Abstract
A novel double-antigen sandwich enzyme-linked immunosorbent assay (ELISA) was developed to measure rabies antibodies in dogs. In contrast to the 4 days required for detecting rabies antibody with conventional rabies antibody virus neutralization assays, this ELISA can be completed in hours, without using live virus, in routine laboratories.
Rabies is recognized as one of the oldest diseases of humankind throughout the world and is still very serious, especially in developing countries (4). Simple methods for titrating antibodies from different species are needed for seroepidemiologic studies of rabies. The mouse neutralization test, rapid fluorescent focus inhibition test, and fluorescent antibody virus neutralization (FAVN) test are not suitable for processing large numbers of serum specimens, because they all require a specialized laboratory. In the present study, we investigated the applicability of the double-antigen sandwich enzyme-linked immunosorbent assay (ELISA), in which an antibody is sandwiched by two antigens.
Antigenic determinants of rabies virus G and N proteins have been mapped, and a chimeric peptide (G5-24-31D) containing a linear epitope of the G and N protein was synthesized and found to be immunogenic in mice (3), suggesting that the chimeric peptide derived from rabies virus may be used as a diagnostic antigen for detecting rabies antibodies.
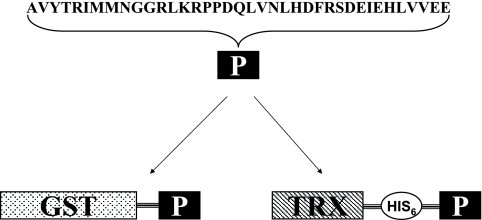
Two recombinant plasmids, pGEX4T2/ep and pET32a/ep, were constructed through the in-frame fusion of a chimeric peptide (AVYTRIMMNGGRLKRPPDQLVNLHDFRSDEIEHLVVEE) representing rabies G (amino acids 253 to 275) and N (amino acids 404 to 418) proteins to the C-terminal coding sequence of glutathione S-transferase (GST) or thioredoxin (Trx) (Fig. 1). Sequence encoding rabies chimeric peptide was synthesized using overlapping PCR. The primers (P1, 5′CAGGATCCGCAGTTTATACCCGTATTATGATGAACGGTGGTCGTCTGAAACGTCCG3′; P2, 5′GGTCGTCTGAAACGTCCGCCGGACCAGCTGGTGAACCTGCATGACTTCCGTTCGGAT3′; and P3, 5′ CCACTCGAGTTATTCTTCCACAACCAGGTGTTCGATTTCATCCGAACGGAAGTCATG3′) used for overlapping PCR contain complementary regions and serve as templates for each other. The resulting PCR product, with introduced BamHI and XhoI restriction sites (5′ and 3′, respectively), was cloned into pGEX-4T-2 and pET32a vectors. The recombinant plasmids pGEX4T2/ep and pET32a/ep were transformed into Rosetta (Novagen) for expression of the fusion proteins. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) results indicated the desired recombinant proteins were expressed in soluble form.
FIG. 1.
Schematic representation of fusion protein GST/GN-epitope and Trx/GN-epitope. P represents rabies virus G and N protein chimeric peptide. GST and Trx represent glutathione S-transferase and thioredoxin, respectively, which were fusion expressed with rabies chimeric peptide.
The purifications of the fusion proteins GST/GN-epitope and Trx/GN-epitope were performed using glutathione Sepharose 4B and nickel-chelating Sepharose (Amersham Biosciences) individually. The final yields were about 16 mg/liter and 19 mg/liter culture, with at least 95% purity (Fig. 2A). Western blot analysis of the purified proteins showed bands of about 34 kDa and 25 kDa (Fig. 2B), which were consistent with the predicted molecular mass of chimeric rabies peptide (4.5 kDa) plus GST (28.7 kDa) or TRX (14 kDa) as well as additional amino acids.
FIG. 2.
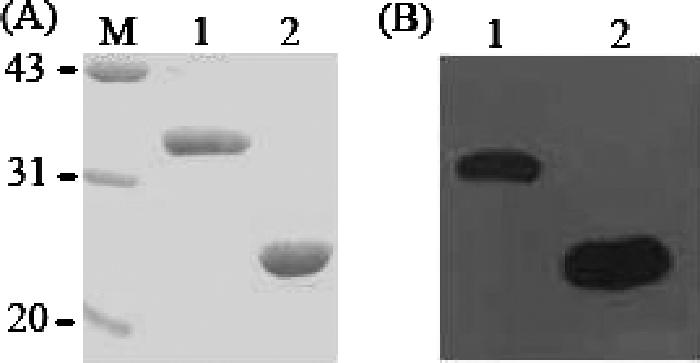
SDS-PAGE and Western blot analysis of purified recombinant fusion proteins GST/GN-epitope and Trx/GN-epitope expressed in Escherichia coli. (A) Coomassie brilliant blue-stained SDS-PAGE. Lane M indicates molecular mass standards in kilodaltons, lane 1 represents purified recombinant GST/GN-epitope, and lane 2 represents purified recombinant Trx/GN-epitope. (B) Western blot analysis of the same samples represented in panel A and run on a separate gel. The labeling is the same as that for panel A. The primary antibody used to visualize recombinant fusion proteins was a canine rabies antiserum, and the secondary antibody was a sheep anti-canine immunoglobulin G antibody linked to HRP.
Horseradish peroxidase (HRP)-conjugated GST/GN-epitope antigen was prepared according to the procedure developed by Nakane and Kawaoi and modified by Wilson and Nakane (7). High binding microtiter plates (Corning) were sensitized with 100 ng of recombinant Trx/GN-epitope protein in each well of the plates. The plates were then blocked and dried at room temperature. Five hundred serum samples (100 μl) collected from both vaccinated (n = 400) and unvaccinated (n = 100) dogs and equally prediluted with sample diluent buffer (phosphate-buffered saline buffer, pH 7.4, including 4% [wt/vol] polyethylene glycol 6000, 3% [wt/vol] NaCl, 0.05% [vol/vol] Tween 20), were added to the wells in duplicate. The negative sera (collected from unvaccinated dogs that tested negative by neutralization test) were added in duplicate at the same dilution. After 30 min of incubation at 37°C, the serum samples were removed and the plates were washed five times. HRP-conjugated GST/GN-epitope antigens (100 μl), prediluted to 1:5,000, were added to each well. After incubation for 15 min at 37°C followed by washing, 100 μl of peroxidase substrate (Sigma) was added to each well and then incubated for 10 min at room temperature. Absorbance at 450 nm was measured against a blank after stopping the reaction by adding 50 μl of 4N sulfuric acid to each well. All ELISAs were repeated two or three times, and the results obtained at different times gave similar results, indicating the assay was consistent. The serum samples were also investigated using the gold standard FAVN test, which was performed in accordance with the protocols (6, 8).
For sample titer calculation, a series of diluted positive reference sera (61.5 IU/ml; National Institute for the Control of Pharmaceutical and Biological Products, Beijing, China) was included in parallel in each measurement (equivalent to FAVN titers of 6, 4, 2, 1, 0.5, and 0.25 IU/ml). Serum titers were expressed as equivalent units (EU) per milliliter, corresponding to international units by using the values obtained with the reference serum (1).
The sensitivity and specificity of the sandwich ELISA were evaluated in comparison to those of the FAVN reference method. Based on the 500 serum samples, the sensitivities and specificities of the ELISA at various cutoffs were calculated by receiver operating characteristics (data not shown). The optimal cutoff value, giving the highest index for both specificity and sensitivity, was 0.32, i.e., 0.9 EU/ml. The sensitivity and specificity of ELISA were calculated {sensitivity, [(1 + 252)/ (2 + 271)] × 100 = 92.67%; specificity, [(95 + 121)/(98 + 129)] × 100 = 95.15%}. A total of 253 tested serum samples were positive, and 216 were negative in both assays (Table 1). There was only one positive sample among 100 naïve dog serum samples defined by both assays, indicating the dog may be infected.
TABLE 1.
Comparison of ELISA and FAVN for detection of rabies antibodiesa
| ELISA test result (0.9-EU/ml cutoff) | FAVN test result (0.5-IU/ml cutoff)
|
|||
|---|---|---|---|---|
| Naïve sera (n = 100)
|
Vaccinated sera (n = 400)
|
|||
| No. negative | No. positive | No. negative | No. positive | |
| Negative | 95 | 1 | 121 | 19 |
| Positive | 3 | 1 | 8 | 252 |
| Total | 98 | 2 | 129 | 271 |
A panel of 500 serum samples was parallel investigated using ELISA and a fluorescent antibody virus neutralization (FAVN) test.
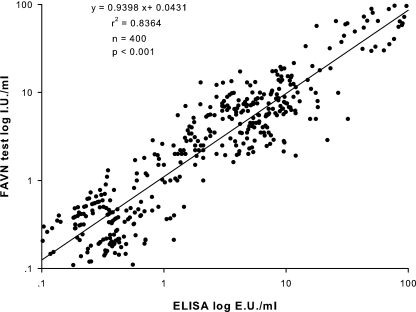
In order to assess the correlation of the FAVN and sandwich ELISA, the vaccinated serum sample results of the two assays were compared using the least-squares linear regression analysis as described previously (2). The correlation coefficient between the ELISA and the FAVN test was 0.92 (n = 400; P < 0.05). A scattered diagram of ELISA and FAVN antibody titers is shown in Fig. 3.
FIG. 3.
Correlation between FAVN and ELISA tests on 400 vaccinated serum samples. The obtained results were converted into the decimal logarithm value. The least-squares linear regression analysis was carried out, which showed a strong correlation between the two assays.
Several papers introduced the use of ELISA for measurement of antibodies to rabies (1, 2, 5). However, inactive virus was used as a coated antigen in some methods. Because rabies virus can infect people through open wounds or mucous membranes if the virus was not inactivated completely, this may pose a safety threat. In addition, the double-antigen sandwich ELISA theoretically can be used to detect rabies antibody from different species. It is a powerful tool for us, because rabies virus has many reservoirs, such as dogs, bats, raccoons, skunks, and foxes.
In conclusion, recombinant rabies virus G and N protein chimeric peptide can be used in double-antigen sandwich ELISA for measuring antibodies following rabies infection, or vaccination in dogs or other species, with high sensitivity, specificity, and correlation with other diagnostic assays. This is the first description of measurement of rabies antibody by double-antigen sandwich ELISA, which has potential clinical significance.
Acknowledgments
This work was supported by the Ministry of Science and Technology of China (grant no. 2004BA519A50) and the 973 Project (grant no. 2005CB523).
REFERENCES
- 1.Cliquet, F., L. M. McElhinney, A. Servat, J. M. Boucher, J. P. Lowings, T. Goddard, K. L. Mansfield, and A. R. Fooks. 2004. Development of a qualitative indirect ELISA for the measurement of rabies virus-specific antibodies from vaccinated dogs and cats. J. Virol. Methods 117:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Cliquet, F., L. Sagne, J. L. Schereffer, and M. F. Aubert. 2000. ELISA test for rabies antibody titration in orally vaccinated foxes sampled in the fields. Vaccine 18:3272-3279. [DOI] [PubMed] [Google Scholar]
- 3.Dietzschold, B., M. Gore, D. Marchadier, H. S. Niu, H. M. Bunschoten, L. Otvos, Jr., W. H. Wunner, H. C. Ertl, A. D. Osterhaus, and H. Koprowski. 1990. Structural and immunological characterization of a linear virus-neutralizing epitope of the rabies virus glycoprotein and its possible use in a synthetic vaccine. J. Virol. 64:3804-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreesen, D. W. 1997. A global review of rabies vaccines for human use. Vaccine 15(Suppl.):S2-S6. [DOI] [PubMed] [Google Scholar]
- 5.Piza, A. S., J. L. Santos, L. B. Chaves, and C. R. Zanetti. 1999. An ELISA suitable for the detection of rabies virus antibodies in serum samples from human vaccinated with either cell-culture vaccine or suckling-mouse-brain vaccine. Rev. Inst. Med. Trop. Sao Paulo 41:39-43. [DOI] [PubMed] [Google Scholar]
- 6.Smith, J. S., P. A. Yager, and G. M. Baer. 1973. A rapid reproducible test for determining rabies neutralizing antibody. Bull. W. H. O. 48:535-541. [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson, M. B., and P. K. Nakane. 1978. Recent developments in the periodate method of conjugating horseradish peroxidase (HRPO) to antibodies, p. 215-224. In W. Knapp, G. Wick, and K. Holubar (ed.), Immunofluorescence and related staining techniques. Elsevier, Amsterdam, The Netherlands.
- 8.Zalan, E., C. Wilson, and D. Pukitis. 1979. A microtest for the quantitation of rabies virus neutralizing antibodies. J. Biol. Stand. 7:213-220. [DOI] [PubMed] [Google Scholar]