Abstract
The var genes encode Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) proteins, a set of highly diverse surface-expressed proteins that mediate adhesion of erythrocytes infected with asexual blood-stage parasites to host endothelium. Switching among expressed PfEMP1 variants in the course of a blood-stage infection is a key component of antigenic variation, and thus immune evasion, by the parasite. The majority of var loci are found in the subtelomeric regions of P. falciparum chromosomes associated with members of other multigene families, including stevor. Both PfEMP1 and STEVOR are expressed in gametocytes, the transmissible parasite stage, but the role of these proteins in the biology of sexual-stage parasites remains unknown. PfEMP1 may continue to mediate antigenic variation in gametocytes, which need to persist in the host for many days before reaching maturity. Using quantitative reverse transcription-PCR and Northern hybridization, we demonstrate that transcription of a defined subset of type C var loci occurs during gametocyte development in vitro. This transcriptional program occurs in gametocytes regardless of the var expression phenotype of their asexual progenitors and therefore is subject to regulatory processes distinct from those that manage antigenic variation in the asexual parasite. In contrast, the same stevor variants are transcribed in both gametocytes and their asexual progenitors. We also provide evidence that for both asexual parasites and gametocytes, var and stevor transcription patterns are not linked to each other.
Plasmodium falciparum gametocytes take 6 to 12 days to reach a mature state that is infective to the mosquito. It is not known whether gametocytes evade the host immune system during this lengthy period of development by employing the strategy of antigenic variation, which characterizes the 48-hour asexual replicative stages. During their development, immature gametocytes sequester in host tissues such as the bone marrow and spleen (27). Whereas a number of host receptors have been implicated in gametocyte adhesion (4, 25), the parasite ligands that mediate this interaction are not known and could be important targets for transmission-blocking immunity. By analogy with adhesion of asexual parasite-infected erythrocytes, these ligands may be variants of the P. falciparum erythrocyte membrane protein 1 (PfEMP1) family of surface adhesins, which are implicated in the severity of disease presentation (2, 12) and also mediate antigenic variation (24, 29). Alternatively, the gametocyte ligands may be either STEVOR variants, known to be expressed in gametocytes and associated with the infected erythrocyte membrane (22), or other, as yet unidentified molecules.
PfEMP1 is a family of clonally variant proteins encoded by approximately 60 var genes per genome. PfEMP1 proteins are known to mediate cytoadhesion of the mature asexual stages and have been demonstrated to bind to a wide array of host receptors (reviewed in reference 16). In both early gametocytes (stages I to IIA) and mature asexual stages, PfEMP1 is localized to knob structures on the surface of the infected erythrocyte. However, in gametocyte stages IIB to V, knob structures are no longer present on the erythrocyte membrane, and PfEMP1 expression appears to be confined to the parasite cytoplasm (4, 10). var transcripts have been detected in both early and late gametocytes in vitro, and RNA dot-blotting studies suggested that the variants transcribed are identical to those expressed by asexual parasite cultures (10).
STEVOR proteins, like PfEMP1, are encoded by a family of subtelomeric genes and are localized in the Maurer's clefts of erythrocytes infected with mature asexual parasites (13). In early gametocytes, STEVOR variants are localized within the parasite cytoplasm, but during gametocyte maturation, STEVOR crosses the parasitophorous vacuole membrane and traffics through the erythrocyte cytoplasm to the plasma membrane, where it is localized in mature gametocytes (22). This process is independent of Maurer's clefts, which are absent from all but the earliest gametocytes. The pattern of STEVOR localization and trafficking in gametocytes is thus distinct from that observed in mature schizonts (13). The role of STEVOR in these two stages of the life cycle is unclear, but STEVOR proteins are also present in sporozoites, suggesting a diversity of functions (22).
Studies of var and stevor expression to date have relied on reagents that recognize conserved features of the two gene families, and thus there are few data on the contributions of specific variants to different life cycle stages. Subgroupings of the var gene family based upon differences in gene structure, chromosomal organization, and untranslated region (UTR) sequence have been proposed (8, 14, 19), and the advent of variant-specific reagents for transcriptional analysis is beginning to reveal the biological significance of these groups. For example, severe malaria has been linked to the upregulation of a specific var subset (12), and malaria during pregnancy has been linked to the upregulation of a single var gene (26). It should therefore be possible to develop a similar set of variant-specific reagents for detailed examination of stevor transcription profiles.
In this study, we used variant-specific quantitative reverse transcription-PCR (qRT-PCR) to investigate the mRNA level of each member of the var and stevor gene families in P. falciparum gametocytes and their asexual progenitors. We set out to test two hypotheses. Firstly, we hypothesized that distinct subgroups of var and stevor genes encoding proteins with stage-specific functions are expressed in gametocytes. Such var and stevor subgroups, should they exist, may be coordinately regulated during development, as observed for distinct multigene families in other developmental systems (30). Secondly, we hypothesized that if stage-specific subgroups of these gene families are expressed in gametocytes, then they will be regulated independently of the variant phenotype displayed by progenitor asexual parasites.
MATERIALS AND METHODS
Parasite strains and asexual parasite culture.
All gametocytes were derived from asexual parasite cultures of the lowest feasible passage number. P. falciparum clone 3D7A parasites were grown from a stabilate received in 1999 directly from David Walliker at the University of Edinburgh (33) and expanded in culture once to generate a large number of vials for cryopreservation. The experimentally derived lines 3D7-Dodowa1, derived from 3D7 by selection with immunoglobulin G (IgG) from semi-immune Ghanian children (28), and 3D7-trHBMEC, derived from 3D7 by repeated panning on the human bone marrow endothelial cell line trHBMEC (12), are both of recent origin, and gametocytes were grown from low-passage stabilates frozen immediately after the first expansion of these lines.
Malaria parasites were cultured using standard protocols (6, 32). Incomplete culture medium consisted of modified RPMI 1640 (with 25 mM HEPES and l-glutamine; Gibco) supplemented with 0.005% hypoxanthine. Before use, the medium was made complete by the addition of pooled AB+ human serum to a final concentration of 10%. Incubation was done at 37°C under a gas phase of 3% O2, 4% CO2, and 93% N2 (BOC Gases). Cultures were synchronized by incubation in 5% d-sorbitol (Sigma) (18). In order to minimize gametocyte production and parasite stress, asexual cultures were kept between 1 and 5% parasitemia at 3% hematocrit.
Parasites used for qRT-PCR analysis were taken at different time points from the same culture. Using the notation stated below, the ring stages examined were harvested on day −2, and the trophozoites were harvested on day −1 from cultures not stimulated for gametocyte production. Ring-stage parasites used for Northern blotting were harvested after incubation with d-sorbitol, and trophozoites were harvested 25 h after the second of two d-sorbitol treatments, which were 33 h apart.
Induction of gametocytogenesis.
Prior to induction of gametocyte development, early passages of P. falciparum clone 3D7a underwent two rounds of synchronization at the ring stage as described above. Synchronized sexual development was stimulated by a sudden increase in hematocrit of a fast-growing ring-stage culture in the presence of partially spent medium (5, 34). Briefly, the partially spent medium was removed from a ring-stage culture at 6 to 8% parasitemia and replaced with fresh medium at a ratio of 2:3 (vol/vol). The final culture flask volumes were ∼50 ml each, with a hematocrit of ∼3%. Without removing the spent medium, each flask culture was split into five flasks ∼32 h later, and fresh complete medium and blood were added to give each culture a final hematocrit of 3.5 to 4.0% in 30 to 40 ml complete medium. The cultures were then incubated with shaking overnight. On the following day, the cultures consisted predominantly of ring-stage parasites, and this time point was denoted day 0.
For the isolation of late-stage gametocytes (stage III or later), the medium was supplemented with N-acetylglucosamine (50 mM) on day 0, and incubation was continued for 5 days to eliminate the asexual stages (23). For the isolation of early-stage gametocytes, the cultures were allowed to undergo another round of invasion with fresh red blood cells and were harvested on day 2, when the culture consisted of early 3D7 gametocyte stages I and II and the majority of asexual parasites were at ring stage. Gametocytes were magnetically separated from uninfected red blood cells and ring-stage parasites by passage through CS magnetic affinity columns (MACs; Miltenyi Biotech, Germany) in incomplete medium supplemented with Albumax type II (Gibco) to a final concentration of 0.5%. Gametocytes containing greater amounts of hemozoin were retained on the magnetic column, while the ring stages and erythrocytes passed through due to iron in these infected erythrocytes being mainly in the form of heme, which is not attracted to magnetic fields (37). The column was thoroughly washed and separated from the magnet, and the retained cells were eluted as an enriched, positively selected early-stage gametocyte cell fraction.
Gametocyte developmental stages were classified according to the method of Carter and Miller (3). The subdivision of stage II gametocytes into the IIA and IIB classes followed the method of Ponnudurai et al. (23).
RNA isolation and RT-PCR.
Parasites were harvested in TRI reagent (Sigma), and total RNA was extracted according to the manufacturer's instructions. Any contaminating genomic DNA was removed by digestion with DNase I (Sigma) before ∼2 μg of RNA was reverse transcribed from random hexamers, using Superscript II (Invitrogen) following the manufacturer's protocol.
Primer design.
The var primer pairs and the control gene seryl-tRNA synthetase primers described previously by Salanti et al. (26) were used, with the exception of var primer pair 58. Primers (Table 1) specific to 37 of the 40 stevor genes identified in the 3D7 genome databases (http://www.plasmodb.org and http://www.genedb.org) and to the var genes PFD0625c, PFD1015c, and PF07_0051 were designed using PRIMER3 (http://www.broad.mit.edu/cgi-bin/primer/primer3.cgi). All primers were manufactured by MWG Biotech.
TABLE 1.
Primer pairs designed to amplify each of 37 stevor genes and 3 var genes
| Primer set | Primer sequence
|
Target gene(s) | |
|---|---|---|---|
| Forward | Reverse | ||
| 1 | TGATGCCCTTGCTAGTTATGC | CAGAAGTTGCAGCAGGTACTGT | PFA0090c |
| 3 | CAGCTATTCAAGCAGGTGCTAA | CAATACCACAACCTCCAGGAA | PFA0705c |
| 5 | GGTTTGGCAAAGGCAAAATA | CAAGCTGAACTACCAGCTTCAA | PFA0105w |
| 7 | GGCTGCCTTTGATACCTTGA | TGTACCGCCTGCAAACATAG | PFA0750w |
| 9 | GCAAAATCTGCTGCCCTTAC | ACCATTTGCACAGGTTCCAC | PFB0065w |
| 11 | AAGGCGCTTGCTGGTATCTA | CTCGCAATACTAGCCATTCTCA | PFB0995w |
| 15 | TGCTGCTGTCACTTCTAGCTTT | CTACACCTCCTGCTGCAGTAAC | PFB1020w |
| 18 | AAAGGTGCTGCTATTTCTACCG | CAGCAGCACAACCTACTGTACC | PFC0025c |
| 20 | TGCATGCTGCTAAAGTTGCT | TTGGTTGCACAAAGAACTGACT | PFC1105w |
| 22 | AAATGCGCATCCTCTATCACT | AGTTCCGACCCCATCACTAA | PFD0065w |
| 24 | TTTATGGTATTAGCGCTGCAAG | GCGCTTTCTATTGATGTTTGAG | PFD0125c |
| 26 | TAGTTGTGTGTGCCCTTTCG | TTCGTCAATTGGGATGATGA | PFD0220c |
| 28 | TGGCAGCTACCAAAGCTACA | TCAGGACCCCAAGCTGTAATA | PFD0035c |
| 30 | GCAAAGGTTCTTGCTGGTGT | GCTATTCTCATAGCATCCCAATG | PFE0030c |
| 34 | GTTGATGCCATCCTTCCTGT | ACGCTTCGGTTGCTTTAAGA | PFF1550w |
| 36 | GGCTGCACTTGCTTACTTTTC | TAGCACATGTTGCACCTCCT | PFI0080w |
| 41 | TGGGGCTCCTACATTAAGTCA | CACGACAATTGCCATACCAT | PF10_0009 |
| 43 | AGGTGCTCTTGCTGAGTATGC | CAAATTTCAGTGCCTGCTTG | PF10_0395 |
| 45 | TGACGTTGCTGCCTTGAATA | ACCAACGCCTGCTTGAATAG | PF11_0516 |
| 47 | AGAACGTTGTGTTGGAGGTGT | GGAAGCCATAAACCAACAGG | PF11_0013 |
| 51 | GCCGTTGCTTCTCTTGTTTATT | ATCAGTAGCACCGGCAAGAG | PF13_0009 |
| 53 | GCAAAAGTTGCTGTCATTGG | GAGCCTGCTGCATTAACTGA | PF14_0767 |
| 55 | CATGAAGGCTGTTGCTGATTAT | ATCAGTAGCACCCTCAACACAT | PF14_0771 |
| 57 | TACGTGCGCATCCCATATTA | TTCCGACTAAATCACCAGCA | MAL7P1.218 |
| 59 | GCAAAAGCTGCTATCCTTGG | ACACCACCTGATGCACTAACTG | MAL8P1.217 |
| 61 | CTGCTAAAACGTGTGCGTCT | ATTTGCAGCAGGCAAAACTA | MAL13P1.505 |
| 63 | AAGTGAGATGTTCCCGTGGT | TGGACAGTAAAGTGCGCAAG | MAL7P1.223 |
| 65 | AATTGTCCCAAACCCTTGTC | AGCCTTCGTAGCCATTGAAC | MAL7P1.310 |
| 67 | TTGCAGAAGTGCCTAAGAATTG | GCACTAACAGTAGTTGCTTCTCCT | PF07_0130 |
| 69 | TGCGTGCAAATCCTCTATCA | AATTGCACCAGCAGAAGTTG | PFL2610w |
| 71 | CTGCTGCCATTGCTACCTTT | AGCAGCACCAGATGCACATA | PFB0050c |
| 73 | CTTCCGTTGGACCACCTTAT | CTTGCAGCAGTTCCGGTTAT | PF14_0007 |
| 77 | GCTAAAACGGCTGCCCTAA | CACCACTTGCACAAAATCCA | PFL2620w/PFL2635w |
| 79 | CTGCACCTTATGATGCTTGTGT | AAACAGTTCCGGCCATATCA | PFF0850c |
| 80 | TTGCAGAAGTGCCTAAGAATTG | CATAGCAGCACTTGCAGCAG | PFI0045c |
| 83 | GCCCTAAGTGCTGTTGCTTC | CACCAACAACACATGCTTTCA | PFB0025c |
| A | TTGTCCGCAAAACGATACAA | TGGAGGGACTTTTTCGTCTG | PFD0625c (CIDR2 non-α) |
| B | GTAGCGTCCTCGGTGAAGAG | TGCTGTTACCCACCAAAATG | PFD1015c (DBL2δ) |
| C | GAGCTGCGAGGTCAGAAAAT | AAGGGGTATCACACGCAGTT | PF07_0051 (CIDR2 non-α) |
DNA cloning.
Primers specific to each stevor variant were used to amplify P. falciparum genomic DNA. PCR products were purified with a QIAquick PCR purification kit (QIAGEN) and ligated into the pGEM-T Easy vector (Promega). Plasmids from at least five independent clones were purified using a QIAprep Spin miniprep kit (QIAGEN) and sequenced to confirm the specificity of each primer pair (all primer pairs except for pair 83 were tested).
RT-PCR and quantification.
Real-time RT-PCR was performed on a Rotorgene 2000 thermal cycler (Corbett Research), using QuantiTect SYBR green PCR master mix (QIAGEN) with primers at 2 μM as previously described (26). The housekeeping gene seryl-tRNase synthetase was used for normalization, as previously described (26). Transcript levels of seryl-tRNase synthetase are relatively similar in trophozoites and gametocyte stages but are up to twofold lower in ring stages (21). We have not corrected for this bias here, and thus any comparisons between life cycle stages may overestimate the proportion of transcripts from ring stages.
To validate the 36 stevor primer pairs, they were first tested on genomic DNA using qRT-PCR, and the median cycle threshold (CT) value was calculated. All primer pairs had CT values within 1.5 cycles of the median CT value, a criterion that has been used previously (26). The melting curves obtained for each primer pair confirmed that they were specific, and the PCR products obtained produced single bands after electrophoresis and ethidium bromide staining. To further validate the specificity of the primers, stevor sequences were amplified from 3D7 genomic DNA, using each of the 36 qRT-PCR primer pairs. Products were cloned into plasmid vectors, and at least five clones were sequenced per primer pair. Four primer pairs were not specific for their target genes and were redesigned.
Estimates of absolute var and stevor copy numbers were calculated from standard curves by correcting for primer bias as previously described (20). In addition, absolute quantification of PFD0625c, PFD1015c, and PF07_0051 transcripts was done with gene-specific standard curves. Triplicate real-time measurements were made for the var primer pairs 27, 34, and 52, the seryl-tRNA synthetase primers (26), and var primers A, B, and C (Table 1) on a dilution series of genomic DNA, and a best-fit standard curve was generated. The standard curve spanned concentrations from 3.1 μg ml−1 to 31 pg ml−1. Correlation coefficients for these primer pairs ranged from 0.99097 to 0.99950. Absolute transcript levels were calculated from these standard curves and normalized by levels of the seryl-tRNase synthetase gene.
Northern blotting and hybridization.
Northern blotting and hybridization were performed as described previously (15), with the exception that blocking reagents, 1% bovine serum albumin, and 100 μg/ml salmon sperm DNA (Gibco) were included in the prehybridization and hybridization buffers. Approximately 10 μg of RNA was loaded for each parasite stage. Northern blots were washed to high stringency at 65°C with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate.
The cloned products of var primers 27 and 52 (26) and pfs16 (LM15PH1, GCGCTCGAGATCAATATTCGAAAGTTCATACC; LM16PH1, GCGAGATCTTAAGAATCATCTACTTAGTCTCC) were amplified by PCR. resa was amplified from 3D7 genomic DNA (ss15L, AGTCAACTTTATGAATCCATCC; ss15R, TTTAGCGACATGTTTGAATATC). Products were purified using a QIAquick kit (QIAGEN) and labeled using a Rediprime II kit (Amersham Biosciences). Both var probes were first tested on Southern blots of P. falciparum chromosomes separated by pulsed-field gel electrophoresis to ensure that they were specific to their target genes. Each hybridized to a single chromosome, as predicted (data not shown).
RESULTS
Gametocytes exhibit a stable var transcript profile unlinked to the phenotype of asexual progenitors.
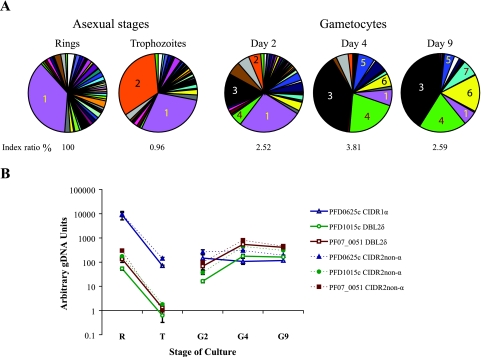
RNAs were taken from 3D7a gametocyte cultures on days 2, 4, and 9, and the proportions of different parasite stages present on each of those days are listed in Table 2. We used primers specific to each var variant in qRT-PCR to compare the level of each var transcript in gametocytes with that in their asexual progenitors. In the ring stages, all 58 var genes examined were transcribed. However, one gene, PFD0625c, accounted for more than one-third of var transcripts (Fig. 1A). Compared to that in the ring stages, var transcript levels decreased dramatically in the trophozoite and gametocyte stages. Total var transcript abundance in trophozoites was just 1.0% of the level in ring stages, and the only gene not down-regulated was PFE1640w (data not shown). This gene was previously shown to be constitutively expressed throughout asexual development (17, 35). The peak transcript level in gametocytes was 3.8% of the level in ring stages (data not shown) and occurred on day 4, when the majority of gametocytes were in stage III (Table 2). Two genes, PF07_0051 and PFD1015c, were the dominant transcripts in the day 4 gametocyte cultures (Fig. 1A), comprising ∼39% and ∼21% of total var transcript levels, respectively. PFD1015c was independently identified as a gametocyte-transcribed var gene by sequence analysis of RT-PCR products from 3D7 gametocyte cultures (data not shown). Transcript abundance of the majority of var genes was lower in gametocytes than in ring stages. However, on days 4 and 9, the transcript levels of PF07_0051 and PFD1015c were the same or higher than those in ring-stage trophozoites (Fig. 1B). Experiments using variant-specific primer pairs located in the CIDR2 non-α domain of each gene, which is proximal to the 3′ splice site of exon I, and experiments with variant-specific primers located further toward the 5′ end provided similar measured patterns of transcription (Fig. 1B). This was true for both PF07_0051 and PFD1015c. Differences between transcript levels in asexual- and sexual-stage parasites were statistically significant irrespective of which primer set was used for either gene (Table 3).
TABLE 2.
Proportions of parasite stages in gametocyte cultures used in this study
| Culture day | % of parasitesa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Asexual stages
|
Gametocyte stages
|
Miscellaneous | |||||||
| Rings | Trophozoites | I | IIa | IIb | III | IV | V | ||
| 2 | 0.4 ± 0.4 | 5.7 ± 3.0 | 2.5 ± 0.6 | 66.8 ± 15.9 | 16.9 ± 10.0 | 2.6 ± 1.9 | 0.1 ± 0.2 | 0.1 ± 0.1 | 4.7 ± 3.6 |
| 4 | 0.0 ± 0.0 | 5.7 ± 2.7 | 1.0 ± 0.3 | 2.7 ± 3.5 | 14.7 ± 8.0 | 72.1 ± 7.8 | 2.3 ± 1.7 | 0.5 ± 0.5 | 1.1 ± 0.3 |
| 9 | 0.0 ± 0.0 | 2.4 ± 2.5 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.2 | 0.4 ± 0.2 | 5.0 ± 5.2 | 91.7 ± 4.9 | 0.2 ± 0.3 |
Parasite preparations were stained with Giemsa stain, and at least 400 infected erythrocytes were counted. Parasites that could not be identified were counted as miscellaneous. The means and standard deviations for three experiments are shown.
FIG. 1.
Changes in var transcript abundance upon differentiation from asexual to sexual reproduction. (A) Transcription levels were measured by real-time PCR using a set of primers that amplify 58 var genes from clone 3D7. The relative abundance of transcript from each var gene is presented as a proportion of total var transcript abundance in the parasite population tested. An index ratio is given under each chart, indicating the var transcript abundance as a percentage of the estimated total var transcript abundance in ring-stage trophozoites, the stage with peak abundance. Numbered sectors of the pie charts correspond to the following genes: 1, PFD0625c; 2, PFE1640w; 3, PF07_0051; 4, PFD1015c; 5, PFL1955w; 6, PFL1960w; and 7, PF08_0103. (B) Transcript levels of PFD0625c, PFD1015c, and PF07_0051 normalized against levels of the control seryl-tRNA synthetase gene. Triplicate real-time measurements were performed for each cDNA, and the mean values are shown. Similar results were obtained when the experiment was repeated with an independent set of 3D7a gametocyte cultures (data not shown). Error bars represent 1 standard deviation. R, ring stages; T, trophozoite stages; G, gametocyte day 2, 4, or 9 cultures; gDNA, genomic DNA. CIDR1 and DBL2 are domains within PfEMP1 proteins.
TABLE 3.
Relative abundance of transcripts of the loci PF07_0051 and PFD1015c in rings and day 4 gametocytes
| Locus | Primer set | Relative transcript abundance (mean ± SD)a
|
P value | |
|---|---|---|---|---|
| Ring-stage trophozoites | Day 4 gametocytes | |||
| PF07_0051 | 52 | 134.5 ± 37.61 | 541.9 ± 148.2 | 0.035 |
| C | 296.7 ± 4.09 | 797.0 ± 54.40 | 0.004 | |
| PFD1015c | 34 | 174.1 ± 7.83 | 446.6 ± 12.56 | 0.009 |
| B | 54.09 ± 7.75 | 176.3 ± 32.33 | 0.018 | |
Values are arbitrary DNA units calculated from standard curves and normalized against the control gene seryl-tRNA synthetase.
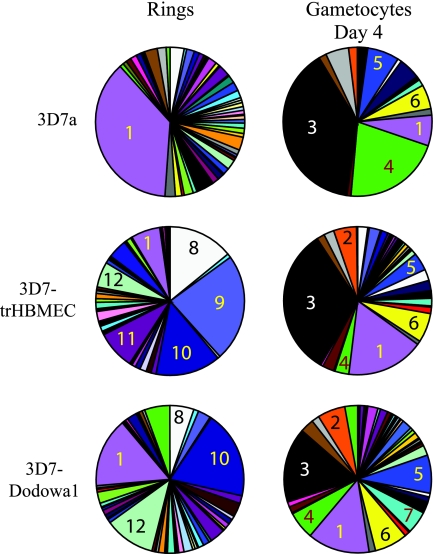
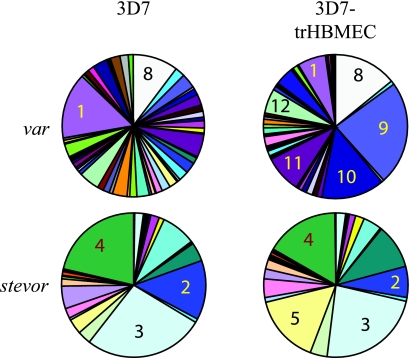
The var transcription profile of the asexual blood stages is dynamic. var genes undergo transcriptional switching, and thus any P. falciparum clone will drift towards a mixed var profile after several generations of in vitro growth. We therefore set out to test whether gametocyte var profiles are determined by the phenotype of their asexual progenitors. Gametocytes were produced from two 3D7 cultures which had been selected to express specific PfEMP1 phenotypes, as previously described (12). These were the 3D7-trHBMEC adhesion-selected line and the 3D7-Dodowa1 line. The ring-stage var transcription profiles of the two selected cultures differed from that of the unselected 3D7 culture and differed from each other in a manner consistent with their previously reported var profiles (Fig. 2) (12). However, gametocytes derived from both selected parasite populations had similar var transcript profiles to those derived from unselected 3D7 parasites, since in all three gametocyte cultures PF07_0051 transcripts were prominent, as well as PFL1960w, PFL1955w, PF08_0103, and PFD0625c transcripts (Fig. 2). PFD1015c transcripts were not as abundant in gametocytes from the selected lines.
FIG. 2.
Change in relative abundance of each var transcript upon differentiation from asexual to sexual reproduction of selected parasite lines with specific phenotypes. Transcript levels were measured by real-time PCR using a set of primers that amplify 58 var genes from clone 3D7. The relative abundance of transcript from each var gene is presented as a proportion of total var transcript abundance in the parasite population tested. Numbered sectors of the pie charts correspond to the following genes: 1, PFD0625c; 2, PFE1640w; 3, PF07_0051; 4, PFD1015c; 5, PFL1955w; 6, PFL1960w; 7, PF08_0103; 8, PF11_0008; 9, PF13_0003; 10, PFD1235w; 11, PF11_0007; and 12, PFF0010w. Transcript levels were measured in an unselected culture of clone 3D7a and in cultures of 3D7 which were either panned on trHBMEC (3D7-trHBMEC) or selected with IgG from semi-immune children (3D7-Dodowa1).
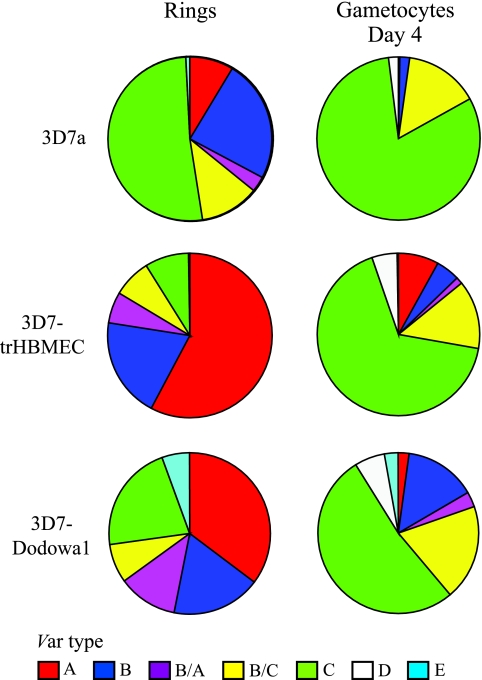
var genes can be divided into seven groups, based upon their upstream and coding sequences, chromosome location, and gene orientation (19). In gametocytes, the most abundant transcripts were those of type C var genes (Fig. 3). Transcripts from type A var genes were not abundant in any of the gametocyte populations tested, even though both the 3D7-Dodowa1 and 3D7-trHBMEC cell lines are characterized by a high prevalence of type A transcripts in ring-stage parasites. Type A genes tend to be located near telomeres, have an UpsA type of 5′ UTR, and tend to be large var genes with complex domain structures. Type C genes, which include PFD0625c, PFD1015c, PF07_0051, and PFL1960w, do not occupy subtelomeric loci and have an UpsC type of 5′ UTR (19). PFL1955w and PF08_0103 have features of both groups B and C, so although they have an UpsB type of 5′ UTR, they are nonsubtelomeric, and at least part of their coding sequence falls into a cluster which mainly consists of group C genes (19).
FIG. 3.
Different var types dominate transcript pools in asexual and sexual parasites. Transcript levels were measured by real-time PCR using a set of primers that amplify 58 var genes from clone 3D7. The relative abundance of transcript from each var gene is presented as a proportion of total var transcript abundance in the parasite population tested. Transcript levels were measured in an unselected culture of clone 3D7a and in cultures of 3D7 which were either panned on trHBMEC (3D7-trHBMEC) or selected with IgG from semi-immune children (3D7-Dodowa1). var genes are arranged according to the groups described by Lavstsen et al. (19).
Full-length var transcripts are present in gametocytes.
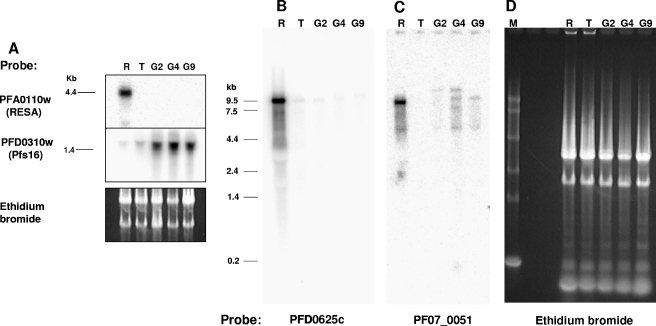
To determine whether var transcripts in gametocytes are full length and not simply products of promiscuous or partial transcription, transcripts of PFD0625c and PF07_0051 were investigated by Northern blotting (Fig. 4B and C). A single PFD0625c transcript of 9.5 kb occurred in ring stages and, although fainter, in trophozoites and the day 2 gametocyte culture. There was also a single full-length transcript of PF07_0051 in ring stages. However, multiple transcripts ranging from 9.5 to 12 kb were detected on the Northern blot by the PF07_0051-specific probe for the day 4 gametocyte culture. It is not clear whether these transcripts represent cross-hybridizing mRNAs or alternatively spliced transcripts of PF07_0051. There was also a faint band of ∼5 kb in both the ring and gametocyte stages, which could be a partial transcript of this gene. The same RNA samples were hybridized with probes to resa and pfs16 as stage-specific controls (Fig. 4A). As expected, transcripts of resa were only present in the ring-stage culture, and high levels of pfs16 transcripts were present in the three gametocyte cultures. pfs16 transcripts were also detected in the ring and trophozoite cultures, which is most likely due to the presence of small numbers of early gametocytes in these cultures (22).
FIG. 4.
Northern blots of ring-stage (R), trophozoite (T), and gametocyte (G) day 2, 4, or 9 cultures. M, 0.24- to 9.5-kb RNA marker. RNAs were size fractionated in either a 1% agarose gel (A) or a 0.8% agarose gel (D). Panels B and C are of the same gel.
The relative abundance of transcripts of PF07_0051 in gametocytes compared to ring stages by Northern blotting (Fig. 4) was lower than that expected from the quantitative RT-PCR results. Since the RNA amounts loaded onto the gel for Northern blotting were normalized by the amount of total RNA present, this discrepancy can be explained if gametocytes have a different ratio of rRNA to mRNA than that of the ring stages. A comparison of estimates of rRNA abundance in ring-stage trophozoites and gametocytes by qRT-PCR supported this explanation (data not shown).
The stevor transcript profile of gametocytes differs from that of their trophozoite progenitors.
Primers specific to each of 37 stevor genes in P. falciparum strain 3D7 were used in qRT-PCR to compare the levels of stevor transcription in gametocytes and their asexual progenitors. After normalization of total transcript levels in gametocytes, it was estimated that stevor mRNA was much less abundant than var mRNA. In asexual parasites, total stevor transcript levels have been shown to peak in mid-stage trophozoites at approximately 28 h postinvasion (13). The level of stevor transcripts in gametocytes was highest in the day 2 culture, when it reached 19% of the level in trophozoite-stage parasites (data not shown). PF10_0395, PFF0850c, and PFD0065w were the dominant transcripts in gametocytes (Fig. 5). However, in contrast to our findings with the var gene family, these stevor transcripts were also present in trophozoites. The major difference between stevor transcription in 3D7 trophozoites and that in gametocytes was the down-regulation of PFI0080w, which accounted for approximately 20% of the stevor transcript pool in trophozoites but was barely detectable (just 2%) in gametocytes. These results suggest that transcription of stevor genes is not coordinated with var expression in developing gametocytes.
FIG. 5.
Changes in stevor transcript upon differentiation from asexual to sexual reproduction. Transcript levels were measured by real-time PCR using a set of primers that amplify 37 stevor genes from clone 3D7. The relative abundance of transcripts from each stevor gene is presented as a proportion of total stevor transcript abundance in either trophozoites or the day 2 gametocyte population. Numbered sectors of the pie charts correspond to the following genes: 1, PFD0065w; 2, PFI0080w; 3, PF10_0395; and 4, PFF0850c.
The stevor transcript profile in trophozoites is unlinked to the var transcript profile.
To test for coordinated transcription of var and stevor genes in asexual parasites, stevor transcription profiles for trophozoites selected for adhesion to trHBMEC were compared with the stevor profiles of their unselected progenitors. Selection for adhesion to bone marrow endothelial cells is not expected to directly impinge on stevor expression, as there is no evidence to date that STEVOR variants are expressed on the surfaces of erythrocytes infected with 3D7 trophozoites and schizonts (13). Specific changes in var transcription did occur in the trHBMEC-selected line (Fig. 6) (12), and therefore indirect changes in stevor transcription may occur if these two families are coregulated due to their shared subtelomeric chromosomal location. However, there was little change in the stevor transcription profile before and after selection (Fig. 6). The only significant change was an increase in transcripts of PF11_0013, which is located on a different chromosome from that carrying the two most upregulated var genes in this line. Therefore, we concluded that there is no coordination of stevor and var transcription in this selected population. stevor transcription also did not change after selection by adhesion to chondroitin sulfate A or by selection with IgG from semi-immune children (3D7-Dodowa1 strain) (data not shown). The proportions of the stevor transcripts present did vary, however, between different unselected parasite lines (Fig. 5 and 6 and data not shown).
FIG. 6.
Levels of stevor transcription in trophozoite-stage parasites either before or after selection on trHBMEC. Transcript levels were measured by real-time PCR using primers specific to each of 37 stevor genes and 58 var genes from clone 3D7. The relative abundance of transcripts from each stevor or var gene is presented as a proportion of total transcript abundance for the relevant gene family in the parasite population tested. Numbered sectors of the pie charts correspond to the following genes: 1, PFD0625c; 8, PF11_0008; 9, PF13_0003; 10, PFD1235w (var genes); 2, PFI0080w; 3, PF10_0395; 4, PFF0850c; and 5, PF11_0013 (stevor genes).
DISCUSSION
In this study, we have quantified the transcriptional profiles of the stevor and var gene families in both the asexual erythrocyte stages and gametocytes of P. falciparum in vitro. The data support two novel conclusions. Firstly, developing P. falciparum gametocytes exhibit a distinct var transcription profile that is independent of the phenotype of their asexual progenitors and consists of nonsubtelomeric type C var genes. Overall, the abundance of var transcripts was lower in cultured gametocytes than in asexual parasites, but transcripts from a number of specific variants were reproducibly found to be more abundant in gametocytes than in asexual-stage parasites. Secondly, transcription of the stevor gene family does not exhibit a distinct profile in gametocytes or in adhesion-selected lines of 3D7 parasites. Therefore, transcription of the var and stevor families is not coordinated in either of these life cycle compartments in the parasite lines studied.
Elucidation of the transcriptional dynamics occurring during sexual-stage parasite development requires reproducible synchronization of gametocyte maturation in vitro. The method described herein is a substantial improvement on previous protocols for stage-specific isolation of P. falciparum gametocytes and is consistently able to produce preparations of each of the classical stages of development with both a high purity and a high yield (Q. L. Fivelman et al., unpublished results).
Earlier studies suggested that the var genes transcribed in early gametocytes (stages I to IIA) of P. falciparum isolate 1776 are also expressed in trophozoites (10). Hayward and colleagues used RT-PCR products (amplified using promiscuous primers) to probe plasmids containing 37 different variant-specific inserts spotted onto nylon membranes. This approach could have missed one-third of the variant repertoire and may have been affected by the primer bias that has dogged previous transcriptional studies of the var family (31). We have taken advantage of the recent elucidation of the full var repertoire in 3D7 (8) to examine 58 var variants in clone 3D7, using qRT-PCR to quantify the dynamics of var transcription in sexual stage development. Our results support the main findings of Hayward et al. in that the most abundant var transcripts in trophozoites were also expressed in the gametocyte sample from day 2. However, quantification revealed that the relative proportions of the transcripts present are quite different between the two stages and that a sexual stage-specific program of var transcription can be defined. It would not have been possible for Hayward et al. to reach this conclusion with the techniques available to them and in the absence of the full genome sequence of the parasite line used.
Quantitative RT-PCR and Northern blotting showed that transcripts of PF07_0051 and PFD1015c were the most abundant var transcripts in day 4 gametocyte cultures and that they were full length. These two transcripts became more abundant as gametocytes matured (Fig. 1), which suggests that they are not products of promiscuous transcription or remnants of asexual-stage transcription. In the asexual stages, var transcription is tightly regulated so that high-level transcription occurs in a brief, 20-h window immediately after red blood cell (RBC) invasion (17). If var transcription in gametocytes is also under tight temporal control, then it is possible that the peak of gametocyte var transcription occurred between the time points analyzed in this study: RNA samples were taken on just 3 of the 9 days from initiation of the gametocyte cultures to maturity. Small differences in the ages of gametocyte populations may have a large impact on transcription levels and thus could explain the small variation observed between two independent experiments (data not shown). However, the difficulty in obtaining highly synchronized gametocyte populations and the long time taken to reach maturity make it difficult to perform a time course that can capture tighter temporal windows of transcription in P. falciparum gametocytes. Furthermore, our measurements were made in bulk parasite cultures, and there is likely to be heterogeneity in var transcription profiles among individual gametocytes.
The occurrence of var transcriptional switching means that the profile of the asexual blood stages is unstable. In vitro experiments have shown that the rate of var switching varies between genes and may be affected by the history of a parasite clone (11). This argues against a simple stochastic process, but there is no evidence to date that var switching in the asexual blood stages follows a preset program, as seen in African trypanosomes. However, we found evidence of a single var transcriptional program in developing gametocytes derived from three phenotypically different asexual populations. This program favors transcription of a subset of type C genes in gametocytes. It can also be envisaged that specific var subgroups may be expressed under particular conditions or in other life cycle compartments, such as ookinetes, oocysts, or sporozoites. Such stage-specific expression has been demonstrated for the P. bergei BIR family, which has been identified in proteomic studies of the erythrocytic, mosquito, and sporozoite stages (9).
What is the function of PfEMP1 in gametocytes? Protein localization suggests a possible cytoadhesion role in early gametocytes, which retain the electron-dense knob structures associated with adhesion in asexual parasites (4). We found that the peak of gametocyte var transcription occurs later, in stage III gametocytes. Evidence that gametocyte var transcripts are translated comes from the identification of PfEMP1 peptides in the proteome of late 3D7 gametocytes by Florens et al. (7). They identified peptides of variants PFD1015c, PFL1955w, and PFL1960w, all of which were found to be prominent transcripts in mid- to late-stage gametocytes (Fig. 1A and 2). Hayward et al. (10) found that PfEMP1 protein expression is limited to the parasite cytoplasm in late-stage gametocytes. However the antiserum used in these experiments was raised against a recombinant polypeptide derived from a single var gene of the Malayan Camp parasite line (1) and may not recognize all PfEMP1 variants. Thus, we cannot rule out the possibility of PfEMP1 being surface expressed in later gametocyte stages and therefore having a role in sequestration. Alternatively, since gametocytes are known to synthesize mRNA (e.g., pfs25/28) and protein (e.g., Pfs230) in preparation for the next stage of development in the mosquito, some var genes transcribed in mid- to late-stage gametocytes may encode PfEMP1 proteins that have a function in the mosquito life cycle compartment and are retained in the parasite cytoplasm until taken up in a blood meal. The possibility that PfEMP1 proteins are expressed in life cycle compartments other than the blood stages is supported by the finding of PfEMP1 tryptic fragments upon proteomic analysis of 3D7 sporozoites (7). Multistage functions have also been postulated for the STEVOR and SURFIN protein families (22, 36).
We found stevor transcript abundance to be highest in early gametocytes, as shown previously (22). We identified three genes, PF10_0395, PFF0850c, and PFD0065w, that dominate the stevor transcript pool in day 2 (stage II) gametocytes. However, in contrast to our findings with the var gene family, the same stevor transcripts were also present in trophozoites, despite the distinct localization of STEVOR protein in asexual- and sexual-stage parasites (22). If these variants are translated in both asexual- and sexual-stage parasites, then the same STEVOR protein may perform different functions in the two life cycle compartments. STEVOR is localized at the infected RBC membrane in gametocytes in stage III (22), which is consistent with an involvement in gametocyte sequestration. In 3D7 trophozoites, STEVOR variants are associated with Maurer's clefts in the erythrocyte cytoplasm, but they have not been shown to localize to the plasma membrane of the infected RBC at any stage in asexual development.
The profile of var transcription during the erythrocytic cycle varies between stages and in response to selection. This does not seem to be the case for stevor; just five variants dominated the transcript pool in each of the parasite lines studied here. The stevor gene family did not respond to changes in adhesion phenotype, which could mean that in contrast to var and rif, they are not clonally variant or that our culture conditions did not favor switching. STEVOR proteins are also present in sporozoites (22), and thus the remaining 35 variants could play a role in this stage or in the mosquito stages. Alternatively, it is possible that transcriptional control of STEVOR expression in cultured 3D7 parasites is deficient in some way and does not accurately reflect the pattern that occurs in vivo.
Members of the var and stevor families tend to cluster together in the genome, which means that coregulation of the two families is possible via mechanisms that take advantage of their close juxtaposition. An analysis of the transcriptional profiles of both families before and after selection did not find any evidence to support this hypothesis. This is, to our knowledge, the first systematic investigation to address the possibility of coordinated expression between two of the subtelomeric multigene families in P. falciparum. Interestingly, the var loci with the most substantial contributions to gametocyte transcript pools, PF07_0051 and PFD1015c, are positioned centrally on chromosomes 7 and 4, respectively, and are not closely linked to stevor genes, almost all of which are subtelomeric. Thus, these genes could not be coordinated with stevor transcription through mechanisms reliant on close proximity between the two families.
In summary, we have described a program of type C var transcription in gametocytes that is unlinked to var transcription in their asexual progenitors. In contrast, a single group of stevor transcripts dominates both life cycle stages. These differences in the expression and regulation of the two families suggest that we cannot understand the functions of the STEVOR family based simply on its subtelomeric proximity to PfEMP1 and other gene families. Rather, variant-specific reagents need to be developed for each family to unravel what may be a complex hierarchy of stage-specific functions.
Acknowledgments
Trine Staalsoe, University of Copenhagen, Denmark, is thanked for providing the 3D7-Dodowa1 parasite line.
S.S. was supported by a studentship from the Medical Research Council, United Kingdom. C.J.S. was supported by the Health Protection Agency, United Kingdom. Q.L.F. and L.M. were supported by Wellcome Trust Grant no. 066742 to D.A.B.
REFERENCES
- 1.Baruch, D. I., B. L. Pasloske, H. D. Singh, B. Xiahui, X. C. Ma, M. Feldman, T. F. Taraschi, and R. J. Howard. 1995. Cloning the Plasmodium falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitised human erythrocytes. Cell 82:77-87. [DOI] [PubMed] [Google Scholar]
- 2.Bull, P. C., M. Berriman, S. Kyes, M. A. Quail, N. Hall, M. M. Kortok, K. Marsh, and C. I. Newbold. 2005. Plasmodium falciparum variant surface antigen expression patterns during malaria. PLoS Pathog. 1:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter, R., and L. H. Miller. 1979. Evidence for environmental modulation of gametocytogenesis in Plasmodium falciparum in continuous culture. Bull. W. H. O. 57:37-52. [PMC free article] [PubMed] [Google Scholar]
- 4.Day, K. P., R. E. Hayward, D. Smith, and J. G. Culvenor. 1998. CD36-dependent adhesion and knob expression of the transmission stages of Plasmodium falciparum is stage specific. Mol. Biochem. Parasitol. 93:167-177. [DOI] [PubMed] [Google Scholar]
- 5.Dyer, M., and K. P. Day. 2003. Regulation of the rate of asexual growth and commitment to sexual development by diffusible factors from in vitro cultures of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 68:403-409. [PubMed] [Google Scholar]
- 6.Fairlamb, A. H., D. C. Warhurst, and W. Peters. 1985. An improved technique for the cultivation of Plasmodium falciparum in vitro without daily medium change. Ann. Trop. Med. Parasitol. 79:379-384. [DOI] [PubMed] [Google Scholar]
- 7.Florens, L., M. P. Washburn, J. D. Raine, R. M. Anthony, M. Grainger, J. D. Haynes, et al. 2002. A proteomic view of the Plasmodium falciparum life cycle. Nature 419:520-526. [DOI] [PubMed] [Google Scholar]
- 8.Gardner, M. J., S. J. Shallom, J. M. Carlton, S. L. Salzberg, V. Nene, A. Shoaibi, et al. 2002. Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14. Nature 419:531-534. [DOI] [PubMed] [Google Scholar]
- 9.Hall, N., M. Karras, J. D. Raine, J. M. Carlton, T. W. Kooij, M. Berriman, et al. 2005. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307:82-86. [DOI] [PubMed] [Google Scholar]
- 10.Hayward, R. E., B. Tiwari, K. P. Piper, D. I. Baruch, and K. P. Day. 1999. Virulence and transmission success of the malarial parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 96:4563-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horrocks, P., R. Pinches, Z. Christodoulou, S. A. Kyes, and C. I. Newbold. 2004. Variable var transition rates underlie antigenic variation in malaria. Proc. Natl. Acad. Sci. USA 101:11129-11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, A. T., P. Magistrado, S. Sharp, L. Joergensen, T. Lavstsen, A. Chiucchiuini, et al. 2004. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J. Exp. Med. 199:1179-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaviratne, M., S. M. Khan, W. Jarra, and P. R. Preiser. 2002. Small variant STEVOR antigen is uniquely located within Maurer's clefts in Plasmodium falciparum-infected red blood cells. Eukaryot. Cell 1:926-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraemer, S. M., and J. D. Smith. 2003. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol. Microbiol. 50:1527-1538. [DOI] [PubMed] [Google Scholar]
- 15.Kyes, S., R. Pinches, and C. Newbold. 2000. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol. Biochem. Parasitol. 105:311-315. [DOI] [PubMed] [Google Scholar]
- 16.Kyes, S., P. Horrocks, and C. Newbold. 2001. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol. 55:673-707. [DOI] [PubMed] [Google Scholar]
- 17.Kyes, S. A., Z. Christodoulou, A. Raza, P. Horrocks, R. Pinches, J. A. Rowe, and C. I. Newbold. 2003. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol. Microbiol. 48:1339-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambros, C., and J. P. Vanderberg. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418-420. [PubMed] [Google Scholar]
- 19.Lavstsen, T., A. Salanti, A. T. Jensen, D. E. Arnot, T. G. Theander, T. Staalsoe, et al. 2003. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar. J. 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavstsen, T., P. Magistrado, C. C. Hermsen, A. Salanti, A. T. Jensen, R. Sauerwein, et al. 2005. Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malar. J. 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, et al. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503-1508. [DOI] [PubMed] [Google Scholar]
- 22.McRobert, L., P. Preiser, S. Sharp, W. Jarra, M. Kaviratne, M. C. Taylor, et al. 2004. Distinct trafficking and localization of STEVOR proteins in three stages of the Plasmodium falciparum life cycle. Infect. Immun. 72:6597-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponnudurai, T., A. H. Lensen, J. F. Meis, and J. H. Meuwissen. 1986. Synchronization of Plasmodium falciparum gametocytes using an automated suspension culture system. Parasitology 93:263-274. [DOI] [PubMed] [Google Scholar]
- 24.Roberts, D. J., A. G. Craig, A. R. Berendt, R. Pinches, G. Nash, K. Marsh, and C. I. Newbold. 1992. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature 357:689-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers, N. J., B. S. Hall, J. Obiero, G. A. Targett, and C. J. Sutherland. 2000. A model for sequestration of the transmission stages of Plasmodium falciparum: adhesion of gametocyte-infected erythrocytes to human bone marrow cells. Infect. Immun. 68:3455-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salanti, A., T. Staalsoe, T. Lavstsen, A. T. Jensen, M. P. Sowa, D. E. Arnot, et al. 2003. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 49:179-191. [DOI] [PubMed] [Google Scholar]
- 27.Smalley, M. E., S. Abdalla, and J. Brown. 1981. The distribution of Plasmodium falciparum in the peripheral blood and bone marrow of Gambian children. Trans. R. Soc. Trop. Med. Hyg. 75:103-105. [DOI] [PubMed] [Google Scholar]
- 28.Staalsoe, T., M. A. Nielsen, L. S. Vestergaard, A. T. Jensen, T. G. Theander, and L. Hviid. 2003. In vitro selection of Plasmodium falciparum 3D7 for expression of variant surface antigens associated with severe malaria in African children. Parasite Immunol. 25:421-427. [DOI] [PubMed] [Google Scholar]
- 29.Su, X. Z., V. M. Heatwole, S. P. Wertheimer, F. Guinet, J. A. Herrfeldt, D. S. Peterson, et al. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89-100. [DOI] [PubMed] [Google Scholar]
- 30.Sutherland, C. J., V. L. Elsom, M. L. Gordon, S. L. Dunwoodie, and E. C. Hardeman. 1991. Coordination of skeletal muscle gene expression occurs late in mammalian development. Dev. Biol. 146:167-178. [DOI] [PubMed] [Google Scholar]
- 31.Taylor, H. M., S. A. Kyes, D. Harris, N. Kriek, and C. I. Newbold. 2000. A study of var gene transcription in vitro using universal var gene primers. Mol. Biochem. Parasitol. 105:13-23. [DOI] [PubMed] [Google Scholar]
- 32.Trager, W., and J. B. Jensen. 1976. Human malaria parasites in continuous culture. Science 193:673-675. [DOI] [PubMed] [Google Scholar]
- 33.Walliker, D., I. A. Quakyi, T. E. Wellems, T. F. McCutchan, A. Szarfman, W. T. London, et al. 1987. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science 236:1661-1666. [DOI] [PubMed] [Google Scholar]
- 34.Williams, J. L. 1999. Stimulation of Plasmodium falciparum gametocytogenesis by conditioned medium from parasite cultures. Am. J. Trop. Med. Hyg. 60:7-13. [DOI] [PubMed] [Google Scholar]
- 35.Winter, G., Q. Chen, K. Flick, P. Kremsner, V. Fernandez, and M. Wahlgren. 2003. The 3D7var5.2 (var COMMON) type var gene family is commonly expressed in non-placental Plasmodium falciparum malaria. Mol. Biochem. Parasitol. 127:179-191. [DOI] [PubMed] [Google Scholar]
- 36.Winter, G., S. Kawai, M. Haeggstrom, O. Kaneko, A. von Euler, S. Kawazu, D. Palm, V. Fernandez, and M. Wahlgren. 2005. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J. Exp. Med. 201:1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young, J. A., Q. L. Fivelman, P. L. Blair, P. de la Vega, K. G. Le Roch, Y. Zhou, et al. 2005. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 143:67-79. [DOI] [PubMed] [Google Scholar]