Abstract
BIBI was designed to automate DNA sequence analysis for bacterial identification in the clinical field. BIBI relies on the use of BLAST and CLUSTAL W programs applied to different subsets of sequences extracted from GenBank. These sequences are filtered and stored in a new database, which is adapted to bacterial identification.
In the medical field, bacterial identification is the main activity of clinical microbiology laboratories. Conventional biochemical methods and phenotypic tests for species differentiation are tedious and time-consuming and may require specialized testing that is beyond the capacity of clinical laboratories. Recent progress in molecular biology and bioinformatics allows the consideration of other methods that are more universal and less time-consuming. Molecular methods using one or several appropriate genes are gaining increasing importance because they yield quick and, in most cases, unequivocal results (2). The increasing number of sequences submitted to GenBank (7) and the data-processing programs already developed led us to think that these techniques will be increasingly developed. Sequence-based identification guarantees a constant response time and may be applied to all microorganisms. Today, sequencing techniques are well controlled, but the identification tasks require the chaining of different programs that are sometimes complex to handle, especially for neophytes. Using BLAST alone without phylogenetic data would not be appropriate to perform bacterial identification.
Thus, we have developed a specific bioinformatics tool dedicated to bacterial identification (BIBI, for Bioinformatics Bacterial Identification) in order to simplify sequences analysis within a bacterial identification framework. BIBI fully automates and speeds up different operations for the treatment of sequences. BIBI, which can be accessed at http://pbil.univ-lyon1.fr/bibi/, enables the identification of a microorganism from a gene fragment sequence of previously described cultured bacteria. This program combines similarity search tools in the sequence databases and phylogeny display programs. Thus, it is possible to easily obtain quick results while preserving great freedom in their interpretation, thanks to the use of phylogenetic tools. In addition, to automate the sequence analysis, BIBI integrates different sequence databases which are specifically adapted to bacterial identification to eliminate inaccuracies related to the direct use of sequences from GenBank.
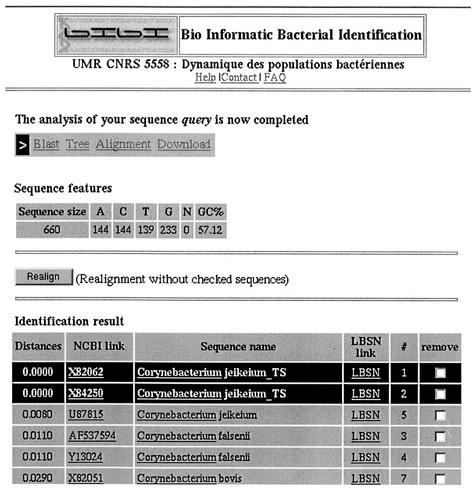
The program implements a chaining of two well-known tools: BLAST (1) and CLUSTAL W (5). CLUSTAL W runs are accelerated by the use of prealigned BLAST results. BIBI is written in standard ANSI C language, and the interface is implemented in HTML-PHP. Analysis of an unknown sequence proceeds in four phases: search for matching sequences, sequence extraction and parsing, sequence alignment, and display of results (Fig. 1). The search for sequences similar to the one submitted is carried out by BLAST. The following stage consists of filtering of the BLAST results, which is, in fact, the key point of the method. Pairwise local alignments from the BLAST output file are extracted and saved in FASTA format. The n similar sequences and the submitted sequence are then multiply aligned with CLUSTAL W, which creates three different files containing (i) a sequence alignment, (ii) a tree in NEWICK format, and (iii) the phylogenetic distances. The use of prealigned sequences produced by BLAST instead of sequences extracted from a database allows an important gain in speed during alignment. Users can also use Dialign (3), another program for multiple-sequence alignment, which builds sequence alignment by comparison of whole segments of the sequences rather than comparison of single residues. The final result corresponds to a sorted table that presents all distinct phylogenetic distances between the query and similar sequences. The results are available within an HTML page (Fig. 2). Phylogenetic alignments and trees are displayed by two Java applets: Jalview (version 1.7 [http://www2.ebi.ac.uk/∼michele/jalview/]) and ATV (8). Bacterial identification is realized by a visual inspection of the tree and/or the multiple alignment. Users can also browse the BLAST output in order to detect possible anomalies in the identification process. It is then possible to remove some sequences to perform a new analysis on a subset of defined sequences. All the files generated are available for direct download through FTP.
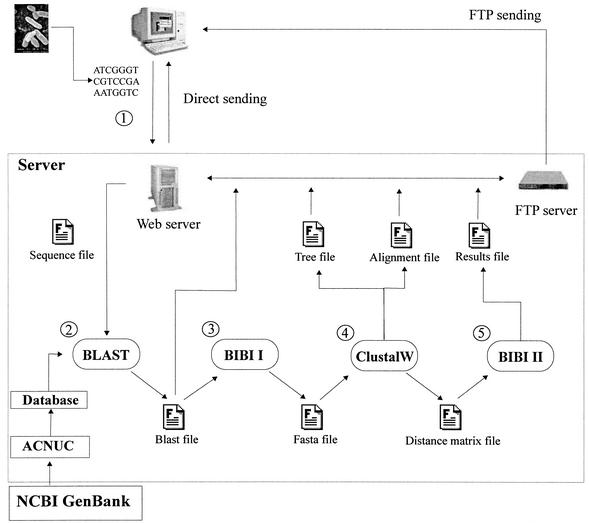
FIG. 1.
Graphical representation of the process of BIBI. The first stage in the process is the submission of the unknown strain sequence, which is stored on the server (step 1). Next, the search for similar sequences is carried out by BLAST on the database selected by the user (step 2). The BLAST results are then filtered by the BIBI program to build a FASTA file containing the similar sequences detected (step 3). These sequences are then multiply aligned with CLUSTAL W (step 4). The results file is generated from the distance matrix file created by CLUSTAL W (step 5). All files created during the whole process are accessible by FTP through links displayed on the HTML page sent to the user by BIBI.
FIG. 2.
Screenshot of BIBI results.
Different sequence databases are designed specifically for bacterial identification. The first contains all of the bacterial sequences of GenBank without sequence checking, while the others are more specific and gather genes belonging to well-known families (rRNA, hsp65, sod, and rpoB genes). Free submission of sequences to general data banks leads to frequent omissions or errors, so inaccuracies related to the direct extraction of the sequences from GenBank may appear (6). Also, many sequences have uninformative definitions. To keep out those inaccuracies, analysis and sequence checking are mandatory. This led to a second type of database. Our improved database results from expertise in crossing the data nomenclature database DSMZ (http://www.dsmz.de/) and a version of GenBank structured with the ACNUC database manager system (4). For each valid species name, an extraction with ACNUC was performed for each gene to build a nomenclature-driven sequence database. We eliminated all the sequences that appeared under uninformative names. Sequences described with basonyms or bacterial names that are usually used without standing in nomenclature are nevertheless extracted thanks to the National Center for Biotechnology Information taxonomy database. All annotations are scanned in order to extract various information related to the sequence. To adapt these databases to the bacterial identification framework, a search of the species type strain numbers in all annotations is performed to identify type strain sequences. All the sequences with varied information are stored in an object-relational database. Thus, we have random access to the inventory of the sequences which exist in a database by genus, species, or genes. For example, users may scan the list of missing species impairing identification of bacteria. This database is regularly updated. Of course, the use of smaller and cleaner gene databases reduces the time required for BIBI searches: several seconds. Two kinds of databases are thus available on BIBI: complete databases and databases adapted to bacterial identification.
The interest of BIBI lies in the integration of well-known tools to automate the bacterial identification process. Homologous segment pairs identified by BLAST are prealigned, allowing faster multiple alignment with CLUSTAL W. The table of sorted phylogenetic distances computed by CLUSTAL W simplifies the reading of the results compared to direct reading of a BLAST file. The clean databases used by BIBI are adapted to bacterial identification. This guarantees unequivocal results. BIBI is a simple and user-friendly data-processing tool, well adapted to the identification of cultured bacteria in a clinical bacteriology laboratory. In the near future, we wish to complete databases for bacteria of medical interest and also to consider the use of a decision-making tool as an aid during identification.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolbert, C. P., and D. H. Persing. 1999. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr. Opin. Microbiol. 2:299-305. [DOI] [PubMed] [Google Scholar]
- 3.Morgenstern, B., K. Frech, A. Dress, and T. Werner. 1998. DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics 14:290-294. [DOI] [PubMed] [Google Scholar]
- 4.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 5.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler, D. L., D. M. Church, A. E. Lash, D. D. Leipe, T. L. Madden, J. U. Pontius, G. D. Schuler, L. M. Schriml, T. A. Tatusova, L. Wagner, and B. A. Rapp. 2001. Databases resources of the National Center for Bio/Technology Information. Nucleic Acids Res. 29:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zmasek, C. M., and S. R. Eddy. 2001. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics 17:383-384. [DOI] [PubMed] [Google Scholar]