Abstract
It has been suggested that enzyme immunoassay (EIA) kits validated in one region may yield variable diagnostic performance results in different regions, possibly due to strain-specific differences in antibody responses in different populations. We tested 13C-urea breath test-characterized serum samples from 109 U.S. patients and 288 Japanese patients using enzyme immunoassay with different preparations of high-molecular-weight cell-associated (HM-CAP) antigens that are conserved across Helicobacter pylori strains. Replicate antigens were prepared from five H. pylori clinical isolates. Eight antigen preparations were evaluated: two of U.S. origin and six of Japanese origin. The accuracies achieved with the eight antigen preparations ranged from 94.4 to 96.3% with the U.S. samples. With the Japanese samples the accuracies achieved ranged from 92.3 to 97.2%. Use of a pool of HM-CAP antigens prepared from isolates from Japan resulted in a higher median enzyme immunoassay value and slightly fewer samples with indeterminate results compared to the results obtained by use of the U.S. standard HM-CAP antigen for H. pylori-positive patients (accuracies, 97.2 and 92.3%, respectively), suggesting that variations in performance between both antigen source and patient population might be reduced by using antigens pooled from several strains.
It has been suggested that enzyme immunoassay (EIA) kits validated in one region may yield variable diagnostic performances in other regions (18, 19, 22, 25, 33). The antigenic heterogeneity of Helicobacter pylori and geographic variations in the immune response to H. pylori antigens have been suggested to be possible contributing factors. Other potential factors include differences in the antigen preparation, the reference method used to determine true H. pylori infection status, and the background rate of spontaneous elimination of H. pylori from the population (4, 12, 14, 29). For example, in Japan and the United States there has been a progressive decline in the incidence of H. pylori in all birth cohorts, such that there is an increasing population of patients without active H. pylori infection but with histologic and serological evidence of past infection (13, 30-32).
The heterogeneity of H. pylori strains has been well documented, with considerable variation in the prevalence of specific strains between isolates from East Asia and Western countries (e.g., between North America and East Asia, in particular, Korea, China, and Japan) (2, 6, 10, 14-17, 23, 28). Heterogeneity related to these geographic areas has also been described with regard to virulence factors such as cagA, vacA, and iceA. Strains from East Asia have been reported to have these virulence factors at a very high frequency, often exceeding 90%, whereas in the United States the frequency of strains with these virulence factors appears to be lower (1, 5). The success of immunoassay depends on the use of antigens present in H. pylori strains that predominate among the population being evaluated (3, 18, 29).
The HM-CAP EIA for immunoglobulin G antibodies to H. pylori (Enteric Products, Inc.) has previously been shown to be an accurate and reliable method for detection of H. pylori infection (21). This assay is based on high-molecular-weight cell-associated antigen HM-CAP (9). The high-molecular-weight antigens are highly conserved complexes with important conformational determinants that are composed of both subunits of urease (UreA and UreB) and the Hsp60 chaperonin (7, 8, 20, 26). This study was designed to determine whether EIAs based on the high-molecular-weight antigens derived from bacterial isolates obtained from the United States and Japan would demonstrate differences in performance between U.S. and Japanese populations.
MATERIALS AND METHODS
Study populations and clinical samples. (i) U.S. samples.
A total of 245 serum samples from U.S. residents were included in this study.
(ii) U.S. samples characterized by UBT.
A total of 109 patients residing in the Houston, Tex., metropolitan area were included in this study. Patients were evaluated at the Texas Medical Center facilities in Houston. This group included asymptomatic volunteers as well as symptomatic patients who had been referred for endoscopy. Enrollment in the study was based on previously described criteria (11). Each patient provided information on a screening questionnaire that included demographic information, medical history, frequency of gastrointestinal symptoms, and history of use of medications within the preceding 2 months, specifically, antibiotics, proton pump inhibitors, H2 blockers, bismuth-containing compounds, or nonsteroidal anti-inflammatory drugs. Subjects were excluded from the study if they regularly used antacids or antibiotics. The H. pylori status of all 109 patients was confirmed by the 13C-urea breath test (UBT; Meretek, Nashville, Tenn.). A result of greater than 2.4‰ was considered positive for H. pylori infection. A blood sample was drawn at the time of breath testing. The serum was separated and frozen at −70°C until use.
(iii) U.S. samples uncharacterized with regard to H. pylori infection status.
An additional 136 serum samples were obtained from a licensed blood center (Millennium Biotech, Fort Lauderdale, Fla.) for testing. These samples had not previously been characterized with regard to H. pylori infection status.
(iv) Characterized Japanese samples.
A total of 288 serum samples from Japanese populations were included in this study, including 179 serum samples from the Hyogo College of Medicine and 109 samples from Hokkaido University. Biopsy for histological analysis and UBT were performed for all Japanese patients. The UBT was performed as follows: 100 mg of 13C-labeled urea was given, breath samples were collected before ingestion of the labeled urea and 20 min after ingestion, and the samples were analyzed by mass spectroscopy by using a positive cutoff of 2.5‰ (27). Patients were considered positive for H. pylori infection if either UBT or histology was positive. Patients were considered negative for infection if both UBT and histology were negative. For both medical centers, patients were excluded from the study if they had a history of use of antibiotics, proton pump inhibitors, H2 blockers, or bismuth-containing compounds within the preceding 2 months. Blood was drawn at the time of UBT, and the serum was separated and frozen at −70°C until use.
H. pylori clinical isolates and culture conditions.
Four clinical isolates from four Japanese patients from Hokkaido University were used for culture. The isolate identification and the disease state of the patients from whom they were obtained were as follows: isolate 159A, gastric ulcer; isolate 193C, gastric cancer; isolate 198C, gastric ulcer; isolate 225C, nonulcer dyspepsia. One clinical isolate from a Caucasian U.S. resident was from the Baylor College of Medicine. The patient from whom this strain was isolated was diagnosed with a gastric ulcer. The bacterial isolate was genotyped and positive for both the vacA and the cagA virulence factors.
All isolates were grown on serum agar plates containing 6.25% horse serum. Confluent cultures were subsequently scaled up in tryptic soy broth containing 10% horse serum. The bacteria were cultured for 2 to 3 days at 37°C under microaerophilic conditions of 10% CO2 and 95% relative humidity. Bacterial colonies were confirmed to be H. pylori on the basis of Gram staining and catalase, oxidase and urease testing performed upon initial culture, at each passage thereafter, and upon final harvest. The culture of each isolate was strictly segregated from the others.
Preparation of antigen.
H. pylori broth cultures were harvested, and the high-molecular-weight cell-associated (HM-CAP) antigen was isolated from each clinical isolate cultured as described by Evans et al. (9). Culture and isolation of all isolates were performed in a single laboratory.
In order to evaluate potential variability in the culture and antigen isolation process and to compare this variability to the variability that might be observed between antigens obtained from the different clinical isolates, replicate batches of HM-CAP antigen were produced. Two batches of antigen were produced from Japanese clinical isolate 159A; these are referred to as batches 159A,1 and 159A,2. All HM-CAP antigens from Japanese isolates were designated JHM-CAP antigen. In addition, two batches of antigen were produced from U.S. isolate 8826. One batch of antigen from U.S. isolate 8826, batch 197SR, was previously used in the manufacture of commercially distributed kits. This batch was designated the U.S. standard HM-CAP antigen lot. The second batch of antigen from U.S. isolate 8826 was manufactured for use in this study exclusively and is referred to as batch 8826A.
In order to compare population antibody responses to antigen derived from a single Japanese H. pylori strain and antigen pooled from all four Japanese isolates, an antigen pool was manufactured, consisting of equal portions of each JHM-CAP antigen isolated from four clinical isolates, isolates 159A, 193C, 198C, and 225C. This pool was designated the JHM-CAP antigen pool.
In summary, a total of 8 H. pylori antigens were manufactured for this study: six JHM-CAP antigen batches (batches 159A,1-J, 159A,2-J, 193C-J, 198C-J, 225C-J, and JHM-CAP pool) and two HM-CAP antigen batches (batches 197SR-US and 8826A-US).
HM-CAP EIA for immunoglobulin G antibodies to H. pylori. (i) Kits.
Eight separate lots of EIA kits were manufactured by using each of the HM-CAP or JHM-CAP antigens described above. These kits were manufactured, and quality control was performed in accordance with the proprietary protocols of Enteric Products, Inc., used for its commercially marketed EIA kit. All kits passed the quality control specifications of Enteric Products, Inc.
(ii) EIA.
All EIAs were performed and interpreted in accordance with the instructions of the manufacturer. In order to eliminate any variability or potential error observed in assay results due to preparation of the initial 1:101 dilution of the serum sample, bulk dilutions were prepared by adding 50 μl of serum to 5.0 ml of kit buffer for all serum samples: 109 U.S. characterized samples, 136 U.S. uncharacterized samples, and 288 Japanese samples. These bulk dilutions were used throughout the study, so that the results for each kit were derived from the same 1:101 dilution of the sample. The EIA kit cutoffs used were those recommended by the manufacturer and were as follows: positive, >2.2 EV (enzyme immunoassay value); indeterminate, 1.8 to 2.2 EV; and negative, <1.8 EV.
Data analysis.
The test results for samples with results in the indeterminate zone were excluded from calculation of EIA performance parameters of sensitivity, specificity, and accuracy. However, the results for these samples were included in the correlation analysis. The performance parameters for each of the eight kits were calculated for U.S. and Japanese populations versus reference method data by using all serum sample results with the exception of the 136 samples from a U.S. blood center, for which no reference method data existed. Correlation analysis was performed by comparing EIA results observed for each kit to those observed for the U.S. standard antigen 197SR-US. In order to obtain EIA performance parameters based on a larger number of U.S. samples, correlation analysis was based on the entire U.S. population of 245 samples.
RESULTS
The rate of H. pylori infection in the U.S. population, as determined by UBT, was 61% (66 of 109 samples). The average rate of infection in the Japanese population, as determined by UBT, was 80% (230 of 288 samples). The performances of the EIAs, including parameters of sensitivity, specificity, and accuracy, for the samples from the U.S. and Japanese populations with all antigens is shown in Tables 1 and 2. For the samples from the U.S. population the U.S. HM-CAP and JHM-CAP antigens performed similarly. The 95% confidence intervals for sensitivity and specificity overlapped for all the antigen isolates. Accuracy exceeded 94% for all.
TABLE 1.
EIA performance with samples from the U.S. populationa
| HM-CAP or JHM-CAP antigen | No. of samples with the following result:
|
% Indeterminate | Sensitivity (%)b | Specificity (%)b | Accuracy (%)b | |||
|---|---|---|---|---|---|---|---|---|
| True positive | True negative | False positive | False negative | |||||
| 197SR-US | 62 | 40 | 2 | 2 | 2.8 | 96.9 (89.2-99.6) | 95.2 (83.8-99.4) | 96.2 (90.6-99.0) |
| 8826A-US | 64 | 40 | 3 | 1 | 0.9 | 98.5 (91.7-100) | 93.0 (80.9-98.5) | 96.3 (90.8-99.0) |
| 159A, 1-J | 64 | 37 | 4 | 0 | 3.7 | 100.0 (94.4-100) | 90.2 (76.9-97.3) | 96.2 (90.5-99.0) |
| 159A, 2-J | 64 | 37 | 3 | 1 | 3.7 | 98.5 (91.7-100) | 92.5 (79.6-98.4) | 96.2 (90.5-99.0) |
| 193C-J | 63 | 39 | 4 | 1 | 1.8 | 98.4 (91.6-100) | 90.7 (77.9-97.4) | 95.3 (89.4-98.5) |
| 198C-J | 64 | 37 | 5 | 1 | 1.8 | 98.5 (91.7-100) | 88.1 (74.4-96.0) | 94.4 (88.2-97.9) |
| 225C-J | 64 | 37 | 5 | 1 | 1.8 | 98.5 (91.7-100) | 88.1 (74.4-96.0) | 94.4 (88.2-97.9) |
| JHM-CAP pool | 67 | 37 | 4 | 0 | 3.7 | 100.0 (94.4-100) | 90.2 (76.9-97.3) | 96.2 (90.5-99.0) |
A total of 109 samples were tested.
The values for samples with Indeterminate values results were excluded from the analysis. Values in parentheses are 95% confidence intervals.
TABLE 2.
EIA performance with samples from the Japanese populationa
| HM-CAP or HM-CAP antigen | No. of samples with the following result:
|
% Indeterminate | Sensitivity (%)b | Specificity (%)b | Accuracy (%)b | |||
|---|---|---|---|---|---|---|---|---|
| True positive | True negative | False positive | False negative | |||||
| 197SR-US | 192 | 58 | 0 | 21 | 5.9 | 90.1 (85.3-93.8) | 100.0 (93.8-100) | 92.3 (88.4-95.1) |
| 8826A-US | 213 | 56 | 0 | 11 | 2.8 | 95.1 (91.4-97.5) | 100.0 (93.6-100) | 96.1 (93.1-98.0) |
| 159A, 1-J | 220 | 54 | 2 | 8 | 1.4 | 96.5 (93.2-98.5) | 96.4 (87.7-99.6) | 96.5 (93.6-98.3) |
| 159A, 2-J | 213 | 57 | 0 | 11 | 2.4 | 95.1 (91.4-97.5) | 100.0 (93.7-100) | 96.1 (93.1-98.0) |
| 193C-J | 201 | 57 | 0 | 18 | 4.2 | 91.8 (87.3-95.1) | 100.0 (93.7-100) | 93.5 (89.9-96.1) |
| 198C-J | 219 | 54 | 3 | 5 | 2.4 | 97.8 (94.9-99.3) | 94.7 (85.4-98.9) | 97.2 (94.5-98.8) |
| 225C-J | 222 | 52 | 3 | 7 | 1.4 | 96.9 (93.8-98.8) | 94.5 (84.9-98.9) | 96.5 (93.6-98.3) |
| JHM-CAP pool | 221 | 54 | 2 | 6 | 1.7 | 97.4 (94.3-99.0) | 96.4 (87.7-99.6) | 97.2 (94.5-98.8) |
A total of 288 samples were tested.
The values for samples with indeterminate results were excluded from analysis. Values in parentheses are 95% confidence intervals.
The accuracies for all samples from the Japanese population were similar in EIAs with both U.S. HM-CAP and JHM-CAP antigens (Table 2). JHM-CAP lot 193C-J and U.S. antigen 197SR-US gave lower sensitivities and higher rates of false-negative and indeterminate results than the other antigens. The results obtained with JHM-CAP antigen 193C-J followed by those obtained with 8826A-US gave the best correlations to the results obtained with U.S. HM-CAP 197SR-US, with correlation coefficients of greater than or equal to 0.975 for isolates from both the U.S. and the Japanese patient populations. Antigen 197SR-US, a standard U.S. HM-CAP antigen, gave the lowest accuracy and the highest rates of false-negative and indeterminate results for the isolates from the Japanese population. The JHM-CAP pool showed the highest accuracy, with lower rates of false-negative and indeterminate results.
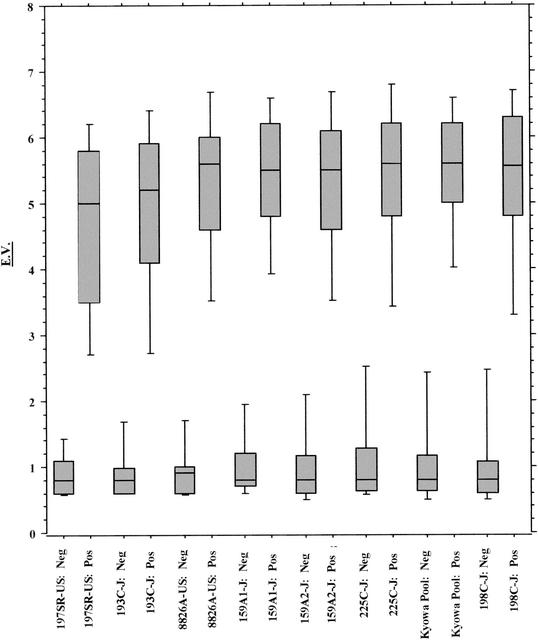
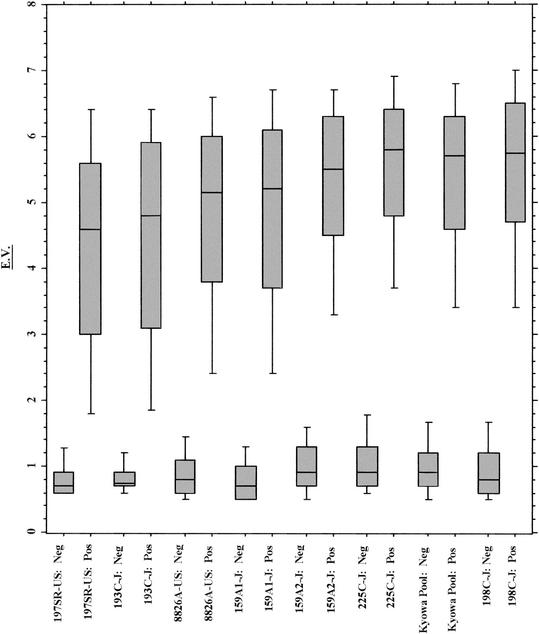
In order to further investigate the trends in the EIA results, a box plot analysis was performed with the EVs obtained with each antigen with isolates from both the U.S. and the Japanese populations (Fig. 1 and 2). The EV is dependent on both the titer of specific antibody and its affinity for the antigens. Box plots were generated for isolates from both groups in order of increasing sensitivity observed with each antigen prepared from isolates from the Japanese population, with U.S. HM-CAP 197SR-US followed by JHM-CAP 193C charting first and second and giving the lowest median positive EVs for samples from both UBT-positive U.S. and Japanese populations. For the Japanese population, a clear trend toward increased EVs for UBT-positive samples was observed as the sensitivity increased, with the JHM-CAP pool, 225C, and 198C antigens giving the highest median EVs. Replicate JHM-CAP lots 159A,1 and 159A,2 gave distributions for positive samples that were between those for the other antigens. In addition, the range of positive results was broader for antigens 197SR-US and 193C and became tighter as the sensitivity and the median EV for the samples from the UBT-positive groups increased. For the samples from the U.S. population, again, HM-CAP antigens 197SR-US and 193C also gave lower median EVs and slightly broader distributions compared to the results obtained with the other antigens.
FIG. 1.
Distribution of EV results by UBT status for 109 U.S. samples.
FIG. 2.
Distribution of EV results by UBT status for 288 Japanese samples.
DISCUSSION
Our study is the first to directly compare the performances of EIAs with samples from U.S. and Japanese populations by using the same antigen preparation from different H. pylori strains. We prepared EIA kits with antigens derived from clinical H. pylori isolates from Japan and compared the performance of those kits with that of the U.S. standard kit, which uses antigens derived from a U.S. isolate. This approach allowed us to address the specific question of whether differences related to H. pylori strain variation exist. We found that the EIA with the high-molecular-weight cell-associated antigen was accurate with samples from both the U.S. and the Japanese populations, independently of the geographic site of origin of the antigen. However, the best overall performance with samples from the Japanese population was observed with the mixture of antigens isolated from Japanese strains. Although this result was data-derived and not the result of hypothesis testing, it suggests that the optimal performance of the EIA depends on the regional origin of the antigens used.
Other studies have reported on the relatively poor performance of Western antigen-based serology with samples from Asian groups. Leung et al. (19) reported on the poor performances of three commercial kits, including the HM-CAP kit, when biopsy and UBT combined was used as the reference method for a group of 80 Chinese patients. Kim et al. (18) in a study of Korean patients, reported on the poor performance of four Western EIA kits compared to that of an in-house EIA when biopsy was used as the reference method. However, additional Western blotting of sera with discrepant results by EIA against sonicates of H. pylori antigen preparations from four separate strains, including strains from Korea, did not explain the variable performances of these samples, suggesting that the observed differences in kit performance were not due to strain variation or were due to conformational structures that could not be determined by Western blotting. As noted before, besides the technical factors related to test kit preparation and performance, the accuracies reported for serological tests are markedly influenced by the histological techniques used to determine the presence or absence of active H. pylori infection as well as those used to estimate the proportion of individuals with prior H. pylori infection (12, 24).
Interpretation of the results of many prior studies is difficult because the evaluations were not standardized in design and did not include a direct comparison of the performance of the assay(s) for the group for which the kit was originally validated and the performance of the assay(s) for the group under evaluation. In general, it has been reported that the in-house EIA was best, but this has rarely been tested in a prospective blinded evaluation. No prior studies have evaluated the performance of the bacterial antigen derived from H. pylori isolates obtained from the locale and the population under study compared with that of the antigen used in the kit under investigation to be able to directly assess whether H. pylori strain differences are indeed responsible for possible differences in the performances observed. We based our evaluations on a commercially available kit using a partially purified, complex, but well-characterized antigen preparation, HM-CAP, comprised virtually exclusively of the UreA and UreB subunits and the associated chaperonin, Hsp60. The HM-CAP antigen complex retains conformational epitopes that cannot be demonstrated by standard Western blotting techniques that denature the tertiary protein structure. Although there is considerable antigenic diversity of H. pylori strains across and within geographic locales, the human serological response has been reported to be equally diverse. However, major H. pylori antigens, including the urease and Hsp60 antigens, are conserved and recognized by sera from all locales, suggesting a core conserved serological response (15). Kim et al. (18) performed sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis with whole-cell sonicates of five H. pylori strains, including Western and Korean isolates, and reported that the major protein bands were present in all strains, including bands at the target molecular weights of UreA, UreB, and Hsp60. However, a pattern of minor protein banding which did vary between strains was also apparent. Our study analyzed antigenic performance at the level of an EIA, thereby preserving additional conformational epitopes not accounted for in Western blotting.
Variations in the performances of tests with different H. pylori antigens for different populations may be reduced by the pooling of antigens from several strains (15, 29). We observed variations in performance with different H. pylori clinical isolates, with one of four of the Japanese strains performing slightly differently than the others. We suggest that the use of antigens pooled from several strains is both a practical and a sound potential approach to resolving this issue, provided the method is validated with samples from the population of interest.
In summary, our results demonstrate the excellent performance of the HM-CAP EIA kit both when the standard U.S. HM-CAP antigens derived from the original U.S. clinical isolate are used to test samples from the U.S. population and when the mixed JHM-CAP pool of antigens derived from Japanese clinical isolates are used to test samples from the Japanese population. Our data suggest that the use of EIAs based on a single local H. pylori isolate will not necessarily improve the diagnostic accuracy locally and that such EIAs are subject to the same pitfalls when assays developed in one region are used in other geographic locales. The variations observed by use of antigens from different strains may be reduced by using a pool of HM-CAP antigens from several H. pylori strains.
REFERENCES
- 1.Atherton, J., and A. Covacci. 1997. Pathogenic properties of Helicobacter pylori. Curr. Opin. Gastroenterol. 13:20-24. [Google Scholar]
- 2.Blaser, M. J. 1994. Helicobacter pylori phenotypes associated with peptic ulceration. Scand. J. Gastroenterol. 29:1-5. [PubMed] [Google Scholar]
- 3.Borba de Arruda, S. M., D. J. Passaro, S. Yang, and J. Parsonnet. 2001. Variability of serologic testing for H. pylori using U.S. and Peruvian antigens. Gastroenterology 120:325-326. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X., K. Haruma, T. Kamada, M. Mihara, K. Komoto, M. Yashihara, K. Sumii, and G. Kajiyama. 2000. Factors that affect results of the 13C urea breath test in Japanese patients. Helicobacter 5:98-103. [DOI] [PubMed] [Google Scholar]
- 5.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans, D. J., and D. G. Evans. 2001. Helicobacter pylori cagA: analysis of sequence diversity in relation to phosphorylation motifs and implications for the role of cagA as a virulence factor. Helicobacter 6:187-198. [DOI] [PubMed] [Google Scholar]
- 7.Evans, D. J., D. G. Evans, L. Engstrand, and D. Y. Graham. 1992. Urease-associated heat shock protein of Helicobacter pylori. Infect. Immun. 60:2125-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, D. J., D. G. Evans, S. S. Kirkpatrick, and D. Y. Graham. 1991. Characterization of Helicobacter pylori urease and purification of its subunits. Microb. Pathog. 10:15-26. [DOI] [PubMed] [Google Scholar]
- 9.Evans, D. J., Jr., D. G. Evans, D. Y. Graham, and P. D. Klein. 1989. A sensitive and specific serologic test for detection of Campylobacter pylori infection. Gastroenterology 96:1004-1008. [DOI] [PubMed] [Google Scholar]
- 10.Go, M. F., V. Kapur, D. Y. Graham, and J. M. Musser. 1996. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J. Bacteriol. 178:3934-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, D. Y., H. M. Malaty, D. G. Evans, D. J. Evans, P. D. Klein, and A. E. 1991. Epidemiology of Helicobacter pylori in an asymptomatic population in the United States. Gastroenterology 100:1495-1501. [DOI] [PubMed] [Google Scholar]
- 12.Graham, D. Y., and S. Reddy. 2001. Rapid detection of anti-Helicobacter pylori IgG in urine using immunochromatography. Aliment. Pharmacol. Ther. 15:699-702. [DOI] [PubMed] [Google Scholar]
- 13.Graham, D. Y., and Y. Yamaoka. 2000. Disease-specific Helicobacter pylori virulence factors: the unfulfilled promise. Helicobacter 4:S3-S9. [DOI] [PubMed] [Google Scholar]
- 14.Herbrink, P., and L. J. van Doorn. 2000. Serological methods for diagnosis of Helicobacter pylori infection and monitoring of eradication therapy. Eur. J. Microbiol. Infect. Dis. 19:164-173. [DOI] [PubMed] [Google Scholar]
- 15.Hook-Nikanne, J., G. I. Perez-Perez, and M. J. Blaser. 1997. Antigenic characterization of Helicobacter pylori strains from different parts of the world. Clin. Diagn. Lab. Immunol. 4:592-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshino, F. B., K. Katayama, K. Watanabe, S. Takahashi, H. Uchimura, and T. Ando. 2000. Heterogeneity found in the cagA gene in Helicobacter pylori from Japanese and non-Japanese isolates. J. Gastroenterol. 35:890-897. [DOI] [PubMed] [Google Scholar]
- 17.Ito, Y., T. Azuma, S. Ito, H. Miyaji, M. Hirai, Y. Yamazaki, F. Sato, T. Kato, Y. Kohli, and M. Kuriyama. 1997. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J. Clin. Microbiol. 35:1710-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, S. Y., J. S. Ahn, Y. J. Ha, H. J. Doh, M. H. Jang, S. I. Chung, and H. J. Park. 1998. Serodiagnosis of Helicobacter pylori infection in Korean patients using enzyme-linked immunosorbent assay. J. Immunoassay 19:251-270. [DOI] [PubMed] [Google Scholar]
- 19.Leung, W. K., E. K. W. Ng, F. K. L. Chan, S. C. S. Chung, and J. J. Y. Sung. 1999. Evaluation of three commercial enzyme-linked immunosorbent assay kits for diagnosis of Helicobacter pylori in Chinese patients. Diagn. Microbiol. Infect. Dis. 34:13-17. [DOI] [PubMed] [Google Scholar]
- 20.Macchia, G., A. Massone, D. Burroni, A. Covacci, S. Censini, and R. Rappuoli. 1993. The HPS60 protein of Helicobacter pylori: structure and immune response in patients with gastroduodenal diseases. Mol. Microbiol. 9:645-652. [DOI] [PubMed] [Google Scholar]
- 21.Marchildon, P. A., L. M. Ciota, F. A. Zamaniyan, J. S. Peacock, and D. Y. Graham. 1996. Evaluation of three commercial enzyme immunoassays compared with the 13C urea breath test for detection of Helicobacter pylori infection. J. Clin. Microbiol. 34:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuo, K., N. Hamajima, S. Tominaga, T. Suzuki, T. Nakamura, and A. Matsuura. 2000. Helicobacter pylori IgG antibody test established in the United States showed a substantially lower sensitivity for Japanese population. Am. J. Gastroenterol. 95:1597-1598. [DOI] [PubMed] [Google Scholar]
- 23.Miehlke, S., K. Kibler, N. Figura, S. M. Small, D. Y. Graham, and M. F. Go. 1996. Allelic variation in the cagA gene of Helicobacter pylori obtained from Korea compared to the United States. Am. J. Gastroenterol. 91:1322-1325. [PubMed] [Google Scholar]
- 24.Misawa, K., T. Kumagai, T. Shimizu, K. Furihata, H. Ota, T. Akamatsu, and T. Katsuyama. 1998. A new histological procedure for re-evaluation of the serological test for Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 17:14-19. [DOI] [PubMed] [Google Scholar]
- 25.Miwa, H., S. Kikuchi, K. Ohtaka, O. Kobayashi, A. Ogihara, M. Hojo, A. Nagahara, and N. Sato. 1998. Insufficient diagnostic accuracy of imported serological kits for Helicobacter pylori infection in Japanese population. Diagn. Microbiol. Infect. Dis. 36:95-99. [DOI] [PubMed] [Google Scholar]
- 26.Mobley, H. L. T., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohara, S., M. Kato, M. Asaka, and T. Toyoto. 1998. Studies of 13C-urea breath test for diagnosis of Helicobacter pylori infection in Japan. J. Gastroenterol. 33:6-13. [DOI] [PubMed] [Google Scholar]
- 28.Rudi, J., C. Kolb, M. Maiwald, D. Kuck, A. Sieg, P. R. Galle, and W. Stremmel. 1998. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J. Clin. Microbiol. 36:944-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widmer, M., J. D. de Korwin, P. Aucher, J. M. Thiberge, S. Suerbaum, A. Labigne, and J. L. Fauchere. 1999. Performance of native and recombinant antigens for diagnosis of Helicobacter pylori infection. Eur. J. Microbiol. Infect. Dis. 18:823-826. [DOI] [PubMed] [Google Scholar]
- 30.Yamaoka, Y., H. M. Malaty, M. S. Osato, and D. Y. Graham. 2000. Conservation of Helicobacter pylori genotypes in different ethnic groups in Houston, Texas. J. Infect. Dis. 181:2083-2086. [DOI] [PubMed] [Google Scholar]
- 31.Yamaoka, Y., E. Orito, M. Mizokami, O. Gutierrez, N. Saitou, T. Kodama, M. S. Osato, J. G. Kim, F. C. Ramirez, V. Mahachai, and D. Y. Graham. 2002. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 517:180-184. [DOI] [PubMed] [Google Scholar]
- 32.Yamaoka, Y., M. S. Osato, A. R. Sepulveda, O. Gutierrez, N. Figura, J. G. Kim, T. Kodama, K. Kashima, and D. Y. Graham. 2000. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol. Infect. 124:91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan, J.-M., M. C. Yu, W.-W. Xu, M. Cockburn, Y.-T. Gao, and R. K. Ross. 1999. Helicobacter pylori infection and risk of gastric cancer in Shanghai, China: updated results based upon a locally developed and validated assay and further follow-up of the cohort. Cancer Epidemiol. Biomarkers Prevent. 8:621-624. [PubMed] [Google Scholar]