Abstract
Listeria monocytogenes strains that were isolated from 314 human listeriosis cases in Finland during an 11-year period were analyzed by O:H serotyping and pulsed-field gel electrophoresis (PFGE). Serotyping divided the isolates into five serotypes, the most common being 1/2a (53%) and 4b (27%). During the study period, the number of cases caused by serotype 1/2a increased from 22% in 1990 to 67% in 2001, and those caused by serotype 4b decreased from 61 to 27%, respectively. PFGE with restriction enzyme AscI divided the strains into 81 PFGE genotypes; among strains of serotypes 1/2a and 4b, 49 and 18 PFGE types were seen, respectively. PFGE type 1 (serotype 1/2a) was the most prevalent single type (37 strains). Together with six other, closely related PFGE types, PFGE type 1 formed a group of 71 strains, representing 23% of all 314 strains. Strains of PFGE type 1 have also been isolated from cold smoked fish, suggesting a source of human infections caused by this type. Moreover, PFGE type 24 (serotype 1/2c) was significantly associated with gender: 5% of 180 male subjects but none of 132 female subjects (P = 0.012). An electronic database library was created from the PFGE profiles to make possible the prompt detection of new emerging profiles and the tracing of potential infection clusters in the future.
Listeria monocytogenes is an important food-borne pathogen which is widely distributed in the environment and which may contaminate foodstuffs at any point of the food chain. It causes an infection, listeriosis, in certain well-defined high-risk groups, including pregnant women, neonates, and immunocompromised adults (10, 36). Usually the infection causes sepsis and may affect the central nervous system, causing meningitis, or may lead to abortion or stillbirth. Occasionally persons without any predisposing conditions may also have gastroenteritis (9, 19, 21, 29, 31, 34). The clinical symptoms take from a few days to several weeks to appear, making the source of the infection difficult to trace. Unlike most other common food-borne infections, listeriosis has a mortality rate of 20 to 30% (8, 12, 15, 26, 35); this fact emphasizes the need for timely epidemiological surveillance.
In Finland, since the 1980s clinical microbiology laboratories have sent their L. monocytogenes findings to the Laboratory of Enteric Pathogens (LEP), National Public Health Institute (KTL); in 1994, such submission became obligatory. Also, physicians have been obligated to report any culture-confirmed cases of human listeriosis to the National Infectious Disease Registry since 1994.
From 1990 through 2001, the annual number of cases of invasive listeriosis in Finland varied between about 20 and 50 (M. Jahkola, unpublished data; http://www.ktl.fi/ttr). On the basis of only limited O serotyping of L. monocytogenes strains, listeriosis cases, excluding an outbreak caused by a rare serotype, 3a (26), have been thought to be sporadic.
Serotyping is a classic phenotypic tool for epidemiological studies (11, 12, 32). Thirteen serotypes of L. monocytogenes have been identified. However, most isolates belong to only three serotypes, 1/2a, 1/2b, and 4b (10, 24). Although the epidemiological benefit of serotyping is limited, it has provided rapid information for the screening of isolates during suspected outbreaks. Various genotyping methods have been used successfully in more detailed epidemiological studies (2, 13, 18, 22, 26, 28, 31). Pulsed-field gel electrophoresis (PFGE) is one of these genotyping methods, and it has proven to be highly discriminating and reproducible (1, 6, 22).
To study retrospectively the potential occurrence of infection clusters caused by L. monocytogenes in Finland since 1990, both systematic serotyping for O and H antigens and genotyping by PFGE were initiated in 1997 and have been continued prospectively since then. An electronic database library of PFGE profiles and serotypes was created to make possible prompt comparison of the PFGE profiles of different strains and rapid identification of infection clusters.
MATERIALS AND METHODS
Strains.
All human L. monocytogenes isolates (n = 314; one isolate per patient) available at LEP from 1990 to 2001 were studied. The median age of the patients was 56, ranging from 1 day to 93 years. Twenty-five of these isolates were connected with an outbreak of listeriosis (26). Most of the isolates (n = 289) originated in blood, cerebrospinal fluid, or other sterile sites (Table 1). Of the isolates, 180 (57%) were from male subjects and 132 (42%) were from female subjects; for 2 isolates (1%) information on gender was no longer available. Thirty-two isolates were reported to be from pregnancy-associated cases, and 14 of these isolates were from seven mother-child pairs. The strains were isolated in routine clinical microbiology laboratories and were subsequently submitted, with information regarding the patient, to LEP for verification and serotyping. Identification of the strains was carried out by standard methods (5, 37). All strains were stored at −70°C in sterilized skim milk.
TABLE 1.
Origins of L. monocytogenes isolates
| Category | No. (%) of isolates (total n, 314) | |
|---|---|---|
| Site of infection | ||
| Blood or cerebrospinal fluid | 278 (88) | |
| Other sterile sitesa | 11 (4) | |
| Other sitesb | 14 (4) | |
| Not known | 11 (4) | |
| Gender | ||
| Male | 180 (57) | |
| Female | 132 (42) | |
| Not known | 2 (1) | |
| Pregnancy associatedc | 32 (10) |
Brain, bursa, hip prosthesis, joint, pericardium, peritoneum, and pleural fluid.
Gastrointestinal tract, genital mucosa, neck abscess, placenta, upper respiratory tract, and wound swabs.
Fourteen were from seven mother-child pairs.
Serotyping.
All strains were serotyped by using antisera against O and H antigens according to the instructions of the manufacturer (Denka Seiken Co., Ltd, Tokyo, Japan), with minor modifications. The strains were taken from storage at −70°C, revived on sheep blood agar (Oxoid, Hampshire, England) overnight at 37°C, and inoculated onto brain heart infusion (BHI) agar (Difco, Detroit, Mich.) for the determination of O antigens. Furthermore, the bacterial suspension in 0.2% NaCl was heated at 100°C for 1 h instead of 121°C for 30 min. For the determination of H antigens, the strains were passed at 25°C through semiliquid BHI medium in Craigie's tubes (0.2% agar) four times instead of three times.
PFGE.
DNA from L. monocytogenes strains isolated from 1990 to 2000 was prepared as described earlier (27), with the following modifications. The strains were grown on blood agar overnight at 37°C and then for 17 to18 h at 37°C in BHI broth. Two milliliters of this broth culture was mixed with 5 ml of cold PIV buffer (10 mM Tris [pH 7.5], 1 M NaCl). The mixture was centrifuged at 3,000 rpm for 15 min at 4°C in a Midispin 2160 (LKB, Bromma, Sweden), and the cell pellet was suspended in 750 μl of cold PIV buffer. This cell suspension was mixed in equal parts with molten 2% low-melting-point agarose (SeaPlaque agarose; FMC BioProducts, Rockland, Maine), and the mixture was pipetted into plug molds. The plugs were incubated overnight at 37°C in a buffer containing 6 mM Tris-HCl (pH 7.5), 1 M NaCl, 100 mM EDTA, 0.5% Brij 58, 0.2% sodium deoksilate, and 0.5% sodium lauroylsarcosine and supplemented with 1 mg of lysozyme per ml and again overnight at 55 to 57°C in a buffer containing 0.5 M EDTA (pH 9.5) and 1% sodium lauroylsarcosine and supplemented with 0.3 mg of proteinase K per ml. The washing of the plugs and the conditions for restriction endonuclease digestion and PFGE were as described previously (25). Chromosomal DNA was digested overnight with 5 U of AscI (New England BioLabs Inc., Beverly, Mass.).
In 2001, the shorter protocol described by Graves and Swaminathan (17) for the preparation of genomic DNA was brought into use, with slight modifications. Bacterial cells were suspended in 2 ml of CBS (100 mM Tris, 100 mM EDTA, pH 8.0) to an optical density at 450 nm of 0.7 to 0.8. When the cell suspension containing lysozyme was mixed with low-melting-point agarose, 2% SeaPlaque agarose and 0.2 mg of proteinase K/ml were used without 1% sodium dodecyl sulfate. Chromosomal DNA was digested for 4 h with 10 U of AscI.
Electrophoresis was performed at 210 V with 1.0% Pronadisa D-5 agarose gels (Hispanlab, Madrid, Spain) by using a CHEF Mapper or CHEF-DR system (Bio-Rad Laboratories, Richmond, Calif.). Running conditions for AscI-digested DNA were 1 to 28 s for 10 h, followed by 28 to 30 s for 10 h. Low-range PFG markers (New England BioLabs Inc.) were used as molecular weight standards. The gels were visualized on a UV transilluminator and were photographed by using AlphaImager 1220 (Alpha Innotec Corporation, San Leandro, Calif.). The TIFF images were analyzed by using BioNumerics software (Applied Maths, Kortrijk, Belgium) and were normalized by using the low-range PFG marker standards on each gel. Any difference between two PFGE profiles was considered sufficient to distinguish these profiles. The different PFGE profiles were marked with numbers based on the coding agreed upon with the National Veterinary and Food Research Institute (L. Rantala, S. Lukinmaa, A. Siitonen, and T. Honkanen-Buzalski, Abstr. ISOPOL XIV Int. Symp. Problems Listeriosis, p. 155, 2001). Similarity values were calculated by the unweighted pair-group method with arithmetic averages and the Dice coefficient by using BioNumerics software.
Groups of PFGE types.
BioNumerics software was used to compare all PFGE types with PFGE types containing five or more strains. When the similarity value was over 80% and the number of fragment differences between the profiles was three or less, the PFGE type was regarded as being closely related to the one with which it was compared (40). Groups of closely related types were designated with the letter G or with the letters GT when a type containing at least five strains was not related to any other type; the group number indicates the PFGE type with which all of the other PFGE types were compared.
Statistical methods.
Fisher's exact two-tailed test (Epi-Info 6.04 software; World Health Organization, Geneva, Switgerland, and Centers for Disease Control and Prevention, Atlanta, Ga.) was used for statistical analysis. A P value of <0.05 indicated statistical significance.
RESULTS
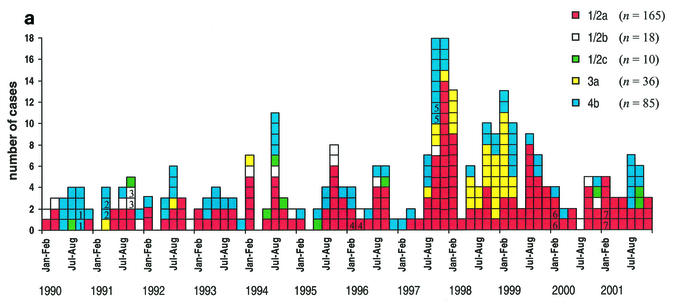
Serotyping divided the 314 isolates into five O:H serotypes (Table 2 and Fig. 1a): serotypes 1/2a (53%), 1/2b (6%), 1/2c (3%), 3a (11%), and 4b (27%). From 1990 through 2001, the percentages of serotype 1/2a strains varied between 22 and 67%, those of serotype 1/2b varied between 0 and 24%, and those of serotype 1/2c varied between 0 and 12%. Strains of serotype 3a were rare, except in 1997, 1998, and 1999, when the percentages were 9, 44, and 22%, respectively. The percentages of serotype 4b strains varied between 12 and 61%. In 1990, most of the strains were of serotype 4b. Since then, strains of serotype 1/2a have been the most common.
TABLE 2.
Annual distributions of serotypes of L. monocytogenes strains isolated from infections in human subjects
| Sero-type | No. (%) of isolates in the following yr:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1990 (n = 18) | 1991 (n = 17) | 1992 (n = 16) | 1993 (n = 16) | 1994 (n = 25) | 1995 (n = 20) | 1996 (n = 21) | 1997 (n = 47) | 1998 (n = 43) | 1999 (n = 45) | 2000 (n = 19) | 2001 (n = 27) | Total (n = 314) | |
| 1/2a | 4 (22) | 6 (35) | 8 (50) | 9 (56) | 14 (56) | 12 (60) | 12 (57) | 26 (55) | 19 (44) | 25 (56) | 12 (63) | 18 (67) | 165 (53) |
| 1/2b | 2 (11) | 4 (24) | 1 (6) | 0 (0) | 3 (12) | 2 (10) | 2 (10) | 1 (2) | 0 (0) | 0 (0) | 3 (16) | 0 (0) | 18 (6) |
| 1/2c | 1 (6) | 1 (6) | 0 (0) | 0 (0) | 3 (12) | 1 (5) | 1 (5) | 0 (0) | 0 (0) | 0 (0) | 1 (5) | 2 (7) | 10 (3) |
| 3a | 0 (0) | 1 (6) | 1 (6) | 0 (0) | 1 (4) | 0 (0) | 0 (0) | 4a (9) | 19a (44) | 10a (22) | 0 (0) | 0 (0) | 36 (11) |
| 4b | 11 (61) | 5 (29) | 6 (38) | 7 (44) | 4 (16) | 5 (25) | 6 (29) | 16 (34) | 5 (12) | 10 (22) | 3 (16) | 7 (26) | 85 (27) |
At least 25 of 33 isolates (76%) were connected with an outbreak (26).
FIG. 1.
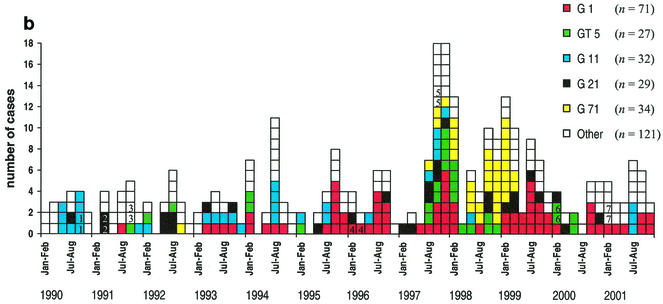
Cases of L. monocytogenes infections by month from 1990 to 2001. The serotypes of the strains (a) and the five most common groups (G1, GT5, G11, G21, and G71) of closely related PFGE types (b) are indicated by colors. The seven mother-child pairs are indicate by numbers (1 to 7).
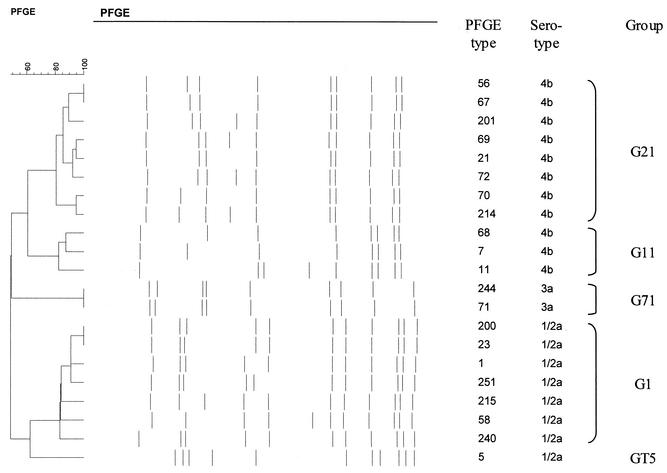
PFGE divided the isolates into 81 PFGE types (Table 3). Each type correlated with one serotype, except for PFGE types 2, 71, and 207, each of which included one strain belonging to a serotype different from the others. PFGE type 2 included strains of serotype 1/2a (8 strains) and 3a (1 strain), PFGE type 71 included strains of serotype 3a (32 strains) and 1/2a (1 strain), and PFGE type 207 included strains of serotype 1/2a (2 strains) and 3a (1 strain). PFGE divided serotypes 1/2a, 1/2b, 1/2c, 3a, and 4b into 49, 10, 2, 5, and 18 PFGE types, respectively. Only 11 of the 81 PFGE types contained five or more strains. Compared with the others, 8 of these 11 PFGE types (1, 2, 11, 21, 61, 71, 74, and 96) were closely related to several other PFGE types and consequently were grouped into G1, G2, G11, G21, G61, G71, G74, and G96, respectively (Table 3 and Fig. 2). The remaining three PFGE types (5, 24, and 65) were not closely related to any other PFGE type; therefore, each formed a group of its own, GT5, GT24, and GT65, respectively. Of the five most common PFGE groups, G1 included seven PFGE types (71 strains, 23%), G71 included two (34, 11%), G11 included three (32, 10%), G21 included eight (29, 9%), and GT5 included one (27, 9%).
TABLE 3.
Annual distributions of L. monocytogenes PFGE types
| No. of isolates | Sero- type | PFGE type | Groupa | No. of isolates in the following yr:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1990 (n = 18) | 1991 (n = 17) | 1992 (n = 16) | 1993 (n = 16) | 1994 (n = 25) | 1995 (n = 20) | 1996 (n = 21) | 1997 (n = 47) | 1998 (n = 43) | 1999 (n = 45) | 2000 (n = 19) | 2001 (n = 27) | ||||
| 37 | 1/2a | 1 | G1 | 2 | 5 | 4 | 7 | 3 | 11 | 2 | 3 | ||||
| 20 | 1/2a | 23 | G1 | 1 | 3 | 3 | 3 | 6b | 1 | 3 | |||||
| 6 | 1/2a | 240 | G1 | 3 | 3 | ||||||||||
| 5 | 1/2a | 200 | G1 | 3 | 1 | 1 | |||||||||
| 1 | 1/2a | 58 | G1 | 1 | |||||||||||
| 1 | 1/2a | 215 | G1 | 1 | |||||||||||
| 1 | 1/2a | 251 | G1 | 1 | |||||||||||
| 8 | 1/2a | 2 | G2 | 1 | 4 | 2 | 1 | ||||||||
| 4 | 1/2a | 42 | G2 | 3 | 1 | ||||||||||
| 27 | 1/2a | 5 | GT5 | 1 | 2 | 2 | 1 | 8 | 9 | 4b | |||||
| 7 | 1/2a | 96 | G96 | 1 | 1 | 1 | 1 | 1 | 2 | ||||||
| 1 | 1/2a | 62 | G96 | 1 | |||||||||||
| 1 | 1/2a | 71 | G71 | 1 | |||||||||||
| 4 | 1/2a | 225 | 1 | 1 | 2 | ||||||||||
| 2 | 1/2a | 33 | 1 | 1 | |||||||||||
| 2 | 1/2a | 53 | 1 | 1 | |||||||||||
| 2 | 1/2a | 63 | 1 | 1 | |||||||||||
| 2 | 1/2a | 75 | 1 | 1 | |||||||||||
| 2 | 1/2a | 207 | 1 | 1 | |||||||||||
| 2 | 1/2a | 237 | 2 | ||||||||||||
| 2 | 1/2a | 248 | 2b | ||||||||||||
| 28 | 1/2a | STc | 2 | 4 | 1 | 1 | 2 | 1 | 1 | 3 | 1 | 5 | 2 | 5 | |
| 6 | 1/2b | 74 | G74 | 2 | 2 | 2 | |||||||||
| 2 | 1/2b | 234 | G74 | 2b | |||||||||||
| 3 | 1/2b | 217 | 1 | 1 | 1 | ||||||||||
| 1 | 1/2b | 224 | G74 | 1 | |||||||||||
| 6 | 1/2b | STd | 1 | 1 | 1 | 1 | 2 | ||||||||
| 9 | 1/2c | 24 | GT24 | 1 | 3 | 1 | 1 | 1 | 2 | ||||||
| 1 | 1/2c | 235 | 1 | ||||||||||||
| 32 | 3a | 71 | G71 | 3e | 19e | 10e | |||||||||
| 1 | 3a | 244 | G71 | 1 | |||||||||||
| 1 | 3a | 2 | G2 | 1 | |||||||||||
| 1 | 3a | 207 | 1 | ||||||||||||
| 1 | 3a | 231 | 1 | ||||||||||||
| 15 | 4b | 11 | G11 | 4b | 1 | 1 | 4 | 1 | 1 | 3 | |||||
| 10 | 4b | 68 | G11 | 4 | 1 | 3 | 1 | 1 | |||||||
| 7 | 4b | 7 | G11 | 1 | 1 | 2 | 3 | ||||||||
| 7 | 4b | 21 | G21 | 1 | 2b | 1 | 1 | 1 | 1 | ||||||
| 4 | 4b | 69 | G21 | 3 | 1 | ||||||||||
| 4 | 4b | 72 | G21 | 1 | 3 | ||||||||||
| 4 | 4b | 201 | G21 | 1 | 2 | 1 | |||||||||
| 3 | 4b | 56 | G21 | 1 | 2 | ||||||||||
| 3 | 4b | 67 | G21 | 1 | 1 | 1 | |||||||||
| 3 | 4b | 70 | G21 | 1 | 1 | 1 | |||||||||
| 1 | 4b | 214 | G21 | 1 | |||||||||||
| 9 | 4b | 61 | G61 | 1 | 1 | 1 | 5 | 1 | |||||||
| 3 | 4b | 10 | G61 | 3 | |||||||||||
| 1 | 4b | 206 | G61 | 1 | |||||||||||
| 5 | 4b | 65 | GT65 | 1 | 1 | 3b | |||||||||
| 4 | 4b | 84 | 1 | 1 | 2 | ||||||||||
| 1 | 4b | 211 | 1 | ||||||||||||
| 1 | 4b | 212 | 1 | ||||||||||||
A “G” prefix indicates genetically closely related strains of several PFGE types; a “GT” prefix indicates at least five identical strains not related to other strains.
Including a mother-child pair; both were counted.
Each strain is connected with a single PFGE type, including PFGE types 14, 27, 44, 57, 59, 66, 73, 78, 202, 205, 208, 209, 210, 216, 218, 220, 227, 228, 229, 230, 233, 241, 243, 245, 246, 247, 249, and 250.
Each strain is connected with a single PFGE type, including PFGE types 64, 213, 226, 236, 239, and 242.
At least 25 of 32 isolates (78%) were connected with an outbreak (26).
FIG. 2.
Dendrogram of closely related PFGE types of the five most common groups (G1, GT5, G11, G21, and G71).
Group G1 (71 strains) formed clusters every year from 1993 onward (Fig. 1b). From 1994 to 1996, most of the cases were caused by PFGE types 1 and 23 of group G1 (Table 3). In 1997 and 1999, large clusters were formed by PFGE type 1 of group G1 alone, and strains of this type were the most common. PFGE type 1 caused infections every year from 1994 onward (2 to 11 cases per year).
Group GT5 (27 strains, of type 5) formed clusters in 1997, 1998, and 2000 (Fig. 1b), and four strains in this group were pregnancy associated.
Strains belonging to group G11 (32 strains) formed clusters in 1990, 1993, 1994, 1995, 1997, and 2001 (Fig. 1b). In 1990, 1991, 1992, and 1993, a total of eight strains were pregnancy associated; four of these cases were caused by PFGE type 68 and four were caused by PFGE type 11. All four cases caused by PFGE type 11 in 1994 occurred in the Helsinki metropolitan area.
Group G21 (29 strains) formed small clusters in 1991, 1992, 1997, 1998, 1999, and 2000 (Fig. 1b). Five strains were pregnancy associated; three of these cases were caused by PFGE types 72, 56, and 69 and two (a mother-child pair) were caused by PFGE type 21.
Group G71 (34 strains) formed clusters in 1997, 1998, and 1999 (Fig. 1b). The clusters were formed by serotype 3a, PFGE type 71. Serotype 1/2a, PFGE type 71, caused only one case, which was pregnancy associated, in 1992.
In group G61 (13 strains), PFGE type 61 (9 strains) formed a cluster of five cases in 1999 (Table 3). Four of these patients and one patient in 1998 were from the greater Tampere area. Also, PFGE type 65 (five strains), which belongs to group GT65, formed a cluster of three cases in 1997 in the Helsinki metropolitan area. However, two of these cases involved a mother-child pair.
The 32 pregnancy-associated cases were caused by 17 different PFGE types. From 1990 to 1993, the pregnancy-associated cases were mostly (8 cases) caused by serotype 4b, PFGE type 68 or 11, each belonging to group G11 (Table 3). After 1993, cases caused by PFGE types in group G11 were not detected. PFGE type 24, which belonged to group GT24, was significantly associated with listeriosis in male subjects (9 of 180 male subjects [5%] versus 0 of 132 female subjects) (P = 0.012). Otherwise, PFGE types were distributed evenly between both genders and among different age groups.
DISCUSSION
In Finland, infections caused by L. monocytogenes have been thought to be sporadic. This insight is based on diagnostic tests in which only the O antigen of the strains was occasionally determined, and more discriminating methods, such as genotyping, were not used before the present study was undertaken.
In order to gain new information on infection clusters and on the distributions of subtypes of L. monocytogenes, 314 clinical isolates from an 11-year period in Finland were analyzed by PFGE and serotyping.
It should be mentioned that not all strains in Finland during the 4-year period from 1990 to 1993 were available for this study, since before 1994 physicians and clinical laboratories notified KTL and submitted strains to LEP only on a voluntary basis. However, the strains studied represented almost 60% of all 119 listeriosis cases diagnosed in the early 1990s (Jahkola, unpublished). Also, the proportion of pregnancy-associated cases might have been underestimated, since information on pregnancy was not always required in the laboratory notes.
From 1990 through 2001, the most common L. monocytogenes serotypes were 1/2a and 4b, accounting, respectively, for 53 and 27% of the 314 isolates from cases of human listeriosis. Since 1990, the number of cases caused by serotype 4b has been fairly constant, at about 4 to 7 cases per year, except in 1997, when the number of cases was 16, and in 1999, when it was 10. However, the number of listeriosis cases caused by serotype 1/2a has increased; therefore, the percentage share of this serotype has also increased. These results support findings in the United Kingdom (30), Denmark (14), Switzerland (33), and Sweden (23) suggesting that serotype 1/2a is replacing serotype 4b in human infections.
In the present study, when an electronic database library was constructed by using BioNumerics software, any difference between two PFGE types was considered sufficient to distinguish two different PFGE types. Therefore, PFGE types containing at least five strains were compared with all other PFGE types to approximate whether they were closely related to any other type according to the criteria of Tenover et al. (40). These criteria are stringent and generally appropriate for studies of strains collected over a short period. However, in the present study, grouping of closely related types collected over a long period yielded more information on clusters. For example, PFGE type 1 was the most prevalent single type, with 37 strains. It belonged to a major cluster, group G1, representing 23% of all 314 strains studied. Furthermore, 27 strains of PFGE type 5 were not related to any other type and formed several clusters over a period of 3 years.
The most prevalent single type, PFGE type 1, alone was previously associated with a vacuum-packed cold smoked rainbow trout product, and it caused a small infection cluster of febrile gastroenteritis in 1997 (31). In addition, Johansson et al. (20) found isolates with an indistinguishable (L. Rantala, personal communication) PFGE profile in retail ready-to-eat vacuum-packed fish products from four different producers in Finland in 1996. However, in order to draw conclusions from this kind of genotyping data, results should be linked to available epidemiological data. It was shown by Autio et al. (4) that similar strains were found in different types of food from different food processors and even in different countries. Therefore, identical PFGE profiles for isolates from food and a patient do not prove, without epidemiological data, that the food isolate caused the infection.
The patients in the 1997 diarrheal cluster described by Miettinen et al. (31) had no known underlying diseases. PFGE type 1, causing the symptoms, was common in sporadic cases from 1994 onward but not before 1994. This finding could reflect a change in dietary habits, i.e., vacuum-packed fish products being more favored in diets; this PFGE type might be more pathogenic than other PFGE types; or this PFGE type simply might be more common than others in fish products.
In some countries, as in France from 1987 to 1997, the United States from 1989 to 1993, and the United Kingdom from 1983 to 1996, the incidence of L. monocytogenes infections has decreased (3, 16, 39). In Finland, since 1990, the incidence has been quite stable, the average being 20 cases per year, except in 1997 (47 cases), 1998 (43 cases), and 1999 (45 cases). An outbreak affecting 25 people, caused by serotype 3a, PFGE type 71, explains in part the high incidences in 1998 and 1999 (26). The findings of the present study also suggested that the outbreak had already started in 1997. In 1998 and 1999, several cases were also caused by PFGE types 1 and 5 of serotype 1/2a. When the outbreak caused by serotype 3a, PFGE type 71, began in 1997, the numbers of cases caused by various PFGE types of serotype 4b (34% of the total cases) and serotype 1/2a (55%) were also unusually high.
In our study, some L. monocytogenes strains of different serotypes displayed indistinguishable PFGE profiles. The serotypes (1/2a and 3a) of these strains belonged to the same flagellar H antigen group (AB); in addition, they have been reported to belong to the same genetic subgroup (7, 18, 32). In these cases, serotyping and PFGE were repeated for the same colony, and the identical results obtained confirmed the findings. However, only one restriction enzyme, AscI, which is commonly used for L. monocytogenes in PFGE (4, 6, 17), was used; the use of other enzymes and another genotyping method might have been more discriminatory for these strains.
From 1990 to 1993, the pregnancy-associated cases were most commonly caused by serotype 4b, PFGE type 68 or 11, group G11. However, no specific serotype or PFGE type could be concluded to be associated with pregnancy after 1993. Also, no particular PFGE types could be connected with gender or age group. The only exception was PFGE type 24, which was significantly associated with gender: all nine patients were men.
In this study, a new database library of the PFGE profiles of different strains was constructed by using BioNumerics software. In this new electronic library, PFGE profiles can be compared with each other more rapidly; therefore, clusters can be detected more rapidly and at an early stage. This database library also makes possible continuous surveillance of invasive L. monocytogenes infection clusters in Finland. After the library was constructed, a bilateral computer-based network for comparison of PFGE profiles of L. monocytogenes isolates from human subjects, food, and food production environments was created (Rantala et al., Abstr. ISOPOL XIV Int. Symp. Problems Listeriosis) by the National Veterinary and Food Research Institute and the KTL. This kind of national cooperation during a suspected outbreak will probably help in recognizing sources of infections. It will also enable authorities to track down food production plants that need to improve production hygiene for their food products or to give direct information on food hygiene to specific risk groups of consumers. During the 1990s, clusters of cases of the same PFGE type or clusters of groups of closely related types were seen every year, indicating the need for and the importance of timely typing of human L. monocytogenes strains. Furthermore, on a larger scale, an electronic network of PFGE profiles for human L. monocytogenes strains has been in use as PulseNet since 1996 in the United States (38), where it has demonstrated its value in the early recognition of outbreaks and the rapid identification of their sources.
Acknowledgments
This work was supported by the Technology Development Centre of Finland (TEKES), the Finnish Research Programme on Environmental Health, and an ABS graduate school scholarship to Susanna Lukinmaa.
The skillful technical assistance of Jari Aho, Sirkku Ekström, Tarja Heiskanen, Liisa Immonen, Ritva Taipalinen, and Sirkku Waarala is gratefully acknowledged. We also thank Tiina Autio (DVM) for scientific advice.
REFERENCES
- 1.Aarnisalo, K., T. Autio, A.-M. Sjöberg, J. Lundén, H. Korkeala, and M.-L. Suihko. 2003. Typing of Listeria monocytogenes isolates originating from the food processing industry with automated ribotyping and pulsed-field gel electrophoresis. J. Food Prot. 66:249-255. [DOI] [PubMed] [Google Scholar]
- 2.Allerberger, F., and S. J. Fritschel. 1999. Use of automated ribotyping of Austrian Listeria monocytogenes isolates to support epidemiological typing. J. Microbiol. Methods 35:237-244. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 1997. Listeriosis in England and Wales: 1983-1996. Commun. Dis. Rep. 7:95. [Google Scholar]
- 4.Autio, T., J. Lunden, M. Fredriksson-Ahomaa, J. Björkroth, A.-M. Sjöberg, and H. Korkeala. 2002. Similar Listeria monocytogenes pulsotypes detected in several foods originating from different sources. Int. J. Food Microbiol. 77:83-90. [DOI] [PubMed] [Google Scholar]
- 5.Bille, J., J. Rocourt, and B. Swaminathan. 1999. Listeria, Erysipelothrix, and Kurthia, p. 346-356. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society of Microbiology, Washington, D.C.
- 6.Brosch, R., M. Brett, B. Catimel, J. B. Luchansky, B. Ojeniyi, and J. Rocourt. 1996. Genomic fingerprinting of 80 strains from the W. H. O. multicenter international typing study of Listeria monocytogenes via pulsed-field gel electrophoresis (PFGE). Int. J. Food Microbiol. 32:343-355. [DOI] [PubMed] [Google Scholar]
- 7.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bula, C. J., J. Billie, and M. P. Glauser. 1995. An epidemic of food-borne listeriosis in western Switzerland: description of 57 cases involving adults. Clin. Infect. Dis. 20:66-72. [DOI] [PubMed] [Google Scholar]
- 9.Dalton, G. B., C. C. Austin, J. Sobel, P. S. Hayes, W. F. Bibb, L. Graves, B. Swaminathan, M. E. Proctor, and P. M. Griffin. 1997. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 336:100-105. [DOI] [PubMed] [Google Scholar]
- 10.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filice, G. A., H. F. Cantrell, A. B. Smith, P. S. Hayes, J. C. Feeley, and D. W. Fraser. 1978. Listeria monocytogenes infection in neonates: investigation of an epidemic. J. Infect. Dis. 138:17-23. [DOI] [PubMed] [Google Scholar]
- 12.Fleming, D. W., S. L. Cochi, K. L. MacDonald, J. Brondum, P. S. Hayes, B. D. Plikaytis, M. B. Holmes, A. Audurier, C. V. Broome, and A. L. Reingold. 1985. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N. Engl. J. Med. 312:404-407. [DOI] [PubMed] [Google Scholar]
- 13.Fonnesbech Vogel, B., H. H. Huss, B. Ojeniyi, P. Ahrens, and L. Gram. 2001. Elucidation of Listeria monocytogenes contamination routes in cold smoked salmon processing plants detected by DNA-based typing methods. Appl. Environ. Microbiol. 67:2586-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerner-Smidt, P., M. Weischer, A. Jensen, and W. Fredriksen. 1995. Listeriosis in Denmark—results of a 10-year survey, p.472. InXII International Symposium on Problems of Listeriosis. Promaco Conventions Pty Ltd., Perth, Western Australia, Australia.
- 15.Goulet, V., J. Rocourt, I. Rebiere, C. Jacquet, C. Moyse, P. Dehaumont, G. Salvat, and P. Veit. 1998. Listeriosis outbreak associated with the consumption of rillettes in France in 1993. J. Infect. Dis. 177:155-160. [DOI] [PubMed] [Google Scholar]
- 16.Goulet, V., H. de Valk, O. Pierre, F. Stainer, J. Rocourt, V. Vaillant, C. Jacquet, and J.-C. Desenclos. 2001. Effect of prevention measures on incidence of human listeriosis, France, 1987-1997. Emerg. Infect. Dis. 7:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 18.Graves, L. M., and B. Swaminathan, M. Reeves, S. Hunter, R. Weaver, B. Plikaytis, and A. Schuchat. 1994. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J. Clin. Microbiol. 32:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heitmann, M., P. Gerner-Smidt, and O. Heltberg. 1997. Gastroenteritis caused by Listeria monocytogenes in a private day-care facility. Pediatr. Infect. Dis. J. 16:827-828. [DOI] [PubMed] [Google Scholar]
- 20.Johansson, T., L. Rantala, L. Palmu, and T. Honkanen-Buzalski. 1999. Occurrence and typing of Listeria monocytogenes strains in retail vacuum-packed fish products and in a production plant. Int. J. Food Microbiol. 47:111-119. [DOI] [PubMed] [Google Scholar]
- 21.Kaczmarski, E. B., and D. M. Jones. 1989. Listeriosis and ready-cooked chicken. Lancet i:549. [DOI] [PubMed]
- 22.Kerouanton, A., A. Brisabois, E. Denoyer, F. Dilasser, J. Grout, G. Salvat, and B. Picard. 1998. Comparison of five typing methods for the epidemiological study of Listeria monocytogenes. Int. J. Food Microbiol. 43:61-71. [DOI] [PubMed] [Google Scholar]
- 23.Loncarevic, S., W. Tham, and M.-L. Danielsson-Tham. 1998. Changes in serogroup distribution among Listeria monocytogenes human isolates in Sweden, p. 20. In XIII International Symposium on Problems of Listeriosis. Halifax, Nova Scotia, Canada.
- 24.Low, J. C., F. Wright, J. McLauchlin, and W. Donachie. 1993. Serotyping and distribution of Listeria isolates from cases of ovine listeriosis. Vet. Rec. 133:165-166. [DOI] [PubMed] [Google Scholar]
- 25.Lukinmaa, S., R. Schildt, T. Rinttilä, and A. Siitonen. 1999. Salmonella Enteritidis phage types 1 and 4: pheno- and genotypic epidemiology of recent outbreaks in Finland. J. Clin. Microbiol. 37:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyytikäinen, O., T. Autio, R. Maijala, P. Ruutu, T. Honkanen-Buzalski, M. Miettinen, M. Hatakka, J. Mikkola, V.-J. Anttila, T. Johansson, L. Rantala, T. Aalto, H. Korkeala, and A. Siitonen. 2000. An outbreak of Listeria monocytogenes serotype 3a infections from butter in Finland. J. Infect. Dis. 181:1835-1841. [DOI] [PubMed] [Google Scholar]
- 27.Maslow, J. N., A. M. Slutsky, and R. D. Arbeit. 1993. Application of pulsed-field gel electrophoresis to molecular epidemiology, p. 563-572. In D. H. Persing, T. H. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 28.McLauchlin, J., A. Audurier, A. Frommelt, P. Gerner-Smidt, C. Jacquet, M. J. Loessner, N. van der Mee-Marquet, J. Rocourt, S. Shah, and D. Wilhelms. 1996. W. H. O. study on subtyping Listeria monocytogenes: results of phage-typing. Int. J. Food Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 29.McLauchlin, J., M. H. Greenwood, and P. N. Pini. 1990. The occurrence of Listeria monocytogenes in cheese from a manufacturer associated with a case of listeriosis. Int. J. Food Microbiol. 10:255-262. [DOI] [PubMed] [Google Scholar]
- 30.McLauchlin, J., and L. Newton. 1995. Human listeriosis in England, Wales and Northern Ireland: a changing pattern of infection, p. 177-181. In XII International Symposium on Problems of Listeriosis. Promaco Conventions Pty Ltd., Perth, Western Australia, Australia.
- 31.Miettinen, M. K., A. Siitonen, P. Heiskanen, H. Haajanen, K. J. Björkroth, and H. J. Korkeala. 1999. Molecular epidemiology of an outbreak of febrile gastroenteritis caused by Listeria monocytogenes in cold smoked rainbow trout. J. Clin. Microbiol. 37:2358-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pak, S.-I., U. Spahr, T. Jemmi, and M. D. Salman. 2002. Risk factors for L. monocytogenes contamination of dairy products in Switzerland, 1990-1999. Prev. Vet. Med. 53:55-65. [DOI] [PubMed] [Google Scholar]
- 34.Salamina, G., E. Dalle Donne, A. Niccolini, G. Poda, D. Cesaroni, M. Bucci, R. Fini, M. Maldini, A. Schuchat, B. Swaminathan, W. Bibb, J. Rocourt, N. Binkin, and S. Salmaso. 1996. A foodborne outbreak of gastroenteritis involving Listeria monocytogenes. Epidemiol. Infect. 117:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlech, W. F., P. M Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls, and C. V. Broome. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 36.Schuchat, A., K. A. Deaver, J. D. Wenger, B. D. Plikaytis, L. Mascola, R. W. Pinner, A. L. Reingold, C. V. Broome, et al. 1992. Role of foods in sporadic listeriosis. I. Case-control study of dietary risk factors. JAMA 267:2041-2045. [PubMed] [Google Scholar]
- 37.Seeliger, H. P. R., and D. Jones. 1986. Genus Listeria Pirie 1940, 383AL, p. 1235-1245. In R. G. E. Murray, D. J. Brenner, M. P. Bryant, J. G. Holt, N. R. Krieg, J. W. Moulder, N. Pfenning, P. H. A. Sneath, and J. T. Stanley (ed.), Bergey′s manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md.
- 38.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tappero, J. W., A. Schuchat, K. A. Deaver, L. Mascola, J. D. Wenger, et al. 1995. Reduction in the incidence of human listeriosis in the United States. Effectiveness of prevention efforts? JAMA 273:1118-1122. [DOI] [PubMed] [Google Scholar]
- 40.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]