Abstract
Human Salmonella enterica serotype Enteritidis infections emerged in Chile in 1994. S. enterica serotype Enteritidis phage type 1 isolates predominated in the north, and phage type 4 isolates predominated in the central and southern regions. A study was planned to characterize this epidemic using the best discriminatory typing technique. Research involved 441 S. enterica serotype Enteritidis isolates, including clinical preepidemic samples (n = 74; 1975 to 1993) and epidemic (n = 199), food (n = 72), poultry (n = 57), and some Latin American (n = 39) isolates. The best method was selected based on a sample of preepidemic isolates, analyzing the discriminatory power (DP) obtained by phage typing and randomly amplified polymorphic DNA and pulsed-field gel electophoresis (PFGE) analysis. The highest DP was associated with BlnI PFGE-bacteriophage typing analysis (0.993). A total of 38 BlnI patterns (B patterns) were identified before the epidemic period, 19 since 1994, and only 4 in both periods. Two major clusters were identified by phylogenetic analysis, and the predominant B patterns clustered in the same branch. Combined analysis revealed that specific B pattern-phage type combinations (subtypes) disappeared before 1994, that different genotypes associated with S. enterica serotype Enteritidis phage type 4 had been observed since 1988, and that strain diversity increased before the expansion of S. enterica serotype Enteritidis in 1994. Predominant subtype B3-phage type 4 was associated with the central and southern regions, and subtype B38-phage type 1 was associated with the north (P < 0.0001). Food and poultry isolates matched the predominant S. enterica serotype Enteritidis subtypes, but isolates identified in neighboring countries (Peru and Bolivia) did not match S. enterica serotype Enteritidis subtypes identified in the north of Chile. The results of this work demonstrate that genetic diversity, replacement, and expansion of specific S. enterica serotype Enteritidis subtypes were associated with epidemic changes.
A Salmonella enterica serotype Enteritidis epidemic started in Chile in 1994. Rates increased from <1 case per 100,000 inhabitants before 1994 to 3.5 per 100,000 in that year, with a further increase to 5.4 cases per 100,000 in 1998 (8, 9). These infections have been related to the consumption of inadequately cooked S. enterica serotype Enteritidis-contaminated poultry items (2, 11, 12, 20-22). During surveillance studies in Chile, S. enterica serotype Enteritidis isolates have been found in ∼1 of every 1,000 egg samples and 7% of poultry meat offered at retail markets in the metropolitan area (1).
The S. enterica serotype Enteritidis epidemic in Chile expanded geographically from north to south and rapidly involved most of the Regional Health Services after 4 years. During 1998, >80% of these services reported human S. enterica serotype Enteritidis infections. At present, >70% of all Salmonella isolates identified in Chile belong to this serotype (9). S. enterica serotype Enteritidis was recognized in Chile as an emergent problem more than a decade after other Latin American countries reported the pathogen (25).
Previous work using a representative sample of Chilean S. enterica serotype Enteritidis isolates indicated that most isolates belonged to phage types 1 and 4 (24). These phage types displayed specific regional distributions, with S. enterica serotype Enteritidis phage type 1 isolates predominating in northern regions and phage type 4 isolates predominating in central and southern areas. Moreover, these types appeared a few years before the epidemic and replaced the early S. enterica serotype Enteritidis phage types 8 and 28 (24). Thus, phage typing suggested that two epidemics were running at the same time, but epidemiological implications derived from this technique are limited due to its low discriminatory power, and a better characterization was needed.
A molecular project was designed to analyze historical changes among available S. enterica serotype Enteritidis isolates; to study whether the observed pattern of geographically predominant S. enterica serotype Enteritidis phage types was associated with the expansion of a rather limited number of bacterial subtypes; to explore whether Chilean S. enterica serotype Enteritidis isolates could be related to those identified in northern neighboring countries, such as Peru and Bolivia; and finally, to compare clinical, food, and poultry S. enterica serotype Enteritidis isolates.
Molecular methods have been applied to typing S. enterica serotype Enteritidis isolates with various results, although they improve the discrimination obtained by ancillary methods. Pulsed-field gel electrophoresis (PFGE), arbitrarily primed PCR, and ribotyping have been applied with a wide range of discriminatory results (7, 17, 18, 29). This variability makes the selection of a typing method a matter of empirical work. In this research, the discriminatory powers (DPs) of different strategies were compared before the best molecular approach could be selected.
MATERIALS AND METHODS
Bacterial isolates.
Strains available at the Centro de Referencia de Enterobacterias, Instituto de Salud Publica (ISP), were used in this study. Salmonella isolates of different serotypes have been kept at this institution since 1975 according to a national microbiological surveillance system. Clinical isolates are sent by public or private laboratories to the ISP to be confirmed by standard biochemical and serological methods (6). S. enterica serotype Enteritidis isolates can be recognized only at the ISP in Chile. Strains were defined as preepidemic if they were collected or sent to the ISP before 1994 and as epidemic if they were sent during or after 1994.
The study was performed using clinical, food, and poultry samples. The last were obtained during surveillance studies conducted by the Metropolitan Health Environmental Service or by the National Agriculture and Husbandry Service. Research involved 441 S. enterica serotype Enteritidis isolates, including 74 preepidemic clinical isolates (1975 to 1993), 199 strains obtained from human sources during the epidemic (1994 to 1996), 72 food isolates (1993 to 1997), 57 poultry isolates (1995 and 1997), and 39 strains obtained from Latin American countries (Peru, Bolivia, Colombia, and Venezuela) from 1999 to 2000.
DP.
The DP was obtained by a previously described formula and indicates the average probability that a typing system will assign a different type to two unrelated strains randomly sampled from a population (14, 17). The best typing method was selected by comparing the DPs of different typing strategies. Seventy-four clinical S. enterica serotype Enteritidis strains collected between 1975 and 1993 were included for this purpose. This sample represented 94% of the available strains for this period. The strains were kept frozen at −70°C until use. The DPs of phage typing and different molecular techniques were calculated either independently or in a combined form. The approach with a higher DP value was chosen for strain typing.
Phage typing.
Phage typing was developed according to a standardized methodology (30). Phage collection was kindly provided by the National Centre for Disease Control, Ontario, Canada. The results of S. enterica serotype Enteritidis phage typing have been published previously (24).
PFGE.
Seven hundred microliters of an overnight S. enterica serotype Enteritidis culture in brain heart infusion broth was used initially. A sample was centrifuged at 2,000 × g for 5 min, the supernatant was discarded, and the pellet was dissolved in 500 μl of buffer solution 1 (NaCl, 75 mM; EDTA, 25 mM; pH 7.9). The sample was again centrifuged at 5,000 rpm for 5 min, the supernatant was discarded, and the pellet was dissolved in 250 μl of buffer solution 2 (Tris, 10 mM; NaCl, 20 mM; EDTA, 50 mM; pH 7.5). Plugs were prepared by adding to the sample 250 μl of 2% low-melting-point agarose dissolved in buffer solution 2. The mixture was poured into casts and kept at 4°C. The plugs were transferred to a tube containing 2 ml of 2% N-lauryl sarcosine dissolved in buffer solution 3 (Tris, 10 mM; EDTA, 250 mM; pH 8.0) containing 1 mg of proteinase K/ml. The plugs were incubated at 55°C overnight and washed five times in 2.5 ml of 3× TE buffer for 30 min each time. After the final wash, the buffer was replaced with the same volume of a 3× TE solution. The plugs were stored in 3× TE buffer solution at 4°C.
Digestion was performed for every plug after the sample was washed eight times with distilled water for 20 min each time. The plugs were suspended in 200 μl of 1× restriction buffer for 1 h to allow equilibration. One hundred microliters of restriction buffer containing 20 to 80 U of the specific restriction enzyme was added, and digestion was performed overnight. The restriction buffer was discarded, and 150 μl of 1× loading dye (6× loading dye is 30% glycerol, 0.25% bromophenol, 0.25% Xilencianol FF [Sigma Co.]) was added. The plugs were placed in the PFGE gel slots prepared using 1% 0.5× Tris-borate-EDTA buffer (PFGE agar concentration, 1%). Digestions were performed with the following enzymes: BlnI, XbaI, SpeI, SmaI, EcoRI, XhoI, DraI, and ApaI. PFGE was performed using the following protocol: voltage, 6 V/cm at 14°C for 21 h; initial pulse, 2 s; final pulse, 40 s; angle, 120°. The runs were developed in a CHEF DR III (Bio-Rad) using 0.5× Tris-borate-EDTA buffer. The gels were stained with ethidium bromide, and profiles were captured using a UV transilluminator coupled to a digital camera (Gel Doc 2000; Bio-Rad). Images were prepared using Quantity One software (Bio-Rad).
RAPD analysis.
Seven primers were used for randomly amplified polymorphic DNA (RAPD) analysis. Primers 23L, OPB-4, OPB-6, OPB-15, OPB-17, P1254, and ERIC 2 were used as described previously (13, 18).
Ribotyping.
The ribotyping technique was developed by Southern blotting, using as a probe an EcoRI DNA fragment obtained from plasmid pKK3535 previously cloned in a PGEN vector (Promega) (4, 10). This fragment was amplified by PCR and labeled with digoxigenin (Roche) as recommended by the manufacturer. Blotting was preformed using a nylon membrane according to the manufacturer's instructions, and genomic DNA was digested with SmaI (23).
Phylogenetic analysis.
The unweighted pair group matching analysis (UPGMA) algorithm was applied to construct phylogenetic trees (28). Clustering was obtained with the assistance of Statistica version 4.5 (Statsoft, Inc).
Statistical analysis.
Required comparisons were performed by the chi-square test.
RESULTS
Selection of typing method.
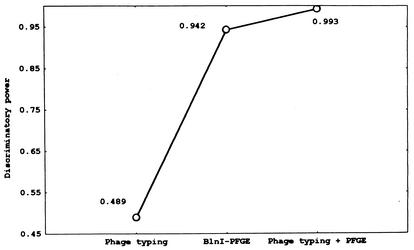
Seventy-four preepidemic isolates (1975 to 1993) were analyzed to select the best discriminatory typing method. A higher DP (0.993) was obtained with the combination of phage typing and BlnI PFGE analysis (Fig. 1). Phage typing was necessary because it increased the BlnI PFGE DP from 0.942 to 0.993, reducing the probability of error from near 6% to <1%.
FIG. 1.
DPs for phage typing, BlnI PFGE analysis, and combined results among clinical preepidemic S. enterica serotype Enteritidis isolates.
The DP for SpeI or XbaI was lower than that obtained with BlnI (0.85 and 0.74, respectively). PFGE-restriction enzyme analysis with SmaI, EcoRI, SalI, XhoI, DraI, or ApaI gave multiple DNA fragments that were difficult to interpret, and they were discarded for further epidemiological analysis (data not shown).
RAPD profiles also showed low DPs (range, 0.04 for primer OPA-4 to 0.68 for primer OPB-17; data not shown). RAPD results did not modify the high DP obtained by PFGE-phage-typing analysis. By combining RAPD with PFGE information and excluding phage-typing data, the DP was raised to 0.983, lower than the 0.993 obtained by PFGE and phage typing. Ribotyping demonstrated similar profiles after genomic-DNA digestion with SmaI and rRNA probing (DP, 0.00).
PFGE results.
Molecular characterization of the 441 isolates by BlnI PFGE analysis identified 6 to 19 DNA fragments per genotype (30 to 550 kbp) and 39 possible DNA band positions along the gel. Minor differences in size or intensity were not considered, but one or more mismatched bands were assigned to different BlnI genotypes. Genotypes from B1 to B57 were arbitrarily assigned according to their order of appearance, and those exclusively identified among international samples were designated IN1 to IN3. Thirty-eight BlnI genotypes (B1 to B38) were identified among 76 clinical or food preepidemic isolates, and 19 types (B39 to B57) were identified among 199 clinical isolates obtained from 1994 to 1996 (epidemic period). Only four genotypes (B3, B22, B36, and B38) were identified in both periods. The combined results for PFGE and bacteriophage typing are presented below.
Phylogenetic analysis.
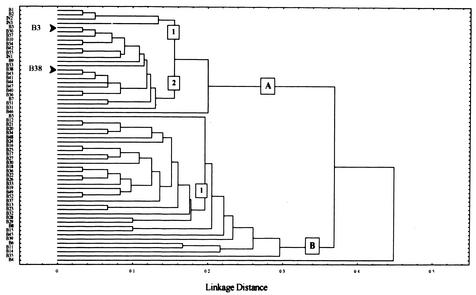
BlnI genotypes were clustered using the UPGMA algorithm, including Chilean and available international isolates. Two major clusters were identified that differed in seven or more DNA fragments (Fig. 2A and B). Group A contained 25 genotypes, and at least two internal branches (1 and 2) could be identified; group B contained 34 genotypes and one branch (Fig. 2). The predominant types B3 and B38 differed in two DNA fragments and clustered in different internal branches within the group A genotypes (branches 1 and 2, respectively). Branch B of the tree contained most of the 38 identified preepidemic genotypes (n = 30). In contrast, group A included most of the epidemic BlnI types (13 out of 19; P = 0.0005 by the chi-square test). International BlnI types clustered only within Group A.
FIG. 2.
Phylogenetic tree of S. enterica serotype Enteritidis genotypes identified by BlnI-PFGE macrorestriction analysis. Clustering was developed by UPGMA, and distances are represented as band mismatching. The predominant BlnI patterns B3 and B38 are indicated.
Combined PFGE and phage-typing analysis.
To enhance epidemiological analysis, phage-typing and BlnI PFGE results were combined to identify specific bacterial subtypes within S. enterica serotype Enteritidis isolates. Eighty combinations were identified using this strategy. Twelve bacterial subtypes accounted for 81.8% of these groups (Table 1). The predominant bacterial subtypes included genotype B3, associated with phage type 4 (B3-pt4), and genotype B38, associated with phage type 1 isolates (B38-pt1) (37.6 and 32.3%, respectively). Other frequent bacterial subtypes are listed in Table 1.
TABLE 1.
Distribution of predominant S. enterica serotype Enteritidis subtypes among clinical, food, and poultry isolates identified in Chile
| Subtype | No. of isolates
|
|||
|---|---|---|---|---|
| Clinical | Food | Poultry | Total (%) | |
| B38-pt1 | 81 | 28 | 21 | 130 (32.3) |
| B3-pt4 | 93 | 31 | 27 | 151 (37.6) |
| B1-pt8 | 9 | 0 | 0 | 9 (2.2) |
| B22-pt4 | 9 | 0 | 0 | 9 (2.2) |
| B56-pt21 | 0 | 4 | 2 | 6 (1.5) |
| B17-pt4 | 5 | 0 | 0 | 5 (1.2) |
| B3-pt1 | 4 | 0 | 0 | 4 (1.0) |
| B56-pt1 | 1 | 1 | 2 | 4 (1.0) |
| B55-pt4 | 1 | 0 | 2 | 3 (0.7) |
| B40-pt1 | 2 | 1 | 0 | 3 (0.7) |
| B36-pt4 | 2 | 1 | 0 | 3 (0.7) |
| B25-pt4 | 1 | 1 | 0 | 2 (0.2) |
| Other subtypes | 65 | 5 | 3 | 73 (18.2) |
| Total | 273 | 72 | 57 | 402 (100) |
(i) Historical changes.
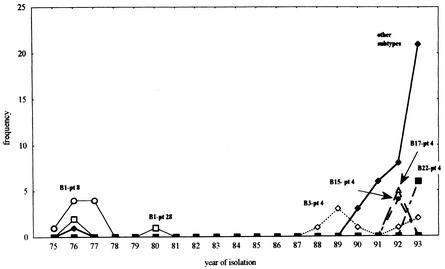
Early S. enterica serotype Enteritidis clinical isolates belonged to subtype B1-pt8 or B1-pt28 (Fig. 3). This combination disappeared, and a prolonged interval without S. enterica serotype Enteritidis isolates was observed from 1981 to 1987. Different genotypes associated with phage type 4 have been observed since 1988, including the predominant subtype B3-pt4. Strain diversity increased before the expansion of S. enterica serotype Enteritidis after this period. S. enterica serotype Enteritidis subtypes B3-pt4, B15-pt4, B17-pt4, and B22-pt4 shared the same phage type, but their genotypes were located in different groups or branches along the phylogenetic tree (Fig. 2). As described above, S. enterica serotype Enteritidis genotype B was allocated to group A (Fig. 2), but genotypes B15, B17, and B22 were allocated to group B.
FIG. 3.
S. enterica serotype Enteritidis subtype distribution from 1975 to 1993. The predominant genotype-phage type combinations are displayed. Subtype B38-pt1 (an important subtype after 1993) and other infrequently observed combinations are included under “other subtypes.”
(ii) Clinical isolates.
The predominant clinical subtypes were studied by analyzing their regional and temporal variation. As shown in Table 2, S. enterica serotype Enteritidis subtype B3-pt4 appeared before subtype B38-pt1. After 1993, both subtypes predominated, although they were concentrated in different regions. Subtype B3-pt4 was highly concentrated in the central or southern area of Chile, whereas subtype B38-pt1 was associated with the north (Table 3) (P < 0.0001 by the chi-square test).
TABLE 2.
Distribution of predominant S. enterica serotype Enteritidis subtypes from clinical sources by year of isolationa
| Yr of isolation | No. of isolates
|
||
|---|---|---|---|
| Subtype B3-pt4 | Subtype B38-pt1 | Total | |
| 1988-1992 | 6 | 0 | 6 |
| 1993-1996 | 87 | 81 | 168 |
| Total | 93 | 81 | 174 |
P = 0.02 by chi-square test.
TABLE 3.
Distribution of S. enterica serotype Enteritidis subtypes obtained from clinical sources according to geographical origina
| Region | No. of isolates
|
||
|---|---|---|---|
| Subtype B3-pt4 | Subtype B38-pt1 | Total | |
| North | 3 | 78 | 81 |
| Central and south | 90 | 3 | 93 |
| Total | 93 | 81 | 174 |
P < 0.0001 by chi-square test.
(iii) Molecular analysis of food and poultry isolates.
Nine S. enterica serotype Enteritidis subtypes were identified among 72 S. enterica serotype Enteritidis food isolates; six of them were detected among clinical isolates (Table 1). As observed with clinical isolates, bacterial subtypes B3-pt4 and B38-pt1 were also predominant, being found in 80% of food isolates (Table 1).
Five bacterial subtypes were identified among poultry isolates, with subtypes B3-pt4 and B38-pt1 also predominant (84.2%) (Table 1). Bacterial subtypes B55-pt4 and B56-pt1 were also observed in human S. enterica serotype Enteritidis isolates. Other groups were observed exclusively among poultry samples (B38-pt2 and B38-pt7; n = 1 each) or matched some food isolates (B56-pt21; n = 2).
Only three bacterial subtypes (B38-pt1, B3-pt4, and B56-pt1) were identified in the three kinds of samples (Table 1).
(iv) Latin American isolates.
Isolates identified in northern neighboring countries, such as Peru and Bolivia, were not associated with the predominant northern subtype, B38-pt1. Only one available international S. enterica serotype Enteritidis isolate, obtained in Colombia, belonged to this subtype. Two infrequently observed northern bacterial subtypes (B3-pt1 and B40-pt1) were detected among samples obtained in neighboring countries. Isolates belonging to bacterial subtype B3-pt4, typically found in central or southern Chile, were also frequently observed in isolates found in Colombia (n = 8), Venezuela (n = 6), or Bolivia (n = 4). Some combinations were only observed in specific countries (data not shown).
DISCUSSION
Acute gastroenteritis provoked by S. enterica serotype Enteritidis is an important cause of morbidity around the world. Recent changes in the poultry industry could explain the expansion of this agent into a major public health problem in Chile.
The results of this work paint a picture that includes genetic diversity, replacement, and expansion of specific S. enterica serotype Enteritidis subtypes during 2 decades of microbiological surveillance. The factors involved in the disappearance of subtypes B1-pt8 and B1-pt28 and the subsequent period without S. enterica serotype Enteritidis isolates are not clear, but these events coincide with a prolonged Salmonella enterica serotype Typhi epidemic that affected the country from 1977 to 1986 (10).
The resurgence of human S. enterica serotype Enteritidis infections was marked by the progressive appearance of new S. enterica serotype Enteritidis subtypes in a short period. Some of these genotypes shared the same phage type, but a common clonal origin for these groups was ruled out by phylogenetic analysis. The Chilean poultry industry satisfies internal demand, and the most suitable explanation for the presence of these new subtypes is the introduction of S. enterica serotype Enteritidis through colonized chickens allocated to reproductive tasks. Genotypic diversity suggests that this happened on several occasions, and it may have been facilitated by important changes that affected the poultry industry during the 1980s. Production changed from numerous small producers to a concentrated industrial pattern. Under these conditions, special reproductive chickens were needed to increase food and egg production, and the import of improved chicken lines was imperative. The potential for the arrival of new S. enterica serotype Enteritidis subtypes was generated. Moreover, new feeding and housing practices were required by this renewed avian industry, facilitating horizontal transfer and dissemination of S. enterica serotype Enteritidis within poultry farms.
During the epidemic, S. enterica serotype Enteritidis subtypes belonging to different phylogenetic clusters were isolated. The predominance of two S. enterica serotype Enteritidis subtypes with specific regional distributions indicates that at least two large coexisting epidemics have occurred since 1994, but the genetic diversity observed within epidemic S. enterica serotype Enteritidis isolates overlaps the outbreaks and suggests multiple infection sources. Zonal differences in S. enterica serotype Enteritidis subtype distribution is probably explained by separate egg providers. Poultry farms located in the north do not supply the central or southern regions, and producers from those regions do not reach markets in the far north. The predominant clinical S. enterica serotype Enteritidis subtypes were also important among food and poultry isolates, confirming the epidemiological link between them.
Although a small sample of nontemporary international S. enterica serotype Enteritidis isolates was analyzed, we were not able to find a combination similar to the predominant northern B38-pt1 subtype among isolates identified in Peru or Bolivia. This fact makes it unlikely that the original or recurrent source of the predominant S. enterica serotype Enteritidis subtypes observed in the north were associated with boundary transmission. Only two infrequent subtypes identified in the north matched Peruvian or Bolivian isolates.
A comprehensive phage-typing or molecular scrutiny of Latin American S. enterica serotype Enteritidis isolates has not yet been developed. S. enterica serotype Enteritidis phage type 4 isolates are now widely distributed around the world, and as reported previously for Brazil and now for Chile, this phage type is displacing phage type 8 S. enterica serotype Enteritidis isolates (15). The broad distribution of this phage type makes it difficult to assign a potential geographical origin for the S. enterica serotype Enteritidis isolates identified in Chile. S. enterica serotype Enteritidis phage type 1 isolates are associated more with European than with North American S. enterica serotype Enteritidis collections, and its presence in Chile may suggest the import of reproductive chickens from that continent.
Comparisons of PFGE BlnI patterns with those found in other locations are limited because most PFGE studies have been developed with the enzymes XbaI or SpeI. We chose BlnI due to its higher DP, a fact that underlines the need for the selection of a proper discriminatory typing technique for a given epidemiological condition. Direct comparison with published BlnI PFGE profiles is difficult and very limited due to different running conditions, equipment, digestion conditions, etc. When compared, BlnI PFGE profiles obtained in Thailand did not reveal a pattern similar to our result (3). In contrast, some profiles observed in our work (B1, B2, and B3) matched results reported by other authors using the same enzyme on a sample of European S. enterica serotype Enteritidis isolates (16). Strains identified in Denmark, Spain, and England had similar BlnI PFGE profiles and were also associated with phage type 4, suggesting a European origin for these isolates in Chile. Comparison with profiles obtained during S. enterica serotype Enteritidis outbreaks observed in Japan were not possible due to the limited photographic resolution of the original publication (22).
In this work, only certain genotypes associated with phage type 4 isolates expanded and predominated. We do not know if this happened for commercial reasons or due to pathogenic properties linked to S. enterica serotype Enteritidis isolates. Some comparative studies have demonstrated that S. enterica serotype Enteritidis phage type 4 isolates are more pathogenic to chickens than phage type 8 strains (5), although to the best of our knowledge, this phenomenon has not been reported for human infections. Some authors have not confirmed virulence differences linked to bacteriophage types (19).
Potential biases for our conclusions are related to the lack of plasmid typing or other typing strategies. Three independent molecular methods were used in this work for typing S. enterica serotype Enteritidis isolates, and diversity could be observed only with macrorestriction analysis. Considering that S. enterica serotype Enteritidis bacteriophage typing is a standardized methodology, has been applied in many research publications, and is a recognized worldwide surveillance method, we preferred to develop our work by including this technique together with macrorestriction analysis instead of combining RAPD results with PFGE information. Moreover, phage typing and BlnI PFGE performed better in this case than RAPD-PFGE analysis (DP, 0.983 versus 0.993). Typing by other methods would probably give modest results after an extensive search. We discarded plasmid typing due to its instability and the low diversity of extrachromosomal DNA harbored among S. enterica serotype Enteritidis isolates (26, 27). Moreover, S. enterica serotype Enteritidis isolates in Chile have not been associated with antibiotic resistance (24).
Acknowledgments
This work was made possible by a grant from the National Council for Research and Technology of Chile (Fondecyt grant 1980912).
REFERENCES
- 1.Alexandre, M., C. Pozo, V. Gonzalez, M. C. Martinez, S. Prat, A. Fernandez, A. Fica, J. Fernandez, and I. Heitmann. 2000. Salmonella Enteritidis detection on avian samples for human consumption in the metropolitan area. Rev. Med. Chile 128:1075-1083. (In Spanish.) [PubMed]
- 2.Alltekruse, S. F., M. L. Cohen, and D. L. Swerdlow. 1997. Emerging food-borne diseases. Emerg. Infect. Dis. 3:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boonmar, S., A. Bangtrakulnonth, S. Pornrunangwong, J. Terajima, H. Watanabe, K. I. Kaneko, and M. Ogawa. 1998. Epidemiological analysis of Salmonella enteritidis isolates from humans and broiler chickens in Thailand by phage typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 36:971-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosius, J., A. Unrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and H. F. Noller. 1981. Construction and fine mapping of recombinant plasmids containing the rrnb ribosomal RNA operon of E. coli. Plasmid 6:112-118. [DOI] [PubMed] [Google Scholar]
- 5.Dhillon, A. S., B. Alisantosa, H. L. Shivaprasad, O. Jack, D. Scharberg, and D. Bandli. 1999. Pathogenicity of Salmonella enteritidis phage types 4, 8, and 23 in broiler chicks. Avian Dis. 43:506-515. [PubMed] [Google Scholar]
- 6.Edwards, P. R., and W. H. Ewing. 1972. The genus Salmonella, p. 146-207. In W. H. Ewing (ed.), Identification of Enterobacteriaceae. Burgess Publishing Company, Minneapolis, Minn.
- 7. Fadl, A. A., A. V. Nguyen, and M. I. Khan. 1995. Analysis of Salmonella enteritidis isolates by arbitrarily primed PCR. J. Clin. Microbiol. 33:987-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fica, A., M. Alexandre, S. Prat, A. Fernandez, J. Fernandez, and I. Heitmann. 2001. Epidemiological changes on salmonellosis in Chile. From Salmonella Typhi to Salmonella Enteritidis. Rev. Chil. Infect. 18:85-93. [Google Scholar]
- 9.Fica, A., A. Fernández, S. Prat, O. Figueroa, R. Gamboa, I. Tsunekawa, and I. Heitmann. 1997. Salmonella Enteritidis, an emergent pathogen in Chile. Rev. Med. Chile 125:544-551. [PubMed] [Google Scholar]
- 10.Fica, A., S. Prat-Miranda, A. Fernandez-Ricci, K. D'Ottone, and F. C. Cabello. 1996. Epidemic typhoid in Chile: analysis by molecular and conventional methods of Salmonella typhi strain diversity in epidemic (1977 and 1981) and nonepidemic (1990) years. J. Clin. Microbiol. 34:1701-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hennesy, T. W., C. W. Hedberg, L. Slustker, K. E. White, J. M. Besser-Wiek, M. E. Moen, J. Feldman, W. W. Coleman, L. M. Edmonson, K. L. MacDonald, and M. T. Osterholm. 1996. A national outbreak of Salmonella Enteritidis infections from ice cream. N. Engl. J. Med. 334:1281-1286. [DOI] [PubMed] [Google Scholar]
- 12.Henzler, D. J., E. Ebel, J. Sanders, D. Kradel, and J. Mason. 1994. Salmonella Enteritidis in eggs from commercial chicken layer flocks implicated in human outbreaks. Avian Dis. 38:37-43. [PubMed] [Google Scholar]
- 13.Hulton, C. S., C. F. Higgins, and P. M. Sharp. 1991. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol. Microbiol. 5:825-834. [DOI] [PubMed] [Google Scholar]
- 14.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irino, K., S. A. Fernandes, A. T. Tavechio, B. C. Neves, and A. M. Dias. 1996. Progression of Salmonella enteritidis phage type 4 strains in Sao Paulo State, Brazil. Rev. Inst. Med. Trop. Sao Paulo 38:193-196. [DOI] [PubMed] [Google Scholar]
- 16.Laconcha, I., D. L. Baggesen, A. Rementeria, and J. Garaizar. 2000. Genotypic characterization by PFGE of Salmonella enterica serotype Enteritidis phage types 1, 4, 6, and 8 isolated from animal and human sources in three European countries. Vet. Microbiol. 75:155-165. [DOI] [PubMed] [Google Scholar]
- 17.Liebana, E., L. Garcia-Migura, M. F. Breslin, R. H. Davies, and M. J. Woodward. 2001. Diversity of strains of Salmonella enterica serotype Enteritidis from English poultry farms assessed by multiple genetic fingerprinting. J. Clin. Microbiol. 39:154-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, W. A., M. A. Usera, T. J. Barrett, and R. A. Goldsby. 1996. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella enteritidis. J. Clin. Microbiol. 34:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, S., A. R. Manges, Y. Xu, F. C. Fang, and L. W. Riley. 1999. Analysis of virulence of clinical isolates of Salmonella enteritidis in vivo and in vitro. Infect. Immun. 67:5651-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishu, B., J. Koehler, L. A. Lee, D. Rodrigue, F. Hickman-Brenner, P. Blake, and R. V. Tauxe. 1994. Outbreak of Salmonella Enteritidis infections in the United States, 1985-1991. J. Infect. Dis. 169:547-552. [DOI] [PubMed] [Google Scholar]
- 21.Mishu, B., P. M. Griffin, R. Tauxe, D. Cameron, R. H. Huthcheson, and W. Schaffner. 1991. Salmonella Enteritidis gastroenteritis transmitted by intact chicken eggs. Ann. Intern. Med. 115:190-194. [DOI] [PubMed] [Google Scholar]
- 22.Murakami, K., K. Horikawa, and K. Otsuki. 1999. Epidemiological analysis of Salmonella enteritidis from human outbreaks by pulsed-field gel electrophoresis. J. Vet. Med. Sci. 61:439-442. [DOI] [PubMed] [Google Scholar]
- 23.Olsen, J. E., M. N. Skov, E. J. Threlfall, and D. J. Brown. 1994. Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J. Med. Microbiol. 40:15-22. [DOI] [PubMed] [Google Scholar]
- 24.Prat, S., A. Fernandez, A. Fica, J. Fernandez, M. Alexandre, and I. Heitmann. 2001. Phage typing of Salmonella enteritidis isolates from clinical, food, and poultry samples in Chile. Pan. Am. J. Public Health 9:7-12. [DOI] [PubMed] [Google Scholar]
- 25.Rodrigue, D. C., R. V. Tauxe, and B. Rowe. 1990. International increase in Salmonella Enteritidis: a new pandemic? Epidemiol. Infect. 105:21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rotgers, R., and J. Casadesus. 1999. The virulence plasmids of Salmonella. Int. Microbiol. 2:177-184. [PubMed] [Google Scholar]
- 27.Sneath, P. H. A., and R. R. Sokal. 1973. Taxonomic structure, p. 188-308. In P. H. A. Sneath and R. R. Sokal (ed.), Numerical taxonomy. W. H. Freeman & Co., San Francisco, Calif.
- 28.Stanley, J., A. P. Burnens, E. J. Threlfall, N. Chowdry, and M. Goldsworthy. 1992. Genetic relationships among strains of Salmonella enteritidis in a national epidemic in Switzerland. Epidemiol. Infect. 108:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thong, K.-L., Y.-F. Ngeow, M. Altwegg, P. Navaratnam, and T. Pang. 1995. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J. Clin. Microbiol. 33:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward, L. R., J. D. H. De Sa, and B. Rowe. 1987. A phagetyping scheme for Salmonella enteritidis. Epidemiol. Infect. 99:291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]