Abstract
This study tested for interaction between two independently evolved mechanisms of fluconazole resistance in Saccharomyces cerevisiae. One set of strains was from a 400-generation evolution experiment, during which the concentration of fluconazole was increased from 16 to 256 μg/ml in four increments. At 100 generations, populations became fixed for resistance mutations in either of two transcriptional regulators, PDR1 or PDR3. At 400 generations, replicate populations became fixed for another resistance mutation in UNK1, an unmapped gene further increasing resistance. Another genotype used in this study came from a population placed initially in 128 μg/ml of fluconazole; this environment selects for resistance through loss of function at ERG3, resulting in altered sterol metabolism. Mutant strains carrying PDR1r or PDR3r were crossed with the erg3r mutant strain, and the doubly mutant, haploid offspring were identified. The double-mutant strains grew less well than the parent strains at all concentrations of fluconazole tested. In genome-wide assays of gene expression, several ABC transporter genes that were overexpressed in one parent and several ERG genes that were overexpressed in the other parent were also overexpressed in the double mutants. Of the 43 genes that were consistently overexpressed in the PDR1r parents at generation 100, however, 31 were not consistently overexpressed in the double mutants. Of these 31 genes, 30 were also not consistently overexpressed after a further 300 generations of evolution in the PDR1r parent populations. The two independently evolved mechanisms of fluconazole resistance are strongly antagonistic to one another.
Resistance to the antifungal drug fluconazole (FLC) can occur by any of several different mechanisms, including increased efflux, alteration of the target enzyme, or alteration in sterol metabolism (15, 19). Predicting trends of antifungal drug resistance in fungal pathogens must take into account the potential for strains with multiple resistance mechanisms to arise, either through the step-wise accumulation of resistance mechanisms during asexual reproduction or through genetic exchange and recombination among strains harboring different resistance mechanisms. The maximum possible level of resistance in a fungal population depends not only on the magnitude of the contribution to resistance by the various individual mechanisms but also on the extent to which the mechanisms interact with one another in combination (2). In this study, we asked how two different mechanisms of fluconazole resistance of independent evolutionary origin interact with one another in Saccharomyces cerevisiae.
In S. cerevisiae, different regimens of selection favor different kinds of resistance in experimental populations (4) that are allowed to evolve. When the concentration of FLC is increased incrementally from 16 to 128 μg/ml over 400 generations, populations of large size behave in a highly predictable manner, first accumulating a mutation in PDR1 (or in the very similar gene, PDR3), a transcriptional regulator that results in increased expression of certain ABC transporters. At this stage, the FLC 80% minimum inhibitory concentration (MIC80) is increased from the initial 16 to 32 or 64 μg/ml, with a sharp transition from high growth to little or no growth at the higher concentrations of FLC in these tests. Beyond 100 generations, a second mutation for resistance in an unmapped gene of unknown function (UNK1) is accumulated, further increasing the FLC MIC80 to ≥256 μg/ml. This evolutionary path, described under “experiment 1” by Anderson et al. (4), provided one set of genotypes for this study.
When cell populations are exposed to 128 μg/ml FLC, resistance also evolves in a highly predictable manner, but it does so through an entirely different molecular mechanism than that observed with the incremental increase in FLC concentration over time. With abrupt exposure to 128 μg/ml FLC, resistance occurs predominantly through loss of function at ERG3. This mechanism of resistance is intimately involved with activation of the stress response. These strains characteristically show an FLC MIC80 of ≥256 μg/ml but with a reduction of growth at 16 μg/ml FLC; above 16 μg/ml, growth is essentially independent of FLC concentration and increases gradually over time. (Scoring MIC at the threshold of 50% reduction in growth is sensitive to time; here, the MIC is initially 16 μg/ml but then abruptly switches to 256 μg/ml FLC as the residual growth crosses the 50% threshold.) The evolution of resistance through loss of function at ERG3 is potentiated by the Hsp90 chaperone; when Hsp90 levels are low, this evolutionary path is effectively blocked, and any existing resistance by this mechanism is abolished (5). In contrast, evolution of resistance through mutation in PDR1 is not sensitive to low levels of Hsp90. This second evolutionary path, described under “experiment 2” by Anderson et al. (4), provided the other genotype for this study.
In this study, we combined the mutations that were favored under two different selection regimens into the same haploid strains. We then measured the growth response at various concentrations of the drug as an indicator of fitness and found that the different mechanisms of independent evolutionary origin were strongly antagonistic with one another, and that the strains with two resistance mechanisms grew less well at certain concentrations of fluconazole than the strains with one or the other mechanism of resistance. We also assayed genome-wide patterns of gene expression in strains from the two different selection regimens and in the strains with combined mechanisms of resistance; here we asked which genes are consistently altered in their expression levels with each regimen of selection and whether the changes in expression carry over from single-mutant to double-mutant strains.
MATERIALS AND METHODS
Strains.
Three PDR1 mutants were from 100 generations of evolution in 16 μg/ml FLC from experiment 1 of Anderson et al. (4). An additional PDR1 mutant and four PDR3 mutants were from 100 generations of evolution in 16 μg/ml FLC from Anderson et al. (3). These mutations were selected to represent a diversity of resistance mutations in each of two regions within these genes, an inhibitory domain toward the N terminus and a transcription activation domain toward the C terminus (12, 18). Multiple mutations were included, because the phenotypic effects of these mutations vary with respect to growth in medium containing FLC. The PDR5 deletion strain (YSC1021-552987) was purchased from Open Biosystems (Huntsville, AL).
Another three strains were from the same three populations of experiment 1 of Anderson et al. (4) but from a later time, 400 generations. Each of these strains carried a PDR1 mutation identical by descent to one of the mutations described above but also carried a resistance determinant in the unidentified gene UNK1.
All of our available erg3r mutations from experiment 2 of Anderson et al. (4) were exact phenocopies of one another and of the yeast strain in which the ERG3 gene is deleted with respect to growth in FLC; in each case, resistance was recessive. We therefore selected one mutant strain, consisting of a nonsense mutation toward the N terminus (Table 1), as the other parent in all crosses.
TABLE 1.
Amplification primers and hybridization probes
| Primer or probe | Description of mutationa | Sequence |
|---|---|---|
| Amplification primers | ||
| PDR1 (5′ end) | TGGAAACAGACGCATACCC | |
| CACTGAGAATGCTTGTGATACC | ||
| PDR1 (3′ end) | GACAACATGTGCATATCCGAC | |
| ACCTGTTGAAACCAAGAGCC | ||
| PDR3 | ACGAGGTTCTCCTTTTGGACA | |
| AGACCGAACAGAAAGGACAGA | ||
| ERG3 | TTTGAAGTGGTTGCAGAGGA | |
| AGACAAACAAGGCAACCGTA | ||
| Allele-specific hybridization probes | ||
| PDR1 | Wild type | TTCCTGACAATCGTC |
| T817K | TTCCTGAAAATCGTC | |
| PDR1 | Wild type | CTCTTGCGCTTATC |
| C862W | CTCTTGGGCTTATC | |
| PDR1 | Wild type | ATATACCTTCACAAG |
| L722P | ATATACCCTCACAAG | |
| PDR3 | Wild type | AGGTATAGGATATTTA |
| G253V | AGGTATAGTATATTTA | |
| PDR1 | Wild type | AAATTATAGATCCAAAT |
| R289I | AAATTATATATCCAAAT | |
| PDR3 | Wild type | GGGCATGTTTGGAC |
| C707F | GGGCATTTTTGGAC | |
| PDR3 | Wild type | GGAGTTTGTCAGCA |
| V954F | GGAGTTTTTCAGCA | |
| PDR3 | Wild type | CGTGAGTTTTTTGAT |
| F815Y | CGTGAGTATTTTGAT | |
| ERG3 | Wild type | CCCATGGATGTCG |
| A148* | CCCATGAATGTCG |
*, nonsense mutation.
Crosses and tetrad dissection.
Standard methods were used for making crosses between mutants and for dissecting tetrads (4). All tetrads were confirmed for 2:2 segregation of mating type. All offspring were assayed for MIC of FLC. Each offspring was then tested for the presence or absence of each resistance determinant using the single-nucleotide polymorphism corresponding to each resistance mutation. These assays were done by using allele-specific oligonucleotides as probes in Southern hybridizations with DNA from each respective gene amplified by the PCR (Table 1) exactly as described previously (7). In each tetrad, resistant and sensitive alleles at PDR1 or PDR3 and at ERG3 all segregated 2:2 (data not shown).
Three additional crosses were constructed between parents that were mutated in PDR1 and in UNK1 and a parent that was mutated in ERG3. The offspring were assayed exactly as in the other eight crosses. In each tetrad, mating types and the alleles for resistance and sensitivity at PDR1 or PDR3 and ERG3 segregated 2:2. Since UNK1 remains unidentified, there were no nucleotide polymorphisms available to assay for the presence or absence of this mutation, which represented a wild card in these crosses.
Resistance to fluconazole, cycloheximide, and rhodamine 6G.
Tests of MIC were done as described by Anderson et al. (4) in 0.5× YPD (1% glucose, 1% Difco peptone, 0.5% Difco yeast extract), except that scoring was at 30 h (Fig. 1 and 2). Care was taken to standardize the inoculum to 500 to 1,000 cells for each well of the 96-well microtiter plates. Optical density was determined with a Multiskan Ascent plate reader (Thermo Labsystems). For each panel in Fig. 1 and 2, each strain was assayed four times on two separate days; standard errors in measurements of optical density at 600 nm (OD600), with a few exceptions for data points at 0 μg/ml FLC, were too small to represent in these figures (mean standard error among four replicate measurements for each data point was 0.015). Replicate assays of MIC80 of cycloheximide (CYH; Sigma C7698) and rhodamine 6G (RHO; Sigma R4127) were exactly like those of FLC, except that for CYH the measurements were from 8 to 0.02 μg/ml in twofold increments, and for RHO they were from 100 to 0.63 μg/ml in twofold increments.
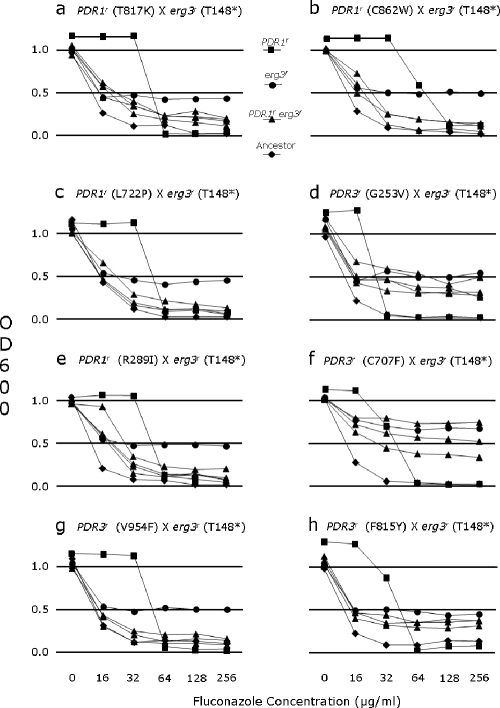
FIG. 1.
Tests for the FLC MIC for the ancestor (diamonds), strains carrying PDR1r or PDR3r from generation 100 of evolution experiment 1 (squares), the mutant strain carrying egr3r (circles), and offspring strains of S. cerevisiae carrying both PDR1r and erg3r (triangles).
FIG. 2.
Tests for FLC MIC for the ancestor (diamonds), strains carrying PDR1r and UNK1r from generation 400 of evolution experiment 1 (squares), the mutant strain carrying erg3r (circles), and offspring strains carrying both PDR1r and erg3r (triangles), among which segregation of UNK1r and UNK1s is expected to occur. In addition, strains carrying the respective PDR1r mutation at generation 100 (stars) are included for comparison.
For CYH and RHO, the sizes of the zones of inhibition of growth on 0.5× YPD agar were also measured. For each strain, a 3-ml top agar layer with 105 cells per ml was poured onto a standard 9-cm petri dish containing 25 ml medium. For each plate, an 8-mm disc of filter paper was spotted with 5 μl of a stock solution (CYH, 2 mg/ml; RHO, 10 mg/ml in ethanol) and then placed on the agar surface. The radius of growth inhibition was measured at 48 h.
Measurement of RHO uptake and efflux.
The RHO uptake and efflux experiments followed the general protocol of Kolaczkowski et al. (13), with the following modifications. Cells from exponentially growing populations in 50 ml of 0.5× YPD were harvested, washed three times in phosphate-buffered saline (PBS), suspended in PBS at 108 per ml, and incubated in 0.5 mM 2-deoxyglucose for 1 h. RHO was next added to 5.0 μM. After another 1.5 h of incubation, cells were washed twice in PBS and resuspended at the same concentration in PBS. Each cell suspension was then divided in two. In one half the glucose concentration was brought to 1 mM, and the other half received no glucose. At various time points, 1-ml samples of the cell suspensions were centrifuged quickly and the RHO concentration was measured by their optical density at 529 nm. No loss of viability, measured as CFU, was seen in any of the strains assayed.
Populations for gene expression assays.
For each of the three populations at 100 and 400 generations of evolution from experiment 1, a single-colony isolate was used for gene expression studies. A single double-mutant, progeny strain from each of the three crosses of the strains from generation 100 of experiment 1 and the erg3r mutant strain were also assayed for gene expression. The laboratory designation of the ancestor strain used as the reference in all gene expression assays was Sce13.
RNA preparation and cDNA synthesis.
As our interest was in constitutive patterns of gene expression, all cells were prepared for RNA extraction in the absence of FLC. A 10-ml culture inoculated from a single colony was grown at 30°C and 250 rpm to saturation (ca. 108 cells/ml) overnight in 0.5× YPD (1% glucose, 1% Difco peptone, 0.5% Difco yeast extract). The culture was diluted to an OD600 of 0.03, used to inoculate 50 ml of fresh medium in a 250-ml flask, and grown to an OD600 of 0.3. Cells were collected by centrifugation at 1,500 rpm for 3 min at 4°C. The cell pellet was rinsed once in 1 ml diethyl pyrocarbonate (DEPC)-treated water, transferred to a 1.5-ml microcentrifuge tube, and spun at maximum speed for 10 s. The supernatant was removed, and the pellet was immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction. RNA preparation and hybridization to yeast Y6.4k4 arrays from the University Health Network Microarray Centre were performed according to a standard hot acid phenol-chloroform extraction method, followed by total RNA purification with QIAGEN RNeasy mini spin columns (catalog no. 74106). Protocols are described at http://transcriptome.ens.fr/sgdb/protocols/preparation_yeast.php. The RNA was quantified by measuring the A260/A280 ratio and was then checked on a 1% agarose gel made with DEPC-treated water in Tris-acetate-EDTA buffer. Direct labeling of cDNA was carried out according to protocols from the University Health Network Microarray Centre at http://www.angiogenesis.ca/protocols/Molecular_Biology/Microarray.html. Reverse transcription used 20 μg of prepared RNA, and the reaction was purified by isopropanol precipitation with hybridization overnight at 37°C in hybridization chambers (2551; Corning).
Scanning and data analyses.
Microarrays were scanned at a 10-μm resolution with a ScanArray 4000 system (GSI Lumonics/Perkin Elmer), using both blue and red lasers. The 16-bit TIFF images were saved, and the fluorescence of individual spots was quantified with QuantArray software (v3.0; Packard Biosciences, Perkin Elmer, Boston, Mass.). The adaptive quantification algorithm was used. Further analysis, including data normalization, was performed with Gene Traffic Duo 2.5 (Iobion Informatics LLC, La Jolla, Calif.). The spot intensity data were exported from QuantArray and uploaded into Gene Traffic analysis software, where Lowess (locally weighted scatter plot smoother) normalization was applied on a Sub-Grid basis. Software filters were applied to flag additional spots whose intensities were less than twice the average background as well as those with absolute fluorescence intensities of less than 100. Once the data were normalized, the following criteria were used for filtering the microarray data and selecting genes that were overexpressed or underexpressed by ≥1.5-fold relative to the ancestor: at least two-thirds of the spots for a given gene in all replicate arrays were unflagged, and the standard deviations expressed as percentages of the means of all unflagged spots from replicate arrays were <100%.
Replication.
For populations 1 and 2, at 100 and 400 generations of the evolution experiment 1 and for each of the double mutants derived from crosses of these strains with the erg3r mutant, four replicate microarrays were used. For population 3, at 100 generations of evolution experiment 1 and the double mutant derived from crosses of this strain with the erg3r mutant, six replicate microarrays were used. For each microarray, the strain assayed and the ancestor strain were cultured independently in preparation for RNA extraction; each microarray represented a fully independent biological replicate.
Data filters.
Our purpose with data filters was to identify consistently altered gene expression in all three populations at generation 100 or generation 400 of the evolution experiment, in all three double mutants assayed, or in the erg3r mutant. For each of the genes meeting one or more of these criteria, all duplicate spot values from each microarray showed the same directionality of expression change relative to the ancestor. Within each of the expression categories in Fig. 3 to 5, genes are listed in order of their chromosomal locations.
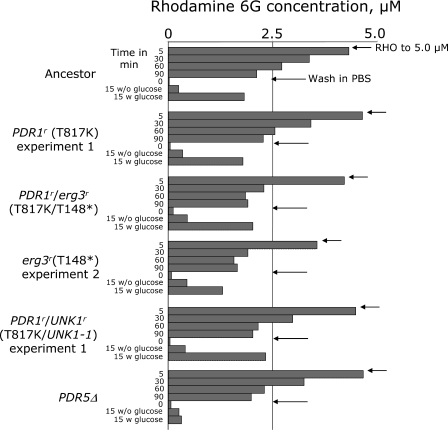
FIG. 3.
Uptake and efflux of rhodamine 6G. The protocol used followed that of Kolaczkowski et al. (13); see Materials and Methods for details. The results are from one of two replicate experiments and are representative of the two. RHO was added at time zero. Note that the erg3r and PDR1r/erg3r strains take up RHO faster initially than any of the other strains, before all converge to the same level of uptake. Glucose-mediated efflux occurred in all strains except the PDR5 deletion strain. Cells of all strains maintained viability throughout the experiment.
FIG. 5.
Gene expression among PDR1r mutant strains from three populations at generation 100 of evolution experiment 1, the erg3r mutant strain, and doubly mutant offspring. Genes and gene expression values are denoted as described in the legend to Fig. 4. Included are (a) genes overexpressed (≥1.5-fold) in all three populations at 100 generations, and in all three respective double-mutant offspring, but not in the erg3r mutant; (b) genes overexpressed in all three populations at 100 generations, in all three respective double-mutant offspring, and in the erg3r mutant; (c) genes overexpressed in all three populations at 100 generations but not in all three respective double-mutant offspring; (d) genes overexpressed (≥1.5-fold) in the erg3r mutant strain, and in all three of the double mutants, but not in all three PDR1r mutants; (e) genes overexpressed (≥1.5-fold) in the erg3r mutant strain but not in all three of the double mutants; (f) genes overexpressed in the three double mutants but not in erg3r, nor in all three of the PDR1r mutants; (g) genes underexpressed (≤1.5-fold) in the erg3r mutant strain but not in all three of the double mutants; and (f) genes underexpressed either in all three PDR1r mutants or in all three double mutants.
Validation of data.
Two levels of validation of the microarray data were used. First, mean expression values for three genes, SNQ2, ERG11, and PDR5, in the three populations at both time points in the evolution from the microarrays were checked against the expression values from Anderson et al. (4) for the same strains, determined by quantitative Northern hybridization relative to YEF3 (Table 2). The values matched well, with an overall R2 of 0.79. Second, many genes known to be overexpressed in the mutants carrying the PDR1-3 and PDR3-7 alleles (8), including key transporter genes such as PDR5, were expected to be overexpressed in this study, and this was the case.
TABLE 2.
Validation of gene expression values from DNA microarrays
| Generation | Test type | Level of expression forc:
|
||
|---|---|---|---|---|
| SNQ2 | ERG11 | PDR5 | ||
| 100 | Arraya | 1.7 ± 0.8 | 0.9 ± 0.0 | 3.4 ± 0.4 |
| Blotb | 2.7 + 0.8 | 1.2 ± 0.1 | 4.8 ± 0.8 | |
| 400 | Arraya | 2.2 ± 0.8 | 1.8 ± 0.2 | 3.3 ± 0.3 |
| Blotb | 2.3 ± 0.8 | 2.5 ± 0.3 | 3.5 ± 0.3 | |
Data are from the present study.
Data from quantitative Northern blot hybridization relative to YEF3 are from Anderson et al. (4).
Fold changes in expression values are means plus or minus standard deviations of the three replicate populations evolved over 400 generations in increasing concentrations of FLC.
Nucleotide sequence accession numbers.
The entire gene expression database is available from Gene Expression Omnibus, National Center for Biotechnology Information, under accession number GSE4261.
RESULTS
Antagonism between two resistance mechanisms.
Eight crosses were made between strains carrying a single resistance determinant, PDR1r or PDR3r, that arose during 100 generations of evolution in 16 μg/ml FLC, and a strain carrying another resistance determinant, erg3r, that was favored during abrupt exposure to 128 μg/ml FLC. From each cross, offspring carrying neither resistance determinant behaved exactly as the ancestor sensitive to FLC, with a MIC80 of 16 μg/ml, and offspring carrying a single resistance determinant behaved exactly as the parent carrying the respective mutation with regard to FLC MIC.
The growth assays for the three to five offspring carrying the resistance determinants of both parents are shown in Fig. 1. The PDR1r and PDR3r mutant parents all showed an FLC MIC of 32 or 64 μg/ml, with a steep reduction in growth at higher concentrations. The parent erg3r mutant showed an FLC MIC of 256 μg/ml but with a characteristic growth reduction above 16 μg/ml FLC that was concentration independent over the range tested. Unlike the PDR1r and PDR3r mutants, the amount of growth of the erg3r mutant increased with time at all drug concentrations. The PDR1r/erg3r double mutants showed substantial reductions in growth at 16 and 32 μg/ml FLC compared with their respective PDR1r or PDR3r mutant parent; this is strong evidence of antagonism with respect to fitness in a given drug concentration. At the high concentrations of FLC, the double mutants showed varying degrees of the residual growth that was prominent in the erg3r mutant parent, with double-mutant offspring showing a greater reduction in growth compared to the erg3r mutant parent; this represents further evidence for antagonism.
Three crosses were done with the three genotypes from 400 generations of evolution in increasing concentrations of FLC and the same erg3r resistance mutation used above. Three tetrads of each of these crosses were assayed for PDR1r or PDR3r and the erg3r mutations as described above. All four genotypic classes of offspring for PDR1 and ERG3 were recovered in each cross. Two different growth responses to FLC in the MIC assays occurred within each genotypic class for PDR1 and ERG3, a result consistent with the segregation of resistance and sensitivity at UNK1 (data not shown). Among the 11 PDR1s ERG3s strains, 7 mimicked the ancestor (FLC MIC, 16 μg/ml), and the FLC MIC for four others was slightly higher (32 μg/ml). Among the seven PDR1r ERG3s offspring, four mimicked the respective PDR1r UNK1r parent (MIC, 256 μg/ml) and three mimicked the PDR1r single-mutant strain (MIC, 64 μg/ml). Among the seven PDR1s erg3r strains, some mimicked the erg3r parent (MIC, 256 μg/ml; however, there was a reduction in growth at 16 μg/ml and above), while others showed somewhat higher growth at the lower FLC concentrations tested (MIC, 32 or 64 μg/ml); in these genotypes, it was not possible to score the presence of the UNK1r allele with confidence. For these three genotypic classes for PDR1 and ERG3, the variation for MIC was consistent with segregation for resistance and sensitivity at UNK1.
Among the 11 PDR1r erg3r offspring shown in Fig. 2, all showed antagonism for resistance, in that none grew as well at all FLC concentrations as the PDR1r UNK1r parent from 400 generations of evolution in FLC, but the growth responses to FLC were more complex than those shown in Fig. 1. In the first cross (Fig. 2a), the PDR1r erg3r mutants grew as much at 32 and 64 μg/ml FLC as the PDR1r single mutant (from 100 generations), which was included here as a reference. At higher concentrations of FLC, the response suggests two classes, one in which two offspring strains grew to a density almost, but not quite, equal to that of the PDR1r UNK1r parent, and the other in which one offspring strain grew to a density that was substantially lower than that of either of the parents. In the second cross (Fig. 2b), the response was different. All four double-mutant, offspring strains showed low levels of growth at the highest concentrations of FLC but showed differences in growth at 32 and 64 μg/ml FLC, with two offspring strains growing at a level similar to that of the PDR1r single mutant and the other two showing less growth. In the third cross (Fig. 2c), there were marked differences at 64 μg/ml and at the higher concentrations of FLC. In each of these three crosses, the differences in growth response among the PDR1r erg3r strains in the MIC assays is probably attributable, at least in part, to the segregation of the UNK1r determinant. In addition, the variability in response of the PDR1r erg3r mutant offspring among the three crosses may reflect differences in the PDR1r and/or the UNK1r alleles and how they interact in concert with the erg3r mutation. Alternatively, as-yet uncharacterized differences in the background genotype of the parents from 400 generations in FLC may have some effect on the expression of the three resistance determinants.
Mechanism of antagonism with respect to PDR5.
Both CYH and RHO are known substrates of Pdr5p. MICs of these drugs were therefore assayed as an indicator of the steady-state, intracellular drug concentration in the various strains studied here (Table 3). The MICs of CYH mirrored those of FLC, except that erg3r, with or without PDR1r, is more highly sensitive (0.02 μg/ml) than the ancestor (0.6 μg/ml). The increase in MIC of CYH from the ancestor to the PDR1r strains after 100 generations (to 0.13 μg/ml) was not seen in the PDR1r/erg3r double mutants. The PDR1r UNK1r strains from 400 generations showed a higher CYH MIC (0.25 μg/ml) than the PDR1r strains from 100 generations. RHO, which is much less inhibitory to growth than CYH, showed three levels of resistance: the PDR5 deletion strain at the lowest level (0.63 μg/ml), the erg3r and double-mutant strains at an intermediate level (13 μg/ml), and all others at the highest level (50 μg/ml). Inhibition assays were consistent with the MICs: the larger the halo the lower the MIC, and vice versa.
TABLE 3.
MIC of cycloheximide and rhodamine 6G and growth inhibitionc
| Strain, mutant status, and expt no. | MIC80
|
Radius of inhibition (mm)a
|
||
|---|---|---|---|---|
| Cyclo- heximide | Rhodamine 6G | Cyclo- heximide | Rhodamine 6G | |
| Ancestor | 0.06 | 50 | 16 | 2 |
| Expt 1, 100 generations | ||||
| Single mutants | ||||
| PDR1r | ||||
| T817 | 0.13 | 50 | 14 | 2 |
| C862 | 0.13 | 50 | 11 | 2 |
| L722 | 0.13 | 50 | 14 | 2 |
| Double mutants | ||||
| PDR1r/erg3r | ||||
| T817K/T148* | 0.02 | 13 | 23 | 5 |
| C862W/T148* | 0.02 | 13 | 23 | 5 |
| L722P/T148* | 0.02 | 13 | 23 | 5 |
| Expt 2 | ||||
| erg3r | ||||
| T148* | 0.02 | 13 | 23 | 5 |
| Expt 1, 400 generations | ||||
| PDR1r/UNK1r | ||||
| T817/UNK1-1 | 0.25 | 50 | 6 | 2 |
| C862/UNK1-2 | 0.25 | 50 | 8 | 2 |
| L722/UNK1-3 | 0.25 | 50 | 8 | 2 |
| PDR5Δb | 0.02 | 0.63 | 23 | 11 |
Halo radius. The response to rhodamine 6G was similar to that observed by Kolackowski et al. (13).
The MIC80 of FLC for the PDR5Δ strain was 0.5 μg/ml, compared with 16 μg/ml for the ancestor.
All tests were done on 0.5× YPD (see Materials and Methods for details).
The next experiment distinguished between uptake and efflux in a subset of the strains (Fig. 3). Glucose-starved cells of the erg3r and double-mutant strains accumulated RHO much faster initially than all others; this effect was prominent visually as a much stronger pink color of pelleted cells. After 1.5 h, however, all strains converged on nearly the same level of accumulation. Strong efflux activity was observed in all strains except the PDR5 deletion strain. Most importantly, the erg3r/PDR1r mutant showed a strong glucose-induced efflux of RHO that was approximately equal to that of the PDR1r single-mutant strain.
Gene expression after evolution in FLC.
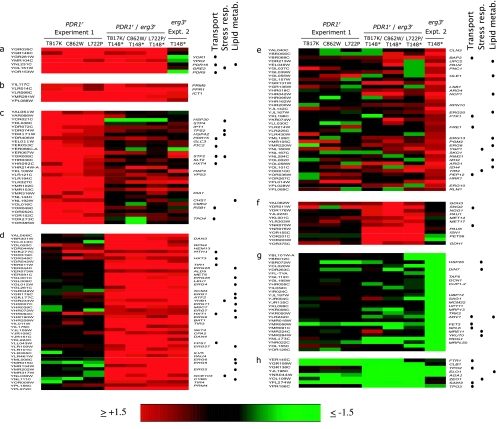
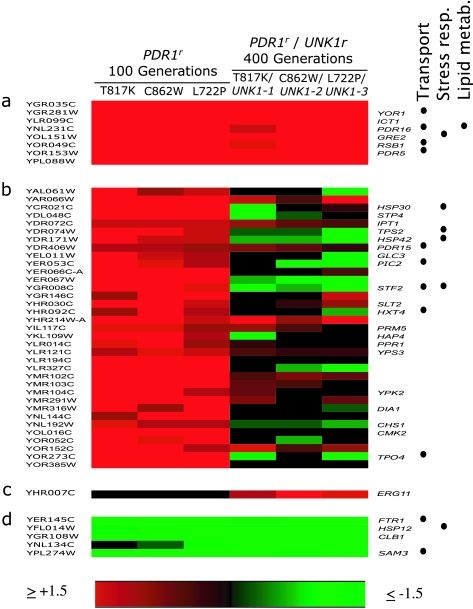
The purpose of the gene expression after evolution in FLC assays was to measure constitutive levels of gene expression in the drug-resistant, evolved populations relative to the drug-sensitive ancestor, with the goal of identifying parallel changes in gene expression in different populations (Fig. 4). Our first search was for genes that were consistently overexpressed at an early stage in the evolution of resistance; our expectation was that this group would include expression changes (i) closely associated with resistance and (ii) peripheral to resistance. At 100 generations, 43 genes were expressed at levels ≥1.5-fold higher than that of the ancestor in all three populations (Fig. 4a and b). The functions of these genes, where known, corresponded predominantly to transport and stress response. Our next search was for genes that remained overexpressed with further evolution; these genes would likely be among those most robustly associated with the resistant phenotype. Interestingly, of the 43 genes overexpressed at generation 100, only 8 remained consistently overexpressed in all three replicate populations at generation 400 (Fig. 4a); all eight were reported as more than twofold overexpressed in response to the mutations PDR1-3 and PDR3-7 by DeRisi and Goffeau (8), and each has a pleiotropic drug resistance element in its promoter region. Three genes, YAR066W, YHR214W-A, and YOR152C, narrowly missed being included in the set of eight genes, because their level of expression was lower than the cutoff of 1.5-fold over that of the ancestor in population 2 (Fig. 4b). Our next search was for genes that were likely to be more peripheral to the resistance. Of the 43 genes consistently overexpressed at generation 100, 35 were not consistently overexpressed at generation 400; only 2 (PDR15 and YOR152C) were identified as responsive to PDR1-3 or PDR3-7 by DeRisi and Goffeau (8).
FIG. 4.
Gene expression among strains from three populations evolving over 400 generations in increasing concentrations of FLC. The systematic gene name appears of the left, and the descriptive gene name appears on the right where available. The functions of genes known to include transport, stress response, and lipid metabolism are marked at the right. The color of boxes represents the constitutive expression of genes, with the scale at the bottom. Included are (a) genes overexpressed (≥1.5-fold) at generations 100 and 400; (b) genes consistently overexpressed in all three populations at generation 100 but not in all three populations at generation 400; (c) genes overexpressed in all three populations at generation 400 but not in any population at generation 100; and (d) genes underexpressed (≤1.5-fold) in all populations, either at generation 100 or generation 400.
Of particular interest were genes that were expressed at normal levels at 100 generations but consistently overexpressed at 400 generations, because these might be functionally associated with the resistance determinant UNK1r and the increased MIC50 of FLC evident at generation 400. Strikingly, there was only one such gene: ERG11 (Fig. 4c). Does ERG11 correspond to the unidentified UNK1 gene? No changes in the nucleotide sequence of the ERG11 open reading frame and entire upstream noncoding region to the adjacent gene YHR007C-A were observed in any of the three populations relative to the S288C standard (data not shown); the sequences of all strains matched the S288C standard exactly. UNK1 could yet be in the general region of ERG11 or could be a trans-acting regulatory gene located elsewhere in the genome. Interestingly, the overexpression of ERG11 at generation 400 was not accompanied by overexpression of any of the other ERG genes (in the erg3r mutant, overexpression of ERG11 was accompanied by overexpression of several other ERG genes; see below).
In addition to the genes that were overexpressed in the 400-generation experiment, five genes were underexpressed at either generation 100, generation 400, or at both time points, but no consistent pattern was obvious (Fig. 4d).
Gene expression changes in the erg3r mutant.
There were 92 genes that were overexpressed in the erg3r mutant (5 are shown in Fig. 5b, 48 in panel d, and 39 in panel e, according to the categories enumerated below). The most common known function of genes that were overexpressed relative to the ancestor was lipid metabolism, especially the ERG genes. Many of these genes were the same as those overexpressed in strains under inhibition with azole drugs (1).
Gene expression changes in the PDR1r erg3r mutants.
The three populations sampled at generation 100 of evolution experiment 1 had two fates, as parents of the double mutants in crosses with the erg3r mutant and continued evolution to generation 400. Our first comparison here was whether or not the overexpressed genes robustly associated with resistance in experiment 1 were also overexpressed in double mutants. Notably, all eight genes overexpressed at generation 100, and at generation 400 (Fig. 4a) they were also consistently overexpressed in each of the double mutants (Fig. 5a and b). Two of these eight genes (YLR099C/ICT1 and YPL088W), however, were also overexpressed in the erg3r mutant (Fig. 5b), and the cause of the overexpression of these genes in the double mutants could reside with the PDR1r mutation, the erg3r mutation, or both.
The next comparison was whether or not the 35 genes peripherally associated with the resistant phenotype in experiment 1 (Fig. 4b) were overexpressed in the double mutants. Of the 31 genes that were overexpressed at generation 100 but not consistently overexpressed in the double mutants (Fig. 5c), 30 were among the set of 35 genes not consistently associated with resistance over the 400 generations of experiment 1 (Fig. 4b).
Unlike the populations at generation 100 of evolution experiment 1, the erg3r mutant was studied solely for its contribution to the double mutants. Like the genes whose expression was altered in evolution experiment 1, our expectation was that these 92 genes of altered expression in evolution experiment 2 would include genes integral to resistance and peripheral to resistance. Of the 92 genes (Fig. 5b, c, and e) overexpressed in the erg3r mutant, 48 genes were also consistently overexpressed in double mutants (Fig. 5b); the predominant function of these genes, including several ERG genes, was lipid metabolism. The remaining 39 genes overexpressed in the erg3r mutant were not consistently overexpressed in double mutants (Fig. 5e); the functions of these genes included lipid metabolism, including two ERG genes, and transport.
Another category of expression included 12 genes that were consistently overexpressed in double mutants but not at 100 and 400 generations of the evolution experiment and not in the erg3r mutant (Fig. 5f). Among these, the SNQ2 transport gene narrowly missed meeting criteria for being included in the set of genes consistently overexpressed at both 100 generations and 400 generations depicted in Fig. 3.
Among the underexpressed genes that passed the predetermined filters for expression values, no consistent pattern was obvious (Fig. 5g and h).
DISCUSSION
The strong antagonism documented here between two mechanisms of resistance to FLC, increased efflux and altered sterol metabolism, has a clear implication for the evolution of drug resistance: multiple drug resistance mechanisms acting together do not always result in increased drug resistance. This kind of antagonism means that genetic exchange and recombination through mating and meiosis would not confer an advantage with respect to these two mechanisms of resistance. It also means that populations derived from a common ancestor can evolve different and mutually exclusive solutions to the problem of growing and reproducing in the presence of an antifungal drug, with a drastic fitness reduction where those mechanisms are combined and the drug is present. If these mutually exclusive adaptive solutions represent different levels of resistance, then the pathogens residing at the lower resistance optimum may be vulnerable to other treatments with antifungal drugs (2). This study represents but one pair of resistance mechanisms. When more combinations of mechanisms are examined in fungal pathogens, it will be clear whether or not the projected disadvantage of sex with respect to the two antifungal resistance mechanisms tested here is general.
The antagonism among resistance mechanisms observed here has another broad implication. The negative interaction corresponds exactly to a speciation mechanism described by the Muller-Dobzhansky model (9, 10, 16, 17). Under this model, populations adapt to different environments, as seen here. At some later time, with renewed contact among the divergent populations, any hybrids formed are of lower fitness, which is not due to any single gene but to negative interactions among genes. This kind of negative interaction represents the beginning of reproductive isolation. We do not imply that speciation actually occurs by this mechanism, only that this kind of genetic interaction could potentially play a role in speciation in fungi, especially fungal pathogens evolving in the presence of antifungal drugs.
The antagonism between the two drug resistance mechanisms likely has two components. First, our observations of the dampening of PDR5-mediated resistance to FLC in an erg3− background is consistent with an earlier report (14). This same effect occurs for CYH (Table 3). This dampening of resistance is likely due to increased uptake characteristics of the erg3− background and not to impairment of pump activity by the altered sterol composition of the membrane. The PDR5-mediated efflux in response to glucose was about the same in all strains except the PDR5 deletion strain. This is consistent with a previous report (11) in which only a small difference in RHO efflux activity was detected between erg3+ and erg3− strains. We speculate that increased uptake characteristics extend to FLC and that the genes involved in increased uptake may be among the 48 genes overexpressed in the erg3r and PDR1r/erg3r strains (Fig. 5d); this possibility remains to be tested.
Second, the presence of increased numbers of efflux pumps may dampen the general stress response seen in the erg3r mutant as the residual growth in FLC that is independent of drug concentration. This hypothesis could be tested by adding a detailed analysis of the activities of known stress response gene products in the strains used here.
The purpose of the gene expression studies was to gain insights into the two kinds of resistance to FLC and to the underlying nature of the interaction in the offspring strains harboring both kinds of resistance, between which there was a strong antagonism. The overall patterns of gene expression were consistent with the carryover of increased efflux, due to the mutant PDR1 and PDR3 transcriptional regulators, and altered sterol metabolism, due to the loss of function at ERG3 in the double mutants in which the negative interactions were caused by the simultaneous presence of these two kinds of alterations in expression. The overall picture for gene expression, however, is somewhat more complicated. In the 400-generation evolution experiment, more genes were overexpressed at generation 100 than 400, a situation reminiscent of compensatory evolution in Candida albicans, where in some replicate populations many genes overexpressed at a given time returned to normal levels of expression later in the evolution experiment (6). Remarkably, the genes that were no longer consistently overexpressed at generation 400 were mostly the same as those that returned to normal levels of expression in the double mutants; these genes are likely more peripheral to resistance to FLC and are more subject to perturbations in background genotype than are the eight genes that are overexpressed both at generation 400 and in the double mutants.
A critical factor distinguishing populations at 100 and 400 generations of evolution experiment 1 is the UNK1 gene. How does the UNK1 gene confer the increase of resistance from moderate to high levels in a PDR1r background? Does the UNK1r allele modulate the expression of genes peripheral to resistance and thereby play a role in managing the fitness costs of resistance? Unfortunately, UNK1 on its own has a subtle phenotype; the FLC MIC for this strain was 32 μg/ml rather than 16 μg/ml in the sensitive ancestor, and it has not yet been identified.
In the erg3r mutant, additional genes were altered in their levels of expression. Overexpression of many genes with known functions in transport or lipid metabolism carried through to the double mutants, but, like those genes in the 400-generation evolution experiment, there were many genes for which overexpression did not carry over to the double mutants and was not robust to perturbation in the background genotype. Possibly more relevant to the negative interaction were the 12 genes that were overexpressed in the double mutants but in neither of the parent types (Fig. 5f). These genes may be involved with the negative interactions measured here, or they may be entirely peripheral to this interaction.
The negative interactions in this study point to potential vulnerabilities in fungal pathogens; an increase in the number of resistance mechanisms does not necessarily translate to a more resistant phenotype. With more resistance mechanisms, the negative effects on phenotype may well be compounded, because the number of potential interactions will increase geometrically with the number of mechanisms. The genes of altered expression identified here can serve as candidates for measuring their contribution to the ability of strains to evolve and maintain resistance. Those genes whose altered expression is closely associated with resistance may be important for the evolution of resistance in the first place. Those genes peripheral to resistance that are shown to confer negative effects on fitness might point to ways of further inhibiting fungal pathogens. The present results may be important beyond antifungal drug resistance, as they show how fitness-enhancing mechanisms can interact in the process of adaptation.
Acknowledgments
This work was supported by an Operating Grant from the Canadian Institutes of Health Research and a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada to J.B.A.
Fluconazole was a gift of Pfizer Canada, Ltd.
REFERENCES
- 1.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998-35015. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. B. 2005. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat. Rev. Microbiol. 3:547-556. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. B., N. Ricker, and C. Sirjusingh. 2004. Ploidy and evolution of antifungal drug resistance. Genetics 168:1915-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, J. B., C. Sirjusingh, A. B. Parsons, C. Boone, C. Wickens, L. E. Cowen, and L. M. Kohn. 2003. Mode of selection and experimental evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics 163:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowen, L. E., and S. Lindquist. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185-2189. [DOI] [PubMed] [Google Scholar]
- 6.Cowen, L. E., A. Nantel, M. S. Whiteway, D. Y. Thomas, D. C. Tessier, L. M. Kohn, and J. B. Anderson. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl. Acad. Sci. USA 99:9284-9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowen, L. E., C. Sirjusingh, R. C. Summerbell, S. Walmsley, S. Richardson, L. M. Kohn, and J. B. Anderson. 1999. Multilocus genotypes and DNA fingerprints do not predict variation in azole resistance among clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 43:2930-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 9.Dobzhansky, T. 1937. Genetics and the origin of species. Columbia University Press, New York, N.Y.
- 10.Dobzhansky, T. 1936. Studies of hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura. Genetics 21:113-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaur, R., and A. K. Bachhawat. 1999. The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology 145:809-818. [DOI] [PubMed] [Google Scholar]
- 12.Kolaczkowska, A., M. Kolaczkowski, A. Delahodde, and A. Goffeau. 2002. Functional dissection of Pdr1p, a regulator of multidrug resistance in Saccharomyces cerevisiae. Mol. Genet. Genomics 267:96-106. [DOI] [PubMed] [Google Scholar]
- 13.Kolaczkowski, M., M. van der Rest, A. Cybularz-Kolaczkowska, J. P. Soumillion, W. N. Konings, and A. Goffeau. 1996. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J. Biol. Chem. 271:31543-31548. [DOI] [PubMed] [Google Scholar]
- 14.Kontoyiannis, D. P. 2000. Efflux-mediated resistance to fluconazole could be modulated by sterol homeostasis in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 46:199-203. [DOI] [PubMed] [Google Scholar]
- 15.Lupetti, A., R. Danesi, M. Campa, M. Del Tacca, and S. Kelly. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8:76-81. [DOI] [PubMed] [Google Scholar]
- 16.Muller, H. J. 1940. Bearing of the Drosophila work on systematics, p. 185-268. In J. Huxley (ed.), The new systematics. Clarendon, Oxford, England.
- 17.Muller, H. J. 1942. Isolating mechanisms, evolution and temperature. Biol. Symp. 6:71-125. [Google Scholar]
- 18.Nourani, A., D. Papajova, A. Delahodde, C. Jacq, and J. Subik. 1997. Clustered amino acid substitutions in the yeast transcription regulator Pdr3p increase pleiotropic drug resistance and identify a new central regulatory domain. Mol. Gen. Genet. 256:397-405. [DOI] [PubMed] [Google Scholar]
- 19.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]