Abstract
Mutations in the human HPD gene (encoding 4-hydroxyphenylpyruvic acid dioxygenase) cause hereditary tyrosinemia type 3 (HT3). We deleted the Aspergillus nidulans homologue (hpdA). We showed that the mutant strain is not able to grow in the presence of phenylalanine and that it accumulates increased concentrations of tyrosine and 4-hydroxyphenylpyruvic acid, mimicking the human HT3 phenotype.
Inborn errors of metabolism corresponding to four of the five sequential enzymatic reactions of tyrosine (Tyr) degradation are known (15). Three such diseases manifest hypertyrosinemia (hereditary tyrosinemia type 1 [HT1], HT2, and HT3). The enzyme 4-hydroxyphenylpyruvic acid dioxygenase (4-HPPD) participates in the oxidation of keto acids of Tyr (10). Homogenizate is produced by this enzyme, and the reaction involves decarboxylation, oxidation, and rearrangement. Mutations in the HPD locus are related to two known distinct diseases, hereditary tyrosinemia type 3 and hawkinsinuria, and HPD activity is markedly reduced in the liver of patients with HT1 (2). HT3 is characterized by elevated blood concentrations of Tyr and massive urinary excretion of 4-hydroxyphenylpyruvic acid (4-HPP), 4-hydroxyphenyllactic acid, and hydrophenylacetic acid (3). Clinical symptoms in some patients include mild mental retardation, ataxia, and convulsions, and these symptoms are likely due to the elevation of 4-HPP and Tyr in body fluids. This enzyme has also been used as a target for inhibiting the normal catabolism of Tyr in patients with HT1, preventing the accumulation of the catabolic intermediates maleylacetoacetate and fumarylacetoacetate. In patients with HT1, these catabolic intermediates are converted to the toxic metabolites succinylacetone and succinylacetoacetate, which are responsible for the observed liver and kidney toxicity. Nitisinone, which acts as an analogue of the hydroxyphenylpyruvic acid substrate, is the single drug used for such treatments (20). However, an understanding of the molecular mechanisms leading to the inhibition of 4-HPPD by nitisinone and derivatives may provide new strategies for designing novel 4-HPPD inhibitors.
Taking its considerable metabolic versatility into consideration, Peñalva previously proposed to use the filamentous fungus Aspergillus nidulans as an alternative to animal models for certain aspects of human metabolic diseases (12). Accordingly, other authors have provided a large amount of data about deficiencies not only in the phenylalanine/Tyr pathway but also in the branched-chain amino acid pathways (4, 5, 13, 14). Here, we extend these studies by characterizing the A. nidulans hpdA gene encoding 4-HPPD. We show that the deletion strain is not able to grow in the presence either of phenylalanine (Phe) or Phe plus lactose as a carbon source. Furthermore, as in the human disease, the deletion strain accumulated and secreted increased concentrations of Tyr and 4-HPP in the intracellular environment and into the culture medium.
Strains used in this work were GR5 (pyrG89 wA3 pyroA4) (FGSC A773), HPPD15, and HPPD21 (pyrG89 wA3 pyroA4 ΔhpdA::pyrGAF). Complete medium (yeast agar glucose medium [YAG]) and minimal medium (MM) were described previously by Kafer (7) (medium composition is available under protocols at http://www.fgsc.net). MC (minimal medium without glucose) with either 25 mM phenylalanine (MC+Phe) or 0.05% lactose (MC+Lac) as a single carbon source was also used. DNA manipulations were performed according to the method described previously by Sambrook and Russell (16). DNA fragment probes for Southern blots were labeled with [α-32P]dCTP using the RTS Rad Prime DNA labeling system kit (Invitrogen). In the deletion construction, the Aspergillus fumigatus pyrG gene was amplified from the pCDA21 plasmid (1) and is referred to as the pyrG cassette. The PCR-mediated construction (9) for the hpdA gene consisted of three first PCRs that amplified a 5′- and a 3′-flanking region of hpdA gene and a final fusion PCR. For the DNA fragments containing the gene flanking regions, genomic DNA was used as a template. The 5′-flanking fragment encompassing about 2,000 bp upstream of the ATG start codon was amplified by using the primers HPPD Anid-1 (5′-ATACGCCCTCAACCTCCTT-3′) and HPPD Anid-2 (5′-CTCAGACAGAATCGTGGCTGTGACATTTGCC-3′). The pyrG cassette was amplified by using plasmid pCDA21 as a template with the following primers: HPPD PyrFw′ (5′-GTCACAGCCACGATTCTGTCTGAGAGGAGGC-3′) and HPPD PyrRev′ (5′-AGGAATTCAAGTGAATTCGCCTCAAACAATGC-3′). The 3′-flanking region encompassing about 2,000 bp downstream of the stop codon was amplified by using primers HPPD Anid-3 (5′-TGAGGCGAATTCACTTGAATTCCTTCAGACTATT-3′) and HPPD Anid-4 (5′-GAGCCTGACGAGCCCAAG-3′). The fusion PCR using the three previous DNA fragments as templates was amplified by using primers HPPD Anid-1 and HPPD Anid-4 and yielded an expected PCR fragment of about 5,902 bp. Transformation of A. nidulans strain GR5 (mutant pyrG recipient strain) was performed according to the procedure described previously by Osmani et al. (11) using 5 μg of linear DNA. Transformants were scored for their ability to grow on YAG medium.
RNA extraction, RNase-free DNase treatment, and real-time reverse transcription-PCRs (RT-PCRs) were done as previously described by Semighini et al. (17). The following primers and Lux fluorescent probes (Invitrogen) were used in this work: 5′-ACCCATTCTCGTCGAAATCAAT-3′ and 5′-CGGATATGAGGACTTTGAGACAATC-6-carboxyfluorescein-G-3′ for the hpdA gene and 5′-GCAGAATGTCTCGTCCGAATG-3′ and 5′-CACTTTATGCCGTCGCCGAAAG-6-carboxyfluorescein-G-3′ for the tubC gene) (GenBank accession number M17520).
For high-performance liquid chromatography (HPLC) analysis, cells were grown on MM for 16 h and then transferred to MC containing 25 mM Phe and incubated for 6 h at 37°C. Aliquots of 1 ml of culture supernatants were subjected to solid-phase extraction (SPE) on Octadecyl (C18) SPE columns (Mallinckrodt Baker). Tyr and 4-hydroxyphenylpyruvate were eluted from the SPE cartridges with 1 ml methanol. The eluate was evaporated, the residues were dissolved in 100 μl of the mobile phase (5% methanol-95% acetic acid [0.02 M]), and a 20-μl aliquot was injected into a LiChrospher RP-8 column with a 5-μm particle size (125 by 4 mm; Merck, Germany) coupled to a 4- by 4-mm guard column of the same material. The HPLC system consisted of two LC-10AD solvent pumps, an SLC-10A system controller, a CTO-10AS column oven, a 7125 Rheodyne injector with a 20-μl loop, and a diode array detector (SPD-M10A) set at 274 nm (Shimadzu, Kyoto, Japan). To confirm the analysis of the samples, the same HPLC system was connected to a triple-stage quadrupole mass spectrometer (Micro-mass, Manchester, United Kingdom). Detection was performed by multiple-reaction monitoring (MRM) of the protonated molecular (precursor) ion ([M − H]+) and their corresponding product ions (182→136 and 181→135 for Tyr and 4-hydroxyphenylpyruvate, respectively).
BLAST searches of the recently completed A. nidulans genome sequence (http://www.genome.wi.mit.edu/annotation/fungi/aspergillus/) (6) were used to identify a homologue of the 4-HPPD gene. These searches resulted in the identification of AN1899.1 (GenBank accession number XP_659503; renamed here hpdA). The hpdA gene encodes a predicted 169-amino-acid protein with 87% identity and 95% similarity to the T-cell-reactive protein Coccidioides immitis 4-HPPD homologue (2e-77) (accession number JC4215) (19) and 77% identity and 88% similarity to the Mycosphaerella graminicola 4-HPPD homologue (9e-68) (accession number O42764) (8).
We examined the mRNA accumulation of the hpdA gene in the presence of different carbon sources by using real-time RT-PCR. Wild-type A. nidulans was grown in MM and transferred to MM, MC+Phe, MM+Phe, or MC+Phe+Lac, and RNA was then isolated and analyzed for the expression of the hpdA gene (Table 1). There is very low expression of the hpdA gene in either MM or MM+Phe; however, in MC+Phe, its expression was increased to about 43- to 70-fold over 8 h of growth. Growth in the presence of lactose as a single carbon source was comparable to that in the presence of glucose; i.e., there is a very low level of expression of the hpdA gene (data not shown). However, when A. nidulans is grown in MC+Phe+Lac, hpdA mRNA accumulation increased 20- to 60-fold over 8 h of growth. Taken together, our data indicate that hpdA mRNA accumulation increases in the presence of Phe and that it is substantially decreased in the presence of glucose; however, lactose cannot act as a repressor for hpdA mRNA production when A. nidulans is growing in MC+Phe+Lac.
TABLE 1.
Increases in hpdA mRNA levels in response to different carbon sources
| Treatmenta | Fold increase in mRNA expression ± SDb | Relative fold increase in transcript levelc |
|---|---|---|
| 0 h | 35.0 ± 6.12 | 1 |
| Glucose, 2 h | 0.03 ± 0.002 | 0.0009 |
| Glucose, 4 h | 0.04 ± 0.002 | 0.0010 |
| Glucose, 6 h | 0.05 ± 0.007 | 0.0013 |
| Glucose, 8 h | 7.88 ± 3.14 | 0.2254 |
| Phe, 2 h | 1,553.10 ± 104.85 | 44.42 |
| Phe, 4 h | 1,985.32 ± 104.35 | 56.79 |
| Phe, 6 h | 2,408.05 ± 147.0 | 68.88 |
| Phe, 8 h | 1,559.27 ± 96.0 | 44.60 |
| Phe+Glu, 2 h | 1.82 ± 0.67 | 0.052 |
| Phe+Glu, 4 h | 0.27 ± 0.02 | 0.008 |
| Phe+Glu, 6 h | 2.34 ± 0.26 | 0.067 |
| Phe+Glu, 8 h | 9.26 ± 0.10 | 0.265 |
| Phe+Lac, 2 h | 630.88 ± 104.5 | 18.04 |
| Phe+Lac, 4 h | 1,089.40 ± 18.39 | 31.16 |
| Phe+Lac, 6 h | 1,095.63 ± 3.61 | 31.34 |
| Phe+Lac, 8 h | 2,096.03 ± 44.0 | 59.95 |
Mycelia were grown in MM for 16 h at 37°C and then transferred to either MM (glucose), MC+Phe, MC+Phe+Glu, or MC+Phe+Lac and grown for 2, 4, 6, or 8 h at 37°C. Real-time RT-PCR was the method used to quantify the mRNA. The measured quantity of the hpdA mRNA in each of the treated samples was normalized using the cycle threshold values obtained for the tubC RNA amplifications run in the same plate. The relative quantitation of all the genes and tubulin gene expression was determined by a standard curve (i.e., cycle threshold, values plotted against logarithm of the DNA copy number).
Number of copies of cDNA divided by the number of copies of β-tubulin cDNA. The results are the averages of four sets of experiments. The results were expressed as means ± standard deviations.
The values represent the number of times the genes are expressed compared to the control (0 h) grown for 16 h in MM (represented absolutely as 1.00).
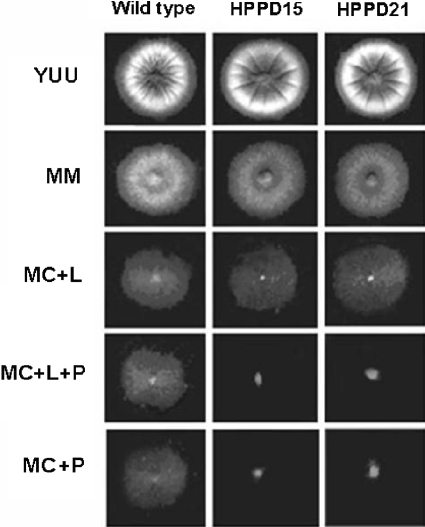
To characterize the function of HpdA, a deletion strain was generated using a fusion PCR-based approach (see above). This 510-bp deletion removes 169 amino acids that encompass the entire open reading frame (data not shown). Allelic replacement of hpdA and single integration of the PCR construct were verified in at least two independent transformants as confirmed by the analysis of Southern blots (data not shown), thereby generating the ΔhpdA strains HPD15 and HPD21. When tested on rich medium (i.e., YAG), the extent of radial colony growth and the production of asexual spores were not affected by the loss of HpdA (Fig. 1). A. nidulans is able to grow on Phe or phenylacetate as the sole carbon source; glucose can repress Phe or phenylacetate utilization, while lactose cannot act as a repressor (4). These strains grew normally on glucose or lactose; however, they were unable to grow on Phe as the sole carbon source (Fig. 1). Interestingly, the deletion strain was also unable to grow on mixtures of a derepressing concentration of lactose and Phe as single carbon sources, suggesting that, analogous to the situation in humans (3, 18), the absence of hpdA directs the possible accumulation of 4-HPP and Tyr as toxic metabolites.
FIG. 1.
Growth phenotypes of the wild-type and ΔhpdA mutant (HPPD15 and HPPD21) strains. Strains GR5 (wild type), HPPD15, and HPPD21 were grown for 72 h at 37°C in YUU, (YAG supplemented with 1.2 g/liter each of uracil and uridine), MM, MC plus lactose (L), MC plus lactose plus phenylalanine (L+P), or MC plus phenylalanine (P). All media except YUU were supplemented with pyridoxin.
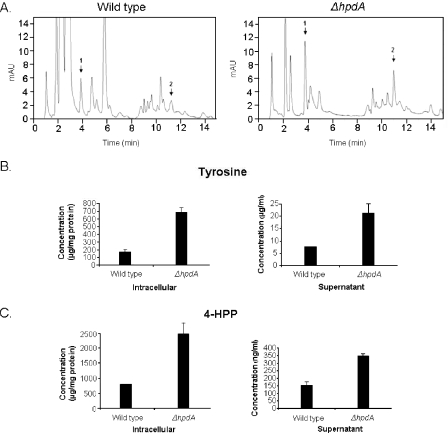
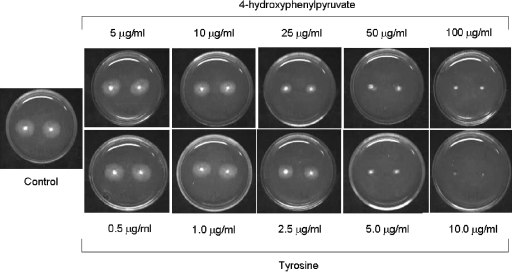
Type 3 tyrosinemia is characterized by elevated blood concentrations of Tyr and massive urinary excretion of 4-HPP, 4-hydroxyphenyllactic, and 4-hydroxyphenylacetic acid (2, 3, 18). Taking into consideration that the growth of A. nidulans ΔhpdA strain was inhibited in MC+Phe+Lac medium, this opened the possibility that 4-HPP and/or 4-HPP metabolites were accumulating as a result of the metabolic block caused by the hpdA deletion. We tested this possible accumulation by applying the mycelium extracts and the supernatants of the wild-type and ΔhpdA mutant strains to HPLC analysis, and we confirmed the identity of the products by mass spectrometry (data not shown). As can be seen in Fig. 2, the ΔhpdA strain secretes about 4.0 and 2.8 times more tyrosine than the wild-type strain in the intracellular environment and in the supernatant, respectively. The mutant strain also secretes more 4-HPP than the wild-type strain (about 3.1 and 2.3 times more in the intracellular environment and in the supernatant, respectively) (Fig. 2). To address the hypothesis that accumulation of Tyr and 4-HPP is enough to inhibit A. nidulans growth, we grew A. nidulans in MC+Lac+Tyr and MC+Lac+4-HPP (Fig. 3). A. nidulans was exposed to increasing concentrations of Tyr and 4-HPP and grown for 48 h at 37°C. A. nidulans growth was inhibited by about 50% at 25 μg/ml of HPP and between 2.5 to 5.0 μg/ml of Tyr and was completely inhibited at 100 μg/ml and 10 μg/ml of 4-HPP and Tyr, respectively. Considering the fact that the ΔhpdA strain secretes about 2.4 mg/mg of protein and 350 ng/ml of 4-HPP (intracellular and supernatant, respectively) and 700 μg/mg of protein and 22 μg/ml of Tyr (intracellular and supernatant, respectively) after 6 h of growth (Fig. 2B and C), these results suggest that the inhibition of the hpdA strain is possibly due to Tyr accumulation.
FIG. 2.
HPLC determination of the Tyr and 4-HPP intracellular and supernatant concentrations. (A) HPLC chromatograms, monitored at 274 nm, of culture supernatants from wild-type and ΔhpdA strains submitted to solid-phase extraction onto Octadecyl (C18) SPE columns. Arrows 1 and 2 represent Tyr and 4-HPP, respectively. The separation was achieved by applying the following gradient with 0.02 M acetic acid (solvent A) and different amounts of HPLC-grade methanol (solvent B) as follows: 3% to 35% solvent B from 0 to 5 min, 35% solvent B from 5 to 12 min, 35% solvent B to 3% solvent B from 12 to 13 min, and 3% solvent B from 13 to 15 min. Standard solutions were also submitted to solid-phase extraction onto Octadecyl (C18) SPE columns (Mallinckrodt Baker) and injected into the C8 column. The intracellular and supernatant concentrations of Tyr (B) and 4-HPP (C) were measured by HPLC determination (A). The concentration of Tyr and 4-hydroxyphenylpyruvate was determined by comparing areas of standards and samples. mAU, milliabsorbance unit.
FIG. 3.
Growth phenotypes of the wild-type strain. Strain GR5 (wild-type strain) was grown for 48 h at 37°C in MC plus lactose with increasing concentrations of either 4-hydroxyphenylpyruvate or Tyr. The control was grown only in MC plus lactose. All the media were supplemented with pyridoxin.
In summary, we identified the A. nidulans hpdA gene that encodes the putative 4-hydroxyphenylpyruvate dioxygenase. This gene is not essential, but growth of the deletion strain is inhibited by Phe. Although the deletion strain produces higher amounts of 4-HPP and Tyr than the wild-type strain, this inhibition seems to be caused by the accumulation of Tyr. Mutations in the HPD locus can cause two distinct genetic diseases, HT3 and hawkinsinuria. It has been suggested that alterations in the structure and activity of HPD are causally related to these two different metabolic disorders (18). Furthermore, HPD activity is markedly reduced in the liver of patients with HT1. A. nidulans can be used as a model system to investigate basic questions related to HT3 and 4-HPPD activity. Hawkinsinuria patients excrete hawkinsin (2-cystein-S-yl-1,4-dihydroxycyclohex-5-en-1-yl acetic acid) in their urine throughout their life; cis- and trans-hydroxycyclohexylacetic acids have also been identified in patients' urine (2, 18). It has been postulated that these are derivatives of products of a partial defect in the HPD enzyme (2). Molecular genetic analyses coupled to structure-function studies of A. nidulans HpdA could be used not only for the investigation of precursor accumulation in hawkinsinuria patients but also for the development of novel 4-HPPD inhibitors that could mitigate the pain of HT1 patients.
Acknowledgments
This research was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil.
REFERENCES
- 1.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97-E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Endo, F., Y. Tanaka, K. Tomoeda, A. Tanoue, G. Tsujimoto, and K. Nakamura. 2003. Animal models reveal pathophysiologies of tyrosinemias. J. Nutr. 133:2063S-2067S. [DOI] [PubMed] [Google Scholar]
- 3.Endo, F., A. Kitano, I. Uehara, N. Nagata, I. Matsuda, T. Shinka, T. Kuhara, and I. Matsumoto. 1983. Four-hydroxyphenylpyruvic acid oxidase deficiency with normal fumarylacetoacetate: a new variant form of hereditary hypertyrosinemia. Pediatr. Res. 17:92-96. [DOI] [PubMed] [Google Scholar]
- 4.Fernández-Cañón, J. M., and M. A. Peñalva. 1995. Molecular characterization of a gene encoding a homogentisate dioxygenase from Aspergillus nidulans and identification of its human and plant homologues. J. Biol. Chem. 270:21199-21205. [DOI] [PubMed] [Google Scholar]
- 5.Fernández-Cañón, J. M., and M. A. Peñalva. 1995. Fungal metabolic model for human type I hereditary tyrosinaemia. Proc. Natl. Acad. Sci. USA 92:9132-9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 7.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 8.Keon, J., and J. Hargreaves. 1998. Isolation and heterologous expression of a gene encoding 4-hydroxyphenylpyruvate dioxygenase from the wheat leaf-spot pathogen, Mycosphaerella graminicola. FEMS Microbiol. Lett. 161:337-343. [DOI] [PubMed] [Google Scholar]
- 9.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushihara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran, G. R. 2005. 4-Hydroxyphenylpyruvate dioxygenase. Arch. Biochem. Biophys. 433:117-128. [DOI] [PubMed] [Google Scholar]
- 11.Osmani, S. A., G. S. May, and N. R. Morris. 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104:1495-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peñalva, M. A. 2001. A fungal perspective on human inborn errors of metabolism: alkaptonuria and beyond. Fungal Genet. Biol. 34:1-10. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez, J. M., P. Ruíz-Sala, M. Ugarte, and M. A. Peñalva. 2004. Fungal metabolic model for 3-methylcrotonyl-CoA. J. Biol. Chem. 279:4578-4587. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez, J. M., P. Ruíz-Sala, M. Ugarte, and M. A. Peñalva. 2004. Fungal metabolic model for type I 3-methylglutaconic aciduria. J. Biol. Chem. 279:32385-32392. [DOI] [PubMed] [Google Scholar]
- 15.Russo, P. A., G. A. Mitchell, and R. M. Tanguay. 2001. Tyrosinemia: a review. Pediatr. Dev. Pathol. 4:212-221. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Semighini, C. P., M. Marins, M. H. S. Goldman, and G. H. Goldman. 2002. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl. Environ. Microbiol. 68:1351-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomoeda, K., H. Awata, T. Matsuura, I. Matsuda, E. Ploechl, T. Milovac, A. Boneh, C. R. Scott, D. M. Danks, and F. Endo. 2000. Mutations in the 4-hydroxyphenylpyruvic acid dioxygenase gene are responsible for tyrosinemia type III and hawkinsinuria. Mol. Gen. Metab. 71:506-510. [DOI] [PubMed] [Google Scholar]
- 19.Wyckoff, E. E., E. J. Pishko, T. N. Kirkland, and G. T. Cole. 1995. Cloning of a gene encoding a T-cell reactive protein from Coccidioides immitis: homology to 4-hydorxyphenylpyruvate dioxygenase and the mammalian F antigen. Gene 161:107-111. [DOI] [PubMed] [Google Scholar]
- 20.Yang, D.-Y. 2003. 4-Hydroxyphenylpyruvate dioxygenase as a drug discovery target. Drug News Perspect. 16:493-496. [DOI] [PubMed] [Google Scholar]