Abstract
The yeast Debaryomyces hansenii has a remarkable capacity to proliferate in salty and alkaline environments such as seawater. A screen for D. hansenii genes able to confer increased tolerance to high pH when overexpressed in Saccharomyces cerevisiae yielded a single gene, named here DhGZF3, encoding a putative negative GATA transcription factor related to S. cerevisiae Dal80 and Gzf3. Overexpression of this gene in wild-type S. cerevisiae increased caffeine and rapamycin tolerance, blocked growth in low glucose concentrations and nonfermentable carbon sources, and resulted in lithium- and sodium-sensitive cells. Sensitivity to salt could be attributed to a reduced cation efflux, most likely because of a decrease in expression of the ENA1 Na+-ATPase gene. Overexpression of DhGZF3 did not affect cell growth in a gat1 mutant but was lethal in the absence of Gln3. These are positive factors that oppose both Gzf3 and Dal80. Genome-wide transcriptional profiling of wild-type cells overexpressing DhGZF3 shows decreased expression of a number of genes that are usually induced in poor nitrogen sources. In addition, the entire pathway leading to Lys biosynthesis was repressed, probably as a result of a decrease in the expression of the specific Lys14 transcription factor. In conclusion, our results demonstrate that DhGzf3 can play a role as a negative GATA transcription factor when expressed in S. cerevisiae and that it most probably represents the only member of this family in D. hansenii. These findings also point to the GATA transcription factors as relevant elements for alkaline-pH tolerance.
Adaptation to conditions of high-salt stress is a biological process of the utmost importance in agriculture and biotechnology due to the increased salinization of irrigated lands. Debaryomyces hansenii is an ascomycetous salt- and high-pH-tolerant yeast found in seawaters and salty food. This yeast is becoming a model for the study of salt tolerance mechanisms in eukaryotic cells (37), and this research will be boosted by the recent release of its complete genomic sequence (21). Although the halotolerant characteristics of this organism were defined a long time ago (33, 34, 36), the molecular basis for the halotolerant phenotype is still obscure. D. hansenii encodes orthologs to most components that have been found to be relevant for salt tolerance in other fungi, particularly in the model yeast Saccharomyces cerevisiae. Examples are the DhHOG1 mitogen-activated protein kinase, the DhHAL2 nucleotidase, and the DhENA1 and DhENA2 sodium ATPase pumps (2, 4, 5). In most cases, these gene products have proven to be functional for complementing the equivalent S. cerevisiae mutants, although they did not confer a particularly strong salt-tolerant phenotype even when overexpressed. However, the fact that D. hansenii is not an easily tractable organism from a genetic point of view and the lack of molecular tools to approach its study have been important disadvantages in attempts to define the basis for its unusual salt tolerance.
The natural ambient for D. hansenii is seawater, which is characterized by high salinity (around 0.5 M NaCl) and an alkaline pH (7.6 to 7.8). S. cerevisiae cannot tolerate high salt and hardly grows at pHs above 7.0. High-salt and high-pH tolerance phenotypes are not functionally dependent upon each other. However, there is evidence that the transcriptional responses to high-salt and high-pH stresses show significant overlap in S. cerevisiae (42). An example is the ENA1 sodium ATPase gene, which is potently induced by both high-salt conditions and ambient alkalinization (23).
In an attempt to identify novel components in D. hansenii that could be important for understanding the ability of this organism to thrive in its specific ambient, we have carried out a screen by transforming an S. cerevisiae wild-type strain with a multicopy D. hansenii genomic library and selecting clones on the basis of increased alkaline-pH tolerance. This screen yielded a unique D. hansenii gene that probably represents the only negative GATA factor encoded by this organism and whose cloning and functional characterization are described in this work.
MATERIALS AND METHODS
Yeast strains and growth conditions.
S. cerevisiae cells were grown, unless otherwise stated, at 28°C in yeast-peptone (YP)-dextrose (YPD) medium or complete minimal medium lacking the appropriate requirements for selection (1). All yeast strains used in this work are listed in Table 1.
TABLE 1.
Yeast strains used in this work
| Strain | Relevant genotype | Reference |
|---|---|---|
| W303-1A | MATα ade-2-1 his3-11 15 ura3-1 leu2-3, 112 trp1 | 45 |
| G19 | W303-1A ena1Δ::HIS3::ena4Δ | 6 |
| R100 | W303-1A nhx1Δ::URA3 | 32 |
| C25 | W303-1A nha1Δ::LEU2 | 7 |
| BY4741 | MATα his3Δ1 leu2Δ met15Δ ura3Δ | 47 |
| BY4741 gzf3::kanMX4 | 47 | |
| BY4741 gln3::kanMX4 | 47 | |
| BY4741 gat1::kanMX4 | 47 | |
| JK9-3da | MATaleu2-3,112 ura3-52 trp1 his4 rme1 HMLa | 16 |
| TB50a | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLa | 16 |
| AS51-1a | JK9-3daure2::URA3 | 16 |
| TB105-3b | TB50agln3::kanMX gat1::HIS3MX | 16 |
| AL11-2c | TB50agln3::kanMX gat1::HIS3MX ure2::URA3 | 16 |
Screen for alkaline-pH tolerance.
Wild-type yeast strain W303-1A was transformed with a genomic multicopy library from D. hansenii strain PYCC2968 (CBS767), which was constructed as described previously (38). Transformants were grown in complete minimal medium plates lacking uracil and replica plated onto the same plates containing 20 mM N-Tris (hydroxymethyl) methyl-3-aminopropanesulfonic acid and adjusted to pH 7.7 with arginine. Plates were incubated for 5 days, and clones that were able to form macroscopic colonies were recovered (under these conditions, wild-type cells harboring an empty plasmid did not grow). About 150 clones were recovered and tested again for alkaline tolerance at a range of pHs from 7.7 to 8.0. Finally, five positive clones were selected and grown, and the plasmid was recovered in Escherichia coli. Plasmids were reintroduced into wild-type strain W303-1A, and their ability to confer alkaline tolerance was confirmed. The molecular nature of the inserts was defined by restriction analysis and DNA sequencing.
Plasmid construction and gene disruption.
The entire open reading frame (ORF) of DhGZF3 was amplified by PCR (nucleotides −4 to 1657 from the initiating Met), cloned into the vector pCR2.1-TOPO (Invitrogen), and digested with XbaI and KpnI. The released insert was cloned into these same sites of plasmid pYPGE15 (11) to yield pDhGZF3.
A genomic fragment containing the entire ORF of S. cerevisiae GZF3 (from nucleotides −505 to +2226) was amplified by PCR, digested with XbaI and HincII, and cloned into plasmid YEplac181 or YCplac111 (25) to produce plasmid YEp-ScGZF3 or YCp-ScGZF3, respectively.
For low-copy centromeric expression of DhGZF3, the 2.8-kbp fragment containing the gene (positions −363 to +2473 from the ATG codon) was recovered from plasmid YEp352-DhGZF3 (clone II) by digestion with EcoRI/SpeI and cloned into the EcoRI/XbaI sites of YCplac111 (LEU2 marker). An identical cloning strategy using plasmid YEplac181 (LEU2 marker) allowed high-copy expression (YEp-DhGZF3).
Determination of Li+ content, efflux, and uptake.
For determination of the Li+ content, cells were grown overnight in yeast nitrogen base (YNB) medium supplemented with different concentrations of LiCl. When growth reached an optical density (A600) of 0.3 to 0.4, cells were collected on Millipore filters and rapidly washed with 20 mM MgCl2. The cells were then extracted with acid and analyzed by atomic emission spectrophotometry (4).
To study lithium uptake, yeast cells were grown overnight in YNB medium. When the A600 reached values of 0.3 to 0.4, cells were harvested by centrifugation, washed with cold water, and resuspended in assay buffer [10 mM MES (2-morpholinoethanesulfonic acid) adjusted to pH 5.8 with Ca(OH)2 and supplemented with 0.1 mM MgCl2-2% (wt/vol) glucose]. At time zero, LiCl (100 mM) was added, and samples were taken periodically, filtered, and treated for determination of intracellular Li+ as described above (20).
For determination of Li+ efflux, cells were grown overnight in YNB medium and incubated in the same medium plus 150 mM LiCl for 4 h. Cells were harvested, washed with cold water, and resuspended in assay buffer supplemented with 50 mM KCl to trigger the efflux process. Samples were taken at intervals, filtered, and treated for determination of intracellular Li+ as described above (20).
β-Galactosidase reporter assays.
Wild-type strain BY4741 was cotransformed with YEplac181 (or YEp-DhGZF3) and the diverse β-galactosidase reporter constructs containing segments of the ENA1 promoter. Cells were grown up to saturation on synthetic medium lacking leucine and uracil and then inoculated into YPD medium to give an A660 of 0.15. Growth was resumed until an A660 of 0.8 was reached, and cells were then centrifuged and resuspended in YPD medium, YPD medium adjusted at pH 8.0, or YPD medium plus 0.2 M LiCl. Growth was resumed for 60 min, cells were recovered by centrifugation, and β-galactosidase levels were measured as described previously (40).
RNA preparation, DNA microarray analysis, and reverse transcription-PCR (RT-PCR).
Strain W303-1A was transformed with plasmid pYPGE15 or pDhGZF3 and grown in synthetic medium lacking uracil (containing 2% glucose as a carbon source and 38 mM ammonium sulfate as a nitrogen source) or in YPD medium up to an A660 of 0.5. Yeast cells were harvested by sedimentation and washed with cold water, and total RNA was extracted using the RiboPure-Yeast kit (Ambion) according to the manufacturer's instructions. RNA quality was assessed by electrophoresis in a denaturing 0.8% agarose gel. Fluorescent Cy3- and Cy5-labeled cDNAs were prepared from 8 μg of purified total RNA by the indirect dUTP-labeling method using the CyScribe Post-Labeling kit (Amersham Biosciences). Aminosilane-coated slides (UltraGAPS; Corning) containing 6,073 yeast ORFs (3) were prehybridized at 42°C for 1 h in a solution containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate, and 1% bovine serum albumin. For hybridization, dried Cy3- and Cy5-labeled probes were resuspended in 35 μl of hybridization solution (50% formamide, 5× SSC, 0.1% sodium dodecyl sulfate) each and mixed. Five micrograms of salmon sperm DNA was added to the mix before denaturalization for 3 min at 95°C. DNA microarrays were hybridized in an ArrayBooster hybridization station (Advalytix AG) for 14 h at 42°C. Microarrays were scanned in a ScanArray 4000 apparatus (Packard) at a 10-μm resolution, and images were processed using GenePix Pro 6.0 software (Molecular Devices). Data correspond to the means of two microarrays, in which dye swapping was performed, for each condition tested.
RNA was prepared for RT-PCR analysis as follows. W303-1A cells were transformed with pYPGE15 or pDhGZF3, grown up to an A600 of 1.0 in YNB without amino acids (except for auxotrophic requirements), and supplemented with proline as nitrogen source and 2% glucose. Cells were collected by filtration, frozen in liquid nitrogen, and ground in a mortar. The powder was resuspended in 1 ml of Tripure (Roche) and centrifuged (12,000 × g for 10 min at 4°C). The supernatant was extracted with 0.2 ml CHCl3 and centrifuged (12,000 × g for 15 min at 4°C), and nucleic acids were precipitated from the aqueous supernatant by adding isopropanol (0.5 ml), followed by centrifugation (12,000 × g for 10 min at 4°C). Pellets were washed with 75% ethanol and dried. RT-PCR was performed with the RetroTools kit (Biotools) using 1 μg of RNA and oligonucleotides 5′-ATGTGTAAAGCCGGTTTTGC-3′ and 5′-GGGGCTCTGAATCTTTCGTTA-3′ for ACT1 amplification and 5′-TCAGGAGCCAATAACCATTCC-3′ and 5′-CTTGTGTCCGATGTACATAACC-3′ for GAP1 amplification (annealing at 55°C for 26 cycles).
Growth assays.
Yeast cells at an initial A660 of 0.05 and the appropriate dilutions (3 μl) were spotted onto YPD or synthetic medium agar plates containing diverse compounds as indicated. Growth was recorded after 2 to 5 days of incubation.
Nucleotide sequence accession number.
The sequence reported in this work has been deposited in GenBank accession no. AM232660.
RESULTS
High-copy expression of the DhGZF3 gene in S. cerevisiae results in increased tolerance to alkaline pH.
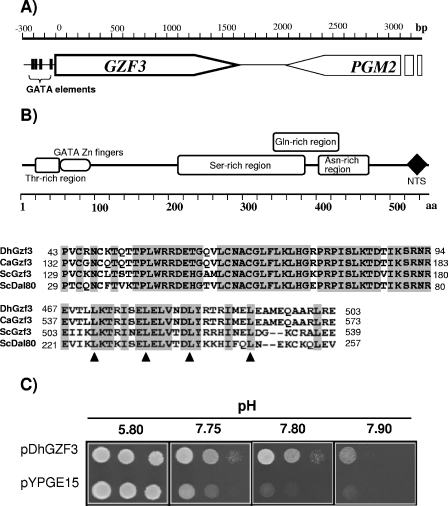
S. cerevisiae strain W303-1A (wild type) was transformed with a multicopy genomic D. hansenii library, and about 40,000 transformants were recovered on selective plates lacking uracil. Colonies were replica plated onto similar plates buffered at pH 7.7 and allowed to grow for 5 days. About 150 putative positive clones were rescreened at pHs ranging from 7.7 to 8.0. Five clones showing the highest tolerance were selected, and the plasmid was recovered. Restriction mapping and DNA sequencing revealed that four of the clones contained the same genomic region, spanning about 3.5 kbp (Fig. 1A), which included the C-terminal half of an open reading frame showing high identity to the S. cerevisiae PGM2 gene, encoding phosphoglucomutase, as well as an entire open reading frame (plus about 400 bp of the 5′ promoter region). A comparison of the sequence of this ORF (GenBank accession number AM232660) with the recently available entire sequence of D. hansenii showed complete identity with ORF DEHA0C05962g. This ORF encodes a 543-residue protein (estimated molecular mass of 59,052.5 Da) rich in Asn and Ser residues that shows structural elements reminiscent of the GATA-related transcription factor family at its amino terminus and a sequence compatible with a leucine zipper motif and a bipartite nuclear targeting sequence at its carboxyl terminus (Fig. 1B). Sequence comparison revealed significant identity with several fungal proteins, principally Gzf3 from Candida albicans and Gzf3 and Dal80 from S. cerevisiae (Table 2). Therefore, we will refer to this ORF as DhGZF3. The entire ORF was amplified by PCR and cloned under the control of the S. cerevisiae PGK1 promoter to yield plasmid pDhGZF3 (see Materials and Methods). This plasmid was introduced into wild-type strain W303-1A and tested for high-pH tolerance (Fig. 1C). The construct was able to fully reproduce the tolerant phenotype of genomic clone II, indicating that it was attributable to the DhGZF3 ORF.
FIG. 1.
The putative D. hansenii GATA-related transcription factor DhGZF3 confers tolerance to high pH to S. cerevisiae. (A) Genomic structure of clone II. (B) Structural elements that characterize the DhGZF3-encoded protein. NTS denotes a bipartite nuclear targeting sequence. The alignment below the diagram shows the similarity between the GATA Zn finger region of DhGzf3 and its putative orthologs in C. albicans and S. cerevisiae as well as a particularly well conserved region near the C terminus of the proteins encoding a putative leucine zipper (relevant Leu residues are indicated with triangles). (C) Wild-type S. cerevisiae strain W303-1A was transformed with plasmid pDhGZF3 as well as with an empty vector (pYPGE15). Tenfold dilutions of the cells were inoculated on synthetic medium plates lacking uracil buffered at the indicated pH and incubated for 5 days.
TABLE 2.
Percent identities between D. hansenii GZF3 and related genes in other fungia
| Gene | % Identity (% conserved residues)
|
||||
|---|---|---|---|---|---|
| DhGzf3 | CaGzf3 | YlQ6CB14 | ScGzf3 | ScDal80 | |
| DhGzf3 | 36.3 (43.6) | 21.9 (27.3) | 19.7 (28.1) | 20.3 (27.9) | |
| CaGzf3 | 18.2 (24.1) | 29.8 (41.1) | 22.5 (30.4) | ||
| Yl_Q6CB14 | 17.0 (23.2) | 14.6 (18.8) | |||
| ScGzf3 | 21.1 (29.6) | ||||
| ScDal80 | |||||
Identities (percentages) were calculated using the EMBOSS pairwise alignment algorithm (global) with the following settings: gap penalty, 10; extended penalty, 0.2. Numbers in parentheses denote percentages of conserved residues. Dh, Debaryomyces hansenii; Ca, Candida albicans; Yl, Yarrowia lipolytica; Sc, S. cerevisiae.
Overexpression of DhGZF3 in S. cerevisiae results in pleiotropic effects.
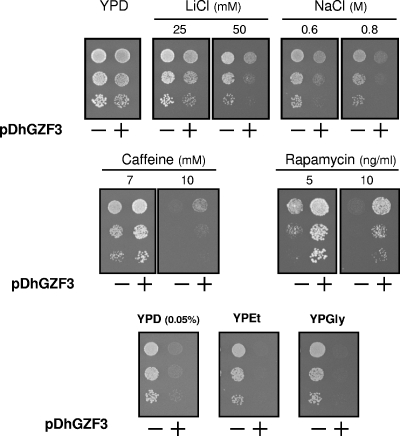
To evaluate the possible functional roles of DhGZF3, we tested the growth of W303-1A yeast cells by overexpressing this gene under a number of conditions. We observed that the overexpression of DhGZF3 had little influence on the growth rate when cells were grown on rich (YPD) medium, although it resulted in a moderately reduced cell growth rate on synthetic medium using ammonium or proline as the nitrogen source (data not shown). Interestingly, as shown in Fig. 2, cells expressing the DhGZF3 gene display a marked sensitivity to the toxic cations lithium and sodium, while growth was not differentially affected by KCl up to concentrations of 2 M (data not shown). The salt-sensitive phenotype was even more prominent when cells were grown in synthetic medium (data not shown). These cells also displayed increased tolerance to drugs such as caffeine and rapamycin (Fig. 2). Tolerance to the cell wall-damaging agent Congo red was not altered, although an increase in the tolerance to this compound and to calcofluor white (another cell wall-damaging agent) was detected in the BY4741 genetic background (data not shown). Furthermore, overexpression of the Debaryomyces gene resulted in greatly impaired growth under glucose-limiting conditions or with nonfermentable carbon sources (Fig. 2). These effects were clearly reproduced when DhGZF3 was expressed in two additional genetic backgrounds, such as strains DBY746 and BY4741 (data not shown).
FIG. 2.
Diverse phenotypic effects of overexpression of DhGZF3 in S. cerevisiae. Dilutions of the strains used in Fig. 1 were spotted and grown for 2 days on YP plates supplemented with the indicated compounds. YPEt, YP medium with 2% ethanol; YPGly, YP medium with 2% glycerol.
We also compared the phenotypic effects of D. hansenii and S. cerevisiae GZF3 genes on a wild-type strain when expressed from their endogenous promoters on a high-copy-number plasmid. Expression of DhGZF3 increased tolerance to caffeine, rapamycin, and alkaline pH, while it conferred a slight sensitivity to LiCl. In contrast, expression of the S. cerevisiae counterpart increased tolerance to only caffeine and rapamycin although much less efficiently than the gene from D. hansenii (data not shown).
Overexpression of DhGZF3 in S. cerevisiae alters cation contents and efflux by affecting ENA1 expression.
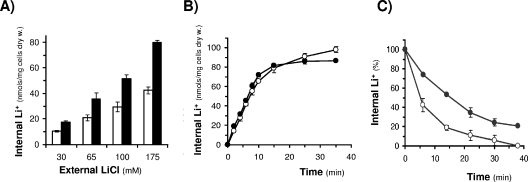
The increased lithium and sodium sensitivity due to the overexpression of DhGZF3 was considered an interesting phenotype that merited further attention. Although decreased salt tolerance can be attributed to many factors, a common circumstance is the failure for the cell to efficiently maintain a harmless concentration of toxic cations. Therefore, we measured the internal content of lithium in cells challenged by various concentrations of external LiCl. As shown in Fig. 3A, in all cases, the expression of DhGZF3 resulted in an increased lithium content, providing a reasonable explanation for the salt-sensitive phenotype. In turn, increased internal content can be produced by an enhanced entry or by a decreased efflux capacity. To evaluate these possibilities, we tested the lithium influx rate and did not observe significant changes (Fig. 3B). In contrast, when cells were loaded with lithium and then transferred to a medium that was free of the cation, those cells expressing DhGZF3 were less effective in removing intracellular lithium cations (Fig. 3C).
FIG. 3.
Analysis of lithium sensitivity in S. cerevisiae cells overexpressing DhGZF3. (A) W303-1A cells transformed with empty vector (pYPGE15) (open bars) or DhGZF3 (filled bars) were grown overnight in synthetic medium lacking uracil and supplemented with the indicated concentrations of LiCl. The internal concentrations of Li+ were measured as described in Materials and Methods. (B) Lithium influx process in S. cerevisiae cells overexpressing DhGZF3 (•) or transformed with the empty vector (○). Cells were grown as described in Materials and Methods and transferred to a buffer supplemented with 100 mM LiCl. (C) Lithium efflux process in S. cerevisiae cells overexpressing DhGZF3 (•) or transformed with the empty vector (○). Cells were loaded with LiCl as described in Materials and Methods and transferred into Li+-free buffer. In all cases, data are means ± standard deviations from three independent experiments.
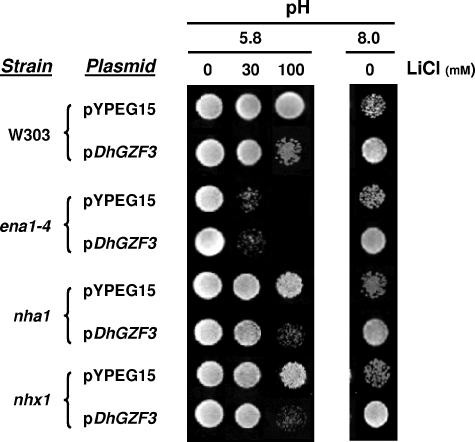
These results suggested that the salt-sensitive phenotype of DhGZF3-expressing cells was due to a defect in cation efflux. To gain information about the molecular nature of the defect, we expressed the DhGZF3 putative transcription factor in cells lacking important elements in cation detoxification, such as the plasma membrane Ena1 sodium ATPase and Nha1 cation/H+ antiporter as well as the prevacuolar Nhx1 antiporter. As shown in Fig. 4, expression of DhGZF3 was unable to increase lithium sensitivity of ena1-4 mutants. In contrast, the overexpression of the Debaryomyces gene in nha1 and nhx1 backgrounds resulted in increased sensitivity. It is worth mentioning that none of these mutations affected the ability to increase tolerance to high pH conferred by the expression of DhGZF3.
FIG. 4.
Effects of overexpression of DhGZF3 on lithium and alkaline-pH sensitivity in S. cerevisiae mutants lacking relevant cation transporters. Strains W303-1A (wild type), G19 (ena1-4), C25 (nha1), and R100 (nhx1) were transformed with the indicated plasmids and spotted onto synthetic medium plates lacking uracil and containing the indicated concentrations of LiCl and/or buffered at the indicated pHs. Growth was recorded after 5 days.
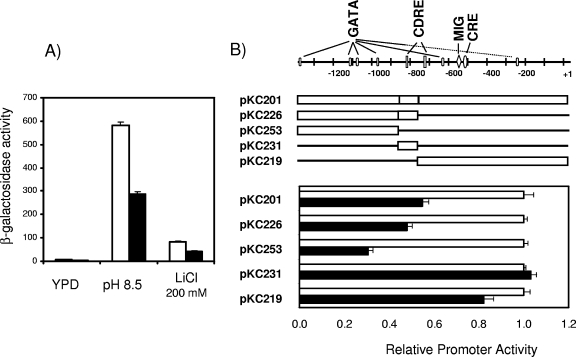
The observation that the expression of DhGZF3 in the absence of the ENA1 gene does not result in increased lithium sensitivity suggested that this effect could be mediated by the function of the ATPase. ENA1 is, by far, the most important component of the ENA1-ENA4 gene cluster, and it is essentially regulated at the expression level. Its expression is almost null in the absence of stress, but the gene is rapidly activated in response to increased salt concentrations or a high pH (23, 35, 42). Therefore, we considered the possibility that the overexpression of the gene might affect the expression levels of ENA1. To test for this possibility, we cotransformed strain BY4741 with plasmid YEp-DhGZF3 and with a β-galactosidase reporter containing the entire ENA1 promoter (pKC201) and measured the expression levels from the promoter under basal conditions as well as under LiCl and high-pH conditions. As shown in Fig. 5A, expression of the Debaryomyces gene resulted in about a 50% decrease in the expression from the ENA1 promoter under all conditions tested. Therefore, the increased sensitivity produced by the overexpression of DhGZF3 on an otherwise wild-type yeast strain can be attributed to a failure in the extrusion of toxic cations because of decreased expression of the ENA1 ATPase.
FIG. 5.
Effects of overexpression of DhGZF3 on the expression levels of the ENA1 ATPase. (A) Wild-type strain BY4741 was cotransformed with plasmid pKC201 and either YEplac181 (empty bars) or YEp-DhGZF3 (filled bars) and subjected to the indicated treatments as described in Materials and methods. β-Galactosidase activities are means ± standard errors of the means of data for six independent clones. (B) The indicated constructs containing different segments of the ENA1 promoter were cotransformed in BY4741 cells with either YEplac181 (empty bars) or YEp-DhGZF3 (filled bars), and β-galactosidase activity was measured. For each case, the activity in cells bearing YEp-DhGZF3 was divided by the activity in cells bearing the empty plasmid (YEplac181), which was considered the unit. Data are means ± standard errors of the means for six independent experiments. CDRE, calcineurin-dependent response element
To further characterize how the expression of DhGZF3 alters the transcriptional response of ENA1, we determined the level of expression driven by different segments of the ENA1 promoter in cells overexpressing the putative transcription factor (Fig. 5B). Interestingly, transcription from the region included in pKC226, which is very strong even under basal conditions (no stress) and is no further activated by pH or LiCl stress (our unpublished results), is reduced by about 50% by expression of DhGZF3. The effect is even more dramatic in pKC253, although the relatively low level of basal expression from this promoter region makes the comparison less accurate. The promoter region included in pKC226 contains several putative GATA boxes and a calcineurin-dependent response element that is insensitive to stress but required for basal expression levels. In contrast, expression from the promoter fragment in pKC231, which is almost as powerful in driving transcription as pKC226 and which includes the calcineurin-dependent response element but no GATA boxes, is not affected at all by expression of DhGZF3. These findings are consistent with the possibility that the overexpression of DhGZF3 would negatively affect ENA1 expression through the GATA sequences present in the promoter of the ATPase gene.
Functional interaction between high levels of DhGzf3 and mutation of GLN3 and GAT1 genes.
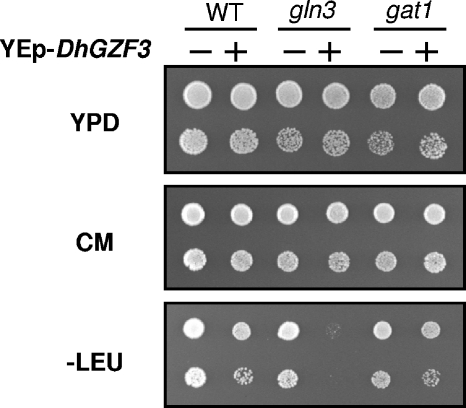
Because of the structural relationship between DhGzf3 and GATA factors related to nitrogen catabolic gene expression, we sought to evaluate the effect of overexpression of DhGZF3 in S. cerevisiae cells lacking Gln3 or Gat1. Interestingly, our attempts to overexpress DhGZF3 in gln3 cells systematically yielded a few very small colonies, suggesting that high levels of DhGzf3 might be harmful for the cell. This was confirmed by two different approaches. First, we selected for plasmid-bearing colonies in synthetic medium lacking leucine and grew them overnight in YPD medium. Samples of the cultures were then spotted onto YPD medium, complete synthetic medium, or synthetic medium lacking leucine. As shown in Fig. 6, growth was insignificant in the selective medium, suggesting that most cells had lost the plasmid during prior nonselective growth. An identical result was obtained when overexpression of DhGZF3 was attempted in gln3 gat1, ure2 gln3, or ure2 gln3 gat1 cells (data not shown). In contrast, overexpression of DhGZF3 in a gat1 background did not negatively affect cell growth (Fig. 6). A second approach consisted of transforming a diploid strain, heterozygous for the deletion of gln3, with different plasmids expressing DhGZF3 at a high copy number from a powerful promoter (pDhGZF3) or from its own promoter at a high copy number (YEp-DhGZF3) or a low copy number (YCp-DhGZF3). Diploids were induced to sporulate, and spores were analyzed for the presence of both the mutation and the plasmid. We recovered colonies carrying the centromeric, low-copy-number plasmid, but we never recovered those colonies that would result in the overexpression of DhGZF3 (data not shown), again supporting the notion that a high level of DhGzf3 is deleterious for cells lacking Gln3.
FIG. 6.
Effect of overexpression of DhGZF3 on Gln3- and Gat1-deficient cells. Wild-type strain BY4741 (WT) and its isogenic gln3 and gat1 derivatives were transformed with YEplac181 (−) or YEp-DhGZF3 (+). Colonies were recovered from transformation plates lacking Leu (−Leu) (very small in the case of gln3 transformants) and grown overnight on liquid medium lacking Leu. Cells were then inoculated into YPD medium and grown overnight. Finally, dilutions were spotted onto the indicated plates and incubated for 48 h. CM, complete medium.
Transcriptional profiling of S. cerevisiae cells overexpressing DhGZF3.
To gain insight into the possible functional role of DhGzf3, we used DNA microarrays to evaluate how the overexpression of DhGZF3 in S. cerevisiae may affect the expression of specific mRNAs. As shown in Table 3, 55 genes were induced at least 1.8-fold in cells overexpressing DhGZF3 (see also the supplemental material). We noted the induction of several genes encoding amino acid transporters, such as the branched-chain amino acid permeases BAP2 and TAT1, the low-affinity methionine permease MUP3, and the high-affinity S-methylmethionine permease MMP1. The list also includes a significant number of genes involved in purine and pyrimidine biosynthesis, such as IMD1-4, encoding the four members of the IMP dehydrogenase family (that catalyzes the first step of GMP biosynthesis from IMP); RNR3, encoding the large subunit of ribonucleotide reductase; and URA1, URA2, and URA4, which define the entire pathways for orotate synthesis from glutamine.
TABLE 3.
Genes induced (>1.8-fold) or repressed (>2.0-fold) in S. cerevisiae strain W303-1A cells overexpressing DhGZH3 and grown in synthetic mediuma
| Gene |
|---|
| Induced genes |
| Amino acid transport |
| MUP3, TAT1, BAP2, MMP1 |
| Energy and carbohydrate metabolism |
| MDH2, GSY1, HAP4, GLC3, TPS2, HXT2, HXT6, CYC7, CYC1 |
| Purine and pyrimidine biosynthesis |
| IMD1, IMD2, IMD3, IMD4, RNR3, URA1, URA2, URA4 |
| Ty element transposition |
| YAR010C, YJR026W, YML040W, YMR051C, YML045W, YJR028W, YMR046C |
| Other |
| TPO4, PHO3, NUP53, KAP123, SSA3, MRK1, SSN3, SKM1, RIB4, CIN5, RPA135, NOP12, AVO2, HSH155, SAS10, SPL2, UTP20 |
| Unknown |
| YOL036w, YKR075c, YER067w, YER076c, YJR115w, FMP48, FMP43, YGR269w, YJL084C, PRM10 |
| Repressed genes |
| Amino acid metabolism and transport |
| LYS1, LYS20, LYS9, LYS12, LYS4, LYS2, PUT1, ARG81, PUT4, SDL1, LYS21, LYS14, ALD2, GAP1, CAN1, VBA3 |
| Transport |
| CTP1, GIT1, MEP2, DAN1, GAL2, MEP1, AVT5, HXT9, QDR2, BTN2, PML39, YIL166C, SIL1, AVT2, VMR1 |
| Transcription and RNA metabolism |
| RPA190, MOT1, GZF3, GAT1, REG2, INO2, SHQ1, AAR2, TIS11 |
| Carbohydrate metabolism and energy |
| INO1, GAL7, PYC2, AMS1, PCK1, JAC1 |
| Other metabolism |
| THI12, YFR055W, POT1 |
| Protein modification and catabolism |
| KSP1, RUB1, RAM1, ERO1, DPH2, RIM8, JJJ3, DER1, BLM10 |
| Cell wall and organelle organization and biogenesis |
| PEX31, PEX21, PCP1, ECM34, YEA4 |
| DNA metabolism and cell cycle |
| YJL113W, STB2, MRC1, MCM10, RIS1, SAE2, DAD1, HOP2, HIR3, SCW11, MFA1, ASG7, PRM6 |
| Sporulation |
| SPR3, ADY4, SPS22, SHC1, OSW2, SPO20 |
| Response to stress |
| HSP26, TSL1, SSA4, FRT2, YJL144W |
| Other |
| DAL3, URA3, ADH7, ATG14, DIG2, FOL3, DCG1, SLI1, NPR1, ATG17, AYT1 |
| Unknown |
| YGR125W, YAR064W, ACO2, YJL195C, YKL070W, YGR031W, YLR458W, YOR387C, RMD6, YOR203W, YDR535C, FMP16, YDR355C, YDR220C, YMR111C, YOL166C, YBR184W, YER085C, YDR090C, YKR105C, VEL1, YHL042W, YJL038C, HSP33, DSF1, FKS3, YBL112C, HSP32, UGX2, YDL196W, YFL066C, YAL018C, YCR095C, YEL041W, YBR285W, CWC23, YBR116C, NIT1, GTO1, YGL138C, YIR041W, YDR290W, APQ13, KRE29, YPR116W, YLR171W, YKL115C, YDR340W, YFL040W, SHE1, COS4, YDR540C, YAL064W, YBR147W, YIL001W, YJL043W, YEL076C, YDR428C, ATG29, YET2, YLR211C, YDR114C, PHM8, YLR004C, YOR300W |
Functional classification was made using Gene Ontology Slim Mapper and Term Finder tools available at SGD (http://www.yeastgenome.org/). Some groups were merged manually.
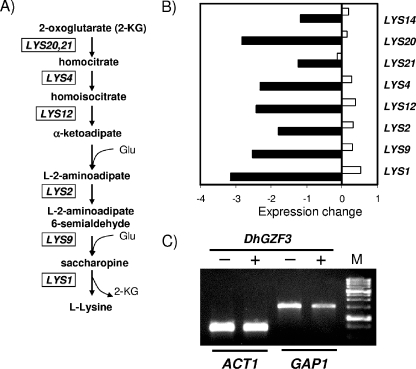
Overexpression of DhGZF3 also resulted in 163 genes whose mRNA levels decreased by at least twofold (Table 3; see the supplemental material). In this case, the strong downregulation of the genes covering the entire pathway that leads to lysine biosynthesis from α-ketoglutarate (aminoadipic pathway) is worth noting (Fig. 7A and B). Remarkably, this pathway was not repressed when the same experiment was repeated using cells grown on YPD medium (data not shown). Repression was not a general effect on amino acid biosynthesis-related genes, although specific genes such as PUT1 or the transporters GAP1, PUT4, CAN1, and VBA3 were also repressed. We noted the enormous amount of repressed genes with no obvious function (labeled “unknown” in Table 3), accounting for 40% of the total number. Figure 7C shows that high levels of DhGzf3 are able to decrease the expression of GAP1, a permease responsible for the uptake of arginine that is induced in poor nitrogen sources such as proline. These results support the role of DhGzf3 as a negative GATA factor acting on nitrogen regulation.
FIG. 7.
Expression profiles of the genes involved in the pathway for l-lysine biosynthesis in cells overexpressing DhGZF3. (A) The l-α-aminoadipate pathway for Lys biosynthesis in S. cerevisiae. (B) The ratio (log2 base) of expression between control cells and cells overexpressing DhGZF3 for genes involved in Lys biosynthesis in synthetic medium (ammonium as nitrogen source) (filled bar) or rich medium (YPD) (empty bars) is represented. See the text for details. (C) Yeast cells bearing the empty plasmid pYPGE15 (−) or expressing DhGZF3 from pDhGZF3 (+) were grown in synthetic medium with Pro as the nitrogen source, and total RNA was prepared. RT-PCR was performed as described in Materials and Methods, and the product was visualized by agarose gel electrophoresis and ethidium bromide staining. M indicates 1-kbp ladder size markers.
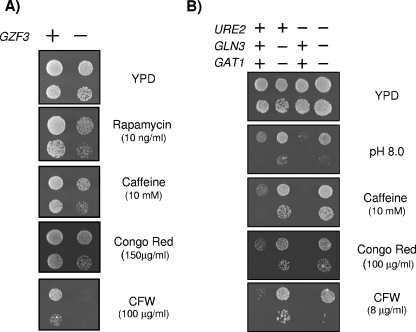
Phenotypic characterization of the S. cerevisiae gzf3 deletion mutant.
Because of the relatively limited amount of knowledge about the function of the GZF3 gene product in S. cerevisiae, we sought to characterize phenotypes derived from its mutation. As shown in Fig. 8A, the gzf3 strain was sensitive to rapamycin and to diverse cell wall-damaging agents such as caffeine, Congo red, and, particularly, calcofluor white but displayed tolerance identical to that of the wild-type strain when confronted with diverse concentrations of LiCl or a wide range of alkaline pHs (data not shown). These phenotypes were abolished by the expression of ScGZF3 on a centromeric plasmid. In contrast, growth of dal80 cells was not affected by caffeine or Congo red and was only very slightly affected by calcofluor white (data not shown).
FIG. 8.
Phenotypes associated with the gzf3, ure2, gln3, and gat1 mutations in S. cerevisiae. (A) Dilutions of wild-type strain BY4741 (+) and its isogenic gzf3 derivative were spotted onto plates containing the indicated compounds. Growth was monitored after 2 days. (B) Strains JK9-3da (WT), AS51-1a (ure2), TB105-3b (gln3 gat1), and AL11-2c (gln3 gat1 ure2) were spotted onto the indicated plates, and growth was monitored after 3 days. CFW, calcofluor white.
We then tested the sensitivity of ure2, gln3 gat1, and ure2 gln3 gat1 mutant strains to alkaline pH and to diverse cell-damaging agents (Fig. 8B). Ure2 acts as an inhibitor of the GATA transcriptional activators Gln3 and Gat1 when optimal sources of nitrogen are available. Remarkably, ure2 cells were sensitive to high pH as well as to Congo red, calcofluor white, and caffeine. In contrast, the gln3 gat1 strain was hypertolerant to the cell-damaging drugs as well as to high pHs. An additional deletion of URE2 was almost without effect under the diverse conditions tested. Therefore, the lack of Gzf3 mimics part of the phenotypes observed upon the disruption of URE2, which are opposite of those observed in a gln3 gat1 mutant strain.
DISCUSSION
In this work, we have cloned and characterized DhGZF3, a putative GATA transcription factor from D. hansenii. As far as we know, this is the first work to report the study of a gene coding for a GATA factor in this organism. In S. cerevisiae, four GATA factors regulate the expression of genes involved in nitrogen catabolite repression in response to the nature of the nitrogen source. While Gln3 and Gat1 are positive regulators, Gzf3 and Dal80 act as negative regulators. When a preferred nitrogen source is available, the entry of Gln3 and Gat1 into the nucleus is prevented (largely due to the control exerted by the TOR pathway), and expression of genes required for the utilization of alternative nitrogen sources is avoided (see references 14 and 31 for reviews). Similarity searches of the recently sequenced D. hansenii genome (21, 29) revealed the presence of likely Gln3 and Gat1 homologs (ORFs DEHA0G21164g and DEHA0E02431g, respectively). However, these searches revealed only a single ORF with almost equal similarity to both ScDal80 and ScGzf3 (Table 2). It is remarkable that the encoded protein maintains a C-terminal sequence comprising a putative leucine zipper domain, which is present in ScGzf3 (43) and ScDal80 (15) and, in the latter, has been shown to be required for its repressive role and for dimerization (15, 44). Therefore, most probably, a unique repressor GATA factor exists in D. hansenii.
The results presented in this work support the notion that DhGzf3 behaves as a negative GATA factor opposed to the action of positive effectors Gln3 and Gat1. For instance, the overexpression of DhGZF3 had little effect on growth in cells cultured in YPD medium but decreased the growth rate when they were grown on less favorable nitrogen sources (ammonium or Pro). Similarly, high levels of DhGzf3 resulted in tolerance to the TOR inhibitor rapamycin, while the deletion of endogenous GZF3 gave rise to sensitivity to this drug (Fig. 2 and 8), a phenotype also observed for gat1 and gln3 mutants (8, 12). Overexpression of DhGZF3 resulted in a mild increase in tolerance to certain cell wall-damaging agents such as caffeine (Fig. 2), Congo red, or calcofluor white (data not shown). In contrast, we show that the gzf3 and ure2 S. cerevisiae strains are sensitive to these treatments and that a double gln3 gat1 mutant is tolerant (Fig. 8A and B), in agreement with the opposed role previously described for Ure2 and Gln3/Gat1 (8). These compounds are known to induce activation of the Slt2 mitogen-activated protein kinase. Interestingly, activation of Slt2 was also observed after inhibition of TOR kinases by the treatment of yeast cells with rapamycin, although the exact nature of this effect is not clear (see reference 30 for a review).
The negative effect of the overexpression of DhGzf3 in the absence of Gln3 on growth (Fig. 6) reinforces the notion that DhGzf3 plays roles that are opposite from those of Gln3 and Gat1. The observation that the overexpression of DhGzf3 is not deleterious in the absence of Gat1 could be explained by the previous observation that functions of Gln3 and Gat1 are not fully overlapping or by a differential competitive behavior of the D. hansenii factor, or it may merely reflect the fact that the lack of Gln3 negatively affects the expression of Gat1 (13). We also observed that wild-type cells overexpressing DhGZF3 cannot grow on limiting glucose or poor carbon sources (Fig. 2). This is remarkable, as a role for Gln3 and Gat1 in mediating signaling in response to poor carbon sources has been postulated in the last few years (9, 27). Finally, it is probably significant that the number of repressed genes found in our DNA microarray experiments is threefold higher than the number of induced genes, a result that agrees with the notion that DhGzf3 acts as a negative transcription factor in S. cerevisiae. It must be noted that high-copy-number expression of ScGZF3 (from its own promoter) does not reproduce the phenotypes obtained by similar expression of the DhGZF3 gene (plasmid YEp-DhGZF3) (data not shown). This could be the result of either unregulated expression of the D. hansenii gene or the ability of DhGzf3 to perform both ScGzf3 and Dal80 functions.
An interesting observation is that the overexpression of DhGZF3 results in S. cerevisiae cells that are less tolerant to lithium or sodium cations. Our data support the idea that the observed decrease in lithium tolerance results from the inability of cells overexpressing DhGZF3 to fully induce the ENA1 cation ATPase in response to lithium stress (Fig. 4 and 5). This is remarkable because the ENA1 promoter contains several putative GATA elements, most of them located far upstream on the promoter, several hundred bases apart from the main regulatory region. Our results would be in agreement with the previously proposed role for the positive regulators Gln3 and Gat1 in the regulation of ENA1 expression under salt stress conditions (16). Those authors observed that a double gln3 gat1 mutant displays a marked growth defect and a substantial decrease of ENA1 expression in high-salt media and postulated that a full response to salt stress requires a modulation of the TOR pathway and the participation of Gat1/Gln3 as positive effectors of ENA1 expression. Therefore, a likely explanation for our results would be that overexpressed DhGzf3 is strongly opposing the positive role of Gln3/Gat1 on the ENA1 promoter. This hypothesis is reinforced by the observation (Fig. 5B) that the overexpression DhGZF3 has a maximum effect on regions of the ENA1 promoter that contain the majority of the six putative GATA motifs. These regions are unaffected by other positive or negative signaling pathways that govern ENA1 expression, such as sodium toxicity, osmotic stress, or glucose availability. Likewise, expression from a region of the promoter that lacks GATA motifs (pKC231) is not affected at all by high levels of DhGzf3. Therefore, these results provide further evidence that DhGzf3 is acting as a negative GATA factor.
On the basis of the indicated hypothesis, one would expect that our genome-wide transcriptional profile of S. cerevisiae cells overexpressing DhGZF3 would reveal changes in the transcription of a number of genes regulated by the quality of the nitrogen source. This is confirmed in many cases, such as the observed downregulation of the ammonium transporters MEP1 and MEP2, the general permease GAP1, or the proline transporter PUT4, which are induced in response to poor nitrogen sources (31). These genes, in addition to DAL3 (which was also repressed in our experiments), were found to be induced in cells under conditions in which Gln3 is activated (17). We also observed the repression of GZF3 and GAT1. This is remarkable, because it has previously been reported that Dal80 is able to antagonize the capability of Gat1 to increase its own expression and to negatively regulate GZF3 expression (13).
A most striking finding in our transcriptomic profiling analysis (Table 3 and Fig. 7) is the intense and general downregulation of genes required for the biosynthesis of Lys through the aminoadipate pathway. Our data also show two functionally related genes, CPT1 (encoding a mitochondrial membrane citrate transporter) and YJL200c (a putative mitochondrial aconitase isozyme), to be strongly repressed (about fivefold). It is very suggestive that both genes have been defined as transcriptionally coregulated with the genes involved in the Lys biosynthetic pathway as a result of a peroxisomal deficiency (10). We detected two downregulated peroxisomal genes in our study, PEX31, which has been shown to participate in the regulation of peroxisome size and number (46), and PEX21, although a relationship between both observations is not obvious. We also observed the downregulation of LYS14, the gene encoding a transcription factor that is specific for the regulation of the expression of genes in this pathway (22, 39). This suggests that the overexpression of DhGZF3 produces the repression of LYS14, which in turn results in the inability to properly express Lys biosynthetic genes. Interestingly, computer analysis of the LYS14 promoter region revealed two GATA boxes separated by 15 nucleotides (nucleotides −56 to −82), a structure that has been shown to be suitable for Dal80 binding (18).
In conclusion, our observations allow us to define DhGzf3 as the unique negative GATA transcriptional factor in D. hansenii. The isolation of a negative GATA factor as a result of a screen for increased alkaline-pH tolerance can be considered surprising because, as far as we know, no previous relationship between this kind of protein and alkaline-pH tolerance has been established. In fact, a recent screen performed in our laboratory to search for S. cerevisiae genes that conferred high-pH tolerance when overexpressed yielded only two genes (FET4 and CTR1), which are apparently unrelated to GATA factors (41). A link between Lys metabolism, its transformation in cadaverine, and pH regulation has been defined in the case of Escherichia coli (19), although most probably, this cannot explain the phenotype that we present in this work. It must be stressed that this phenotype cannot be justified by the effect of the overexpression of DhGZF3 on ENA1, since in this case, one might expect a decrease in tolerance, as deduced from the alkali-sensitive phenotype of ena1 strains (26, 28). In an attempt to identify upregulated or downregulated genes that might plausibly confer alkaline-pH tolerance, we have compared our DNA microarray data with the list of strains sensitive to alkaline pH identified from systematic deletion mutant screens (24, 41). Thus, RIB4, NOP12, and BAP2 are genes whose mutation results in high-pH sensitivity and which appear to be upregulated by DhGZF3 overexpression, while KSP1 and SHE1 (plus YDR290w and YLR170w, encoding hypothetical proteins) are repressed by high DhGZF3 levels. However, the diverse functions of these genes do not provide a reasonable explanation for the tolerance observed in cells overexpressing DhGZF3. In contrast, the link between DhGzf3 and high-pH tolerance established in the present work can be reasonability connected with the identification of a ure2 mutant as being sensitive to high pH by the above-mentioned screens (24, 41). This phenotype is confirmed here in a different genetic background, and moreover, we show that a double gln3 gat1 mutant displays hypertolerance to alkali, similar to what is observed with the overexpression of DhGZF3. Therefore, the identification of DhGZF3 in our screen most likely reflects the function of this gene product as a negative GATA factor and emphasizes the possible relevance of this type of protein in the mechanisms of the response to high-pH stress in S. cerevisiae.
Supplementary Material
Acknowledgments
We thank J. L. Crespo for strains, suggestions, and critical reading of the manuscript. The excellent technical assistance of Anna Vilalta and María Jesús Álvarez is acknowledged.
This work was supported by grants BMC2002-04011-C05-04 and BFU2005-06388-C4-04-BMC to J.A.; BFU2004-00014 to A.C.; BMC2002-04011-C05-02, HP2003-0050, and BFU2005-06388-C4-03-BMC to J.R. (Ministerio de Educación y Ciencia, Spain, and Fondo Europeo de Desarrollo Regional); and POCTI 2000/BIO/32749 (Fundação para a Ciência e a Tecnologia) to M.C.L.-D. J.A. is the recipient of an Ajut de Suport a les Activitats dels Grups de Recerca (2005SGR-00542, Generalitat de Catalunya). A.R. was the recipient of a fellowship from the Generalitat de Catalunya, Spain. A.G. is the recipient of a fellowship from the Spanish Ministry of Education and Science. C.P. is the recipient of a fellowship funded by FCT, Portugal.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Aggarwal, M., P. K. Bansal, and A. K. Mondal. 2005. Molecular cloning and biochemical characterization of a 3′(2′),5′-bisphosphate nucleotidase from Debaryomyces hansenii. Yeast 22:457-470. [DOI] [PubMed] [Google Scholar]
- 3.Alberola, T. M., J. Garcia-Martinez, O. Antunez, L. Viladevall, A. Barcelo, J. Arino, and J. E. Perez-Ortin. 2004. A new set of DNA macrochips for the yeast Saccharomyces cerevisiae: features and uses. Int. Microbiol. 7:199-206. [PubMed] [Google Scholar]
- 4.Almagro, A., C. Prista, B. Benito, M. C. Loureiro-Dias, and J. Ramos. 2001. Cloning and expression of two genes coding for sodium pumps in the salt-tolerant yeast Debaryomyces hansenii. J. Bacteriol. 183:3251-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bansal, P. K., and A. K. Mondal. 2000. Isolation and sequence of the HOG1 homologue from Debaryomyces hansenii by complementation of the hog1Delta strain of Saccharomyces cerevisiae. Yeast 16:81-88. [DOI] [PubMed] [Google Scholar]
- 6.Banuelos, M. A., F. J. Quintero, and A. Rodriguez-Navarro. 1995. Functional expression of the ENA1(PMR2)-ATPase of Saccharomyces cerevisiae in Schizosaccharomyces pombe. Biochim. Biophys. Acta 1229:233-238. [DOI] [PubMed] [Google Scholar]
- 7.Banuelos, M. A., H. Sychrova, C. Bleykasten-Grosshans, J. L. Souciet, and S. Potier. 1998. The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 144:2749-2758. [DOI] [PubMed] [Google Scholar]
- 8.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 9.Bertram, P. G., J. H. Choi, J. Carvalho, T.-F. Chan, W. Ai, and X. F. S. Zheng. 2002. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol. Cell. Biol. 22:1246-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitling, R., O. Sharif, M. L. Hartman, and S. K. Krisans. 2002. Loss of compartmentalization causes misregulation of lysine biosynthesis in peroxisome-deficient yeast cells. Eukaryot. Cell 1:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunelli, J. P., and M. L. Pall. 1993. A series of yeast shuttle vectors for expression of cDNAs and other DNA sequences. Yeast 9:1299-1308. [DOI] [PubMed] [Google Scholar]
- 12.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffman, J. A., R. Rai, D. M. Loprete, T. Cunningham, V. Svetlov, and T. G. Cooper. 1997. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J. Bacteriol. 179:3416-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper, T. G. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26:223-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coornaert, D., S. Vissers, B. Andre, and M. Grenson. 1992. The UGA43 negative regulatory gene of Saccharomyces cerevisiae contains both a GATA-1 type zinc finger and a putative leucine zipper. Curr. Genet. 21:301-307. [DOI] [PubMed] [Google Scholar]
- 16.Crespo, J. L., K. Daicho, T. Ushimaru, and M. N. Hall. 2001. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 276:34441-34444. [DOI] [PubMed] [Google Scholar]
- 17.Crespo, J. L., S. B. Helliwell, C. Wiederkehr, P. Demougin, B. Fowler, M. Primig, and M. N. Hall. 2004. NPR1 kinase and RSP5-BUL1/2 ubiquitin ligase control GLN3-dependent transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279:37512-37517. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham, T. S., and T. G. Cooper. 1993. The Saccharomyces cerevisiae DAL80 repressor protein binds to multiple copies of GATAA-containing sequences (URSGATA). J. Bacteriol. 175:5851-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dell, C. L., M. N. Neely, and E. R. Olson. 1994. Altered pH and lysine signalling mutants of cadC, a gene encoding a membrane-bound transcriptional activator of the Escherichia coli cadBA operon. Mol. Microbiol. 14:7-16. [DOI] [PubMed] [Google Scholar]
- 20.de Nadal, E., F. Calero, J. Ramos, and J. Arino. 1999. Biochemical and genetic analyses of the role of yeast casein kinase 2 in salt tolerance. J. Bacteriol. 181:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 22.Feller, A., E. Dubois, F. Ramos, and A. Pierard. 1994. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol. Cell. Biol. 14:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garciadeblas, B., F. Rubio, F. J. Quintero, M. A. Banuelos, R. Haro, and A. Rodriguez-Navarro. 1993. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol. Gen. Genet. 236:363-368. [DOI] [PubMed] [Google Scholar]
- 24.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 25.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 26.Haro, R., B. Garciadeblas, and A. Rodriguez-Navarro. 1991. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 291:189-191. [DOI] [PubMed] [Google Scholar]
- 27.Kuruvilla, F. G., A. F. Shamji, and S. L. Schreiber. 2001. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc. Natl. Acad. Sci. USA 98:7283-7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb, T. M., W. Xu, A. Diamond, and A. P. Mitchell. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276:1850-1856. [DOI] [PubMed] [Google Scholar]
- 29.Lepingle, A., S. Casaregola, C. Neuveglise, E. Bon, H. Nguyen, F. Artiguenave, P. Wincker, and C. Gaillardin. 2000. Genomic exploration of the hemiascomycetous yeasts. 14. Debaryomyces hansenii var. hansenii. FEBS Lett. 487:82-86. [DOI] [PubMed] [Google Scholar]
- 30.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magasanik, B., and C. A. Kaiser. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1-18. [DOI] [PubMed] [Google Scholar]
- 32.Nass, R., K. W. Cunningham, and R. Rao. 1997. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. Insights into mechanisms of sodium tolerance. J. Biol. Chem. 272:26145-26152. [DOI] [PubMed] [Google Scholar]
- 33.Neves, M. L., R. P. Oliveira, and C. M. Lucas. 1997. Metabolic flux response to salt-induced stress in the halotolerant yeast Debaryomyces hansenii. Microbiology 143:1133-1139. [DOI] [PubMed] [Google Scholar]
- 34.Norkrans, B., and A. Kylin. 1969. Regulation of the potassium to sodium ratio and of the osmotic potential in relation to salt tolerance in yeasts. J. Bacteriol. 100:836-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Posas, F., M. Camps, and J. Arino. 1995. The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J. Biol. Chem. 270:13036-13041. [DOI] [PubMed] [Google Scholar]
- 36.Prista, C., A. Almagro, M. C. Loureiro-Dias, and J. Ramos. 1997. Physiological basis for the high salt tolerance of Debaryomyces hansenii. Appl. Environ. Microbiol. 63:4005-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prista, C., M. C. Loureiro-Dias, V. Montiel, R. Garcia, and J. Ramos. 2005. Mechanisms underlying the halotolerant way of Debaryomyces hansenii. FEMS Yeast Res. 5:693-701. [DOI] [PubMed] [Google Scholar]
- 38.Prista, C., A. Soeiro, P. Vesely, A. Almagro, J. Ramos, and M. C. Loureiro-Dias. 2002. Genes from Debaryomyces hansenii increase salt tolerance in Saccharomyces cerevisiae W303. FEMS Yeast Res. 2:151-157. [DOI] [PubMed] [Google Scholar]
- 39.Ramos, F., E. Dubois, and A. Pierard. 1988. Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Evidence for a regulatory role of gene LYS14. Eur. J. Biochem. 171:171-176. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz, A., L. Yenush, and J. Ariño. 2003. Regulation of ENA1 Na+-ATPase gene expression by the Ppz1 protein phosphatase is mediated by the calcineurin pathway. Eukaryot. Cell 2:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serrano, R., D. Bernal, E. Simon, and J. Arino. 2004. Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 279:19698-19704. [DOI] [PubMed] [Google Scholar]
- 42.Serrano, R., A. Ruiz, D. Bernal, J. R. Chambers, and J. Arino. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46:1319-1333. [DOI] [PubMed] [Google Scholar]
- 43.Soussi-Boudekou, S., S. Vissers, A. Urrestarazu, J. C. Jauniaux, and B. Andre. 1997. Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol. Microbiol. 23:1157-1168. [DOI] [PubMed] [Google Scholar]
- 44.Svetlov, V. V., and T. G. Cooper. 1998. The Saccharomyces cerevisiae GATA factors Dal80p and Deh1p can form homo- and heterodimeric complexes. J. Bacteriol. 180:5682-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 46.Vizeacoumar, F. J., J. C. Torres-Guzman, D. Bouard, J. D. Aitchison, and R. A. Rachubinski. 2004. Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 15:665-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.