments, J. Rocher was in charge of amplifying rep-containing HdIV segments, F. Cousserans and M. Barat-Houari helped with qPCR experiments, G. Devauchelle and P. Cerutti identified the novel rep genes in HdIV genome, P. Fournier assisted in manuscript composition with A.-N. Volkoff.
Acknowledgments
Aknowledgements
The authors are grateful to Bertrand Limier for providing the insects.
Contributor Information
L Galibert, Email: galibert@ensam.inra.fr.
G Devauchelle, Email: devauche@ensam.inra.fr.
F Cousserans, Email: coussera@ensam.inra.fr.
J Rocher, Email: Janick.Rocher@ema.fr.
P Cérutti, Email: cerutti@ensam.inra.fr.
M Barat-Houari, Email: mouna.barat@chu-nimes.fr.
P Fournier, Email: fourniep@ensam.inra.fr.
AN Volkoff, Email: volkoff@ensam.inra.fr.
References
- Balgopal MM, Dover BA, Goodman WG, Strand MR. Parasitism by Microplitis demolitor induces alterations in the juvenile hormone titers and juvenile hormone esterase activity of its host, Pseudoplusia includens. Journal of Insect Physiology. 1996;42:337–345. doi: 10.1016/0022-1910(95)00111-5. [DOI] [Google Scholar]
- Malva C, Varricchio P, Falabella P, La Scaleia R, Graziani F, Pennacchio F. Physiological and molecular interaction in the host-parasitoid system Heliothis virescens-Toxoneuron nigriceps: current status and future perspectives. Insect Biochem Mol Biol. 2004;34:177–183. doi: 10.1016/j.ibmb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Shelby KS, Adeyeye OA, Okot-Kotber BM, Webb BA. Parasitism-linked block of host plasma melanization. J Invertebr Pathol. 2000;75:218–225. doi: 10.1006/jipa.2000.4925. [DOI] [PubMed] [Google Scholar]
- Shelby KS, Webb BA. Polydnavirus infection inhibits synthesis ofan insect plasma protein, arylphorin. J Gen Virol. 1994;75:2285–2292. doi: 10.1099/0022-1317-75-9-2285. [DOI] [PubMed] [Google Scholar]
- Webb BA. Polydnavirus biology, genome structure, and evolution. In: Miller LK, Ball LA, editor. The Insect Viruses. Plenum Publishing Corporation, New York; 1998. pp. 105–139. [Google Scholar]
- Espagne E, Dupuy C, Huguet E, Cattolico L, Provost B, Martins N, Poirie M, Periquet G, Drezen JM. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science. 2004;306:286–289. doi: 10.1126/science.1103066. [DOI] [PubMed] [Google Scholar]
- Webb BA, Strand MR, Dickey SE, Beck MH, Hilgarth RS, Barney WE, Kadash K, Kroemer JA, Lindstrom KG, Rattanadechakul W, et al. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology. 2006;347:160–174. doi: 10.1016/j.virol.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Volkoff AN, Cerutti P, Rocher J, Ohresser MC, Devauchelle G, Duonor-Cerutti M. Related RNAs in lepidopteran cells after in vitro infection with Hyposoter didymator virus define a new polydnavirus gene family. Virology. 1999;263:349–363. doi: 10.1006/viro.1999.9929. [DOI] [PubMed] [Google Scholar]
- Theilmann DA, Summers MD. Physical analysis of the Campoletis sonorensis virus multipartite genome and identification of a family of tandemly repeated elements. Journal of Virology. 1987;61:2589–2598. doi: 10.1128/jvi.61.8.2589-2598.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JGW, Krell P. Polydnavirus genome organization. In: Beckage NE, editor. Parasites and Pathogens of Insects. I. Parasites. Academic Press, Inc; 1993. pp. 189–225. [Google Scholar]
- Hilgarth RS, Webb BA. Characterization of Campoletis sonorensis ichnovirus segment I genes as members of the repeat element gene family. J Gen Virol. 2002;83:2393–2402. doi: 10.1099/0022-1317-83-10-2393. [DOI] [PubMed] [Google Scholar]
- Volkoff AN, Beliveau C, Rocher J, Hilgarth R, Levasseur A, Duonor-Cerutti M, Cusson M, Webb BA. Evidence for a conserved polydnavirus gene family: ichnovirus homologs of the CsIV repeat element genes. Virology. 2002;300:316–331. doi: 10.1006/viro.2002.1535. [DOI] [PubMed] [Google Scholar]
- Kroemer JA, Webb BA. Polydnavirus genes and genomes: emerging gene families and new insights into polydnavirus replication. AnnuRev Entomol. 2004;49:431–456. doi: 10.1146/annurev.ento.49.072103.120132. [DOI] [PubMed] [Google Scholar]
- Theilmann DA, Summers MD. Identification and comparison of Campoletis sonorensis virus transcripts expressed from four genomic segments in the insect hosts Campoletis sonorensis and Heliothis virescens. Virology. 1988;167:329–341. [PubMed] [Google Scholar]
- Galibert L, Rocher J, Ravallec M, Duonor-Cerutti M, Webb BA, Volkoff AN. Two Hyposoter didymator ichnovirus genes expressed in the lepidopteran host encode secreted or membrane-associated serine and threonine rich proteins in segments that may be nested. J Insect Physiol. 2003;49:441–451. doi: 10.1016/S0022-1910(03)00061-1. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) 1993.
- Bonvin M, Kojic D, Blank F, Annaheim M, Wehrle I, Wyder S, Kaeslin M, Lanzrein B. Stage-dependent expression of Chelonus inanitus polydnavirus genes in the host and the parasitoid. J InsectPhysiol. 2004;50:1015–1026. doi: 10.1016/j.jinsphys.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Kroemer JA, Webb BA. Ikappabeta-related vankyrin genes in the Campoletis sonorensis ichnovirus: temporal and tissue-specific patterns of expression in parasitized Heliothis virescens lepidopteran hosts. J Virol. 2005;79:7617–7628. doi: 10.1128/JVI.79.12.7617-7628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkoff AN, Ravallec M, Bossy JP, Cerutti P, Rocher J, Cerutti M, Devauchelle G. The replication of Hyposoter didymator polydnavirus:Cytopathology of the calyx cells in the parasitoid. Biology of theCell. 1995;83:1–13. doi: 10.1016/0248-4900(96)89926-6. [DOI] [Google Scholar]
- Beliveau C, Levasseur A, Stoltz D, Cusson M. Three related TrIVgenes: comparative sequence analysis and expression in host larvae and Cf-124T cells. J Insect Physiol. 2003;49:501–511. doi: 10.1016/S0022-1910(03)00055-6. [DOI] [PubMed] [Google Scholar]
- Volkoff AN, Rocher J, Cerutti P, Ohresser MC, d'Aubenton-Carafa Y, Devauchelle G, Duonor-Cerutti M. Persistent expression of a newly characterized Hyposoter didymator polydnavirus gene in long-term infected lepidopteran cell lines. J Gen Virol. 2001;82:963–969. doi: 10.1099/0022-1317-82-4-963. [DOI] [PubMed] [Google Scholar]
- Glatz RV, Asgari S, Schmidt O. Evolution of polydnaviruses as insect immune suppressors. Trends Microbiol. 2004;12:545–554. doi: 10.1016/j.tim.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Turnbull MW, Volkoff AN, Webb BA, Phelan P. Functional gap junction genes are encoded by insect viruses. Curr Biol. 2005;15:R491–492. doi: 10.1016/j.cub.2005.06.052. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]