Abstract
Gcn5 is a coactivator protein that contributes to gene activation by acetylating specific lysine residues within the N termini of histone proteins. Gcn5 has been intensively studied in the budding yeast, Saccharomyces cerevisiae, but the features of genes that determine whether they require Gcn5 during activation have not been conclusively clarified. To allow comparison with S. cerevisiae, we have studied the genome-wide role of Gcn5 in the distantly related fission yeast, Schizosaccharomyces pombe. We show that Gcn5 is specifically required for adaptation to KCl- and CaCl2-mediated stress in S. pombe. We have characterized the genome-wide gene expression responses to KCl stress and show that Gcn5 is involved in the regulation of a subset of stress response genes. Gcn5 is most clearly associated with KCl-induced genes, but there is no correlation between Gcn5 dependence and the extent of their induction. Instead, Gcn5-dependent KCl-induced genes are specifically enriched in four different DNA motifs. The Gcn5-dependent KCl-induced genes are also associated with biological process gene ontology terms such as carbohydrate metabolism, glycolysis, and nicotinamide metabolism that together constitute a subset of the ontology parameters associated with KCl-induced genes.
Gcn5 was discovered independently as a transcriptional coactivator in Saccharomyces cerevisiae (2) and a histone acetyltransferase in tetrahymena (4), thus linking transcriptional activation to histone acetylation. Gcn5 is found in the evolutionarily conserved Spt-Ada-Gcn5 acetyltransferase (SAGA) coactivator complex, as well as some related complexes (12). Besides Gcn5, which is required for histone acetyltransferase activity, the SAGA complex contains subunits that are required for its structural integrity as well as other subunits, such as Spt3, that functionally interact with the TATA-binding protein (TBP) (28). While mutations affecting the structural-integrity proteins have a very profound effect, the effects of defects in the Gcn5 or Spt3 modules are more subtle. Thus, the SAGA complex contains two or more coactivator activities, one of which involves Gcn5-dependent acetylation of chromatin (29, 30). Recombinant Gcn5 protein shows a strong preference for acetylation of Lys-14 of histone H3 when free histone proteins are used as a substrate (21). However, Gcn5 needs to be present in a coactivator complex in order to acetylate histone proteins in the context of nucleosomes (13). In the context of the SAGA complex, Gcn5 prefers Lys-14, but its substrate specificity is broadened to include other lysine residues in the N-terminal tail of histone H3.
Genes differ in their requirement for coactivators, such as Gcn5, during their activation. For example, Gcn5 is required for activation of the Saccharomyces cerevisiae PHO8 gene but not the PHO5 gene, even though both are strongly activated by the Pho4 activator protein during phosphate starvation (17). The ability of activator proteins to recruit different repertoires of coactivator proteins to different target genes may be a characteristic feature of their activation domains. The activation domains of many activators show a low propensity for secondary and tertiary structure formation but adopt a folded conformation during interaction with target proteins, such as coactivators (11, 16). The structural flexibility of activator proteins provides potential for flexible recruitment of coactivators, and thus, consistent with experimental evidence, the identity of activator proteins cannot be expected to predict the coactivators they recruit to different target genes.
Various approaches have been taken to understand why some genes require the SAGA complex for activation while others do not. Evidence from experiments using fluorescence resonance energy transfer has shown that the SAGA complex is an important target for the S. cerevisiae Gal4 activator protein in vivo (3). DNA microarray studies in S. cerevisiae suggest that the SAGA complex is redundant with the TFIID coactivator complex (22). More recently, it has been suggested that the SAGA complex is important for the activation of highly induced stress-regulated genes while the TFIID complex plays a role in the expression of housekeeping genes whose expression level does not change greatly under different growth conditions (20). Another recent study shows that the Gcn5 component of the SAGA complex is recruited to essentially all expressed genes with an efficiency corresponding to their expression level (27).
Fission yeast exhibits several chromatin features that distinguish it from S. cerevisiae and which are similar to chromatin in mammals. Most notably, the fission yeast genome contains larger regions of heterochromatin that are established via binding of the HP1-like Swi6 protein to methylated histone H3 (lysine 9) and a mechanism involving the RNA interference machinery (8). In Schizosaccharomyces pombe, Gcn5 is involved in histone acetylation and chromatin remodeling events that are important for mitotic and meiotic recombination (32), but its role in gene regulation has not been studied previously.
S. cerevisiae contains several mitogen-activated protein (MAP) kinase signal transduction pathways that together contribute to the specificity of stress responses. S. pombe uses only one MAP kinase pathway and selectivity of the stress response depends on Tup1-like corepressors that mediate stress-specific changes in chromatin structure (18). Indeed, gene duplication has lead to the acquisition of two Tup1-like proteins in S. pombe (Tup11 and Tup12), one of which plays a specific role in the stress response (10). Given the chromatin differences between S. cerevisiae and S. pombe, the apparent key role of Gcn5 in the stress response of S. cerevisiae, and the differences in the mechanisms regulating stress in the two yeasts, S. pombe provides an interesting comparative model system for studies of Gcn5. Here we show that Gcn5 is not important for the growth of S. pombe under normal conditions but that it does play a critical role in the response of cells to specific stress conditions. We describe the use of the physiological adaptation to one of these stresses (KCl) as a model to understand how Gcn5 contributes to reprogramming genome expression in response to external signals.
MATERIALS AND METHODS
Strains and media.
The gcn5− strain was created by replacement of SPAC1952.05 by a kanamycin resistance marker gene (KanMX4) in strain Fy368 (h− leu1-32 ura4-D18 ade6-M210) using a PCR-based approach as previously described (1). The primers used for the disruption and their sequences were as follows: gcn5:KanMX4 forward primer, TTCGTGCTCGTTTCATCTTGTATCGTTCTTGACAATTTCTGTATCTTCACTTTTTGGATTTATTTGTTTGGATGCGTGGCATAGAATAATCCGGATCCCCGGGTTAATTAA; and gcn5:KanMX4 reverse primer, ATGCCAGCATATTTGGAAGGCACAAAAGATATTCGTGTAAAAAATTAAAAGGTGAAATGTATATGTTATAATCAATAAAACTTCGGAATAGACGTTTCGATGGAATTCGAGCTCGTTTTCGACAC. The gcn5 deletion mutant was backcrossed to a wild-type strain, Fy367 (h+ leu1-32 ura4-D18 ade6-M216), following tetrad dissection in which three mutants, Hu798 (h+ leu1-32 ura4-D18 ade6-M216), Hu799 (h− leu1-32 ura4-D18 ade6-M216), and Hu800 (h− leu1-32 ura4-D18 ade6-M210), were selected. All three mutant strains displayed segregation of the stress sensitivity phenotypes with the gcn5 mutation. Hu799 was used in this study.
For growth assays strains were cultivated in YEA medium (0.5% yeast extract, 3% glucose, 0.2% Casamino Acids with 100 mg/liter of adenine, uracil, and leucine). For cell plating assays, YEA medium alone or supplemented with KCl (1 M), CaCl2 (0.1 M), sorbitol (1.2 M), NaCl (0.18 M), MnCl2 (0.25 mM), or FeCl3 (0.1 mM) was used. For microarray experiments cells were cultivated in YES medium (31) and YES medium supplemented with 1 M KCl. The parental strain Fy368 was used as a wild-type control for all experiments.
Microarray analysis.
For expression profiling of the gcn5− strain under nonstress growth conditions three biological replicates were used. For KCl stress experiments, at least two biological replicates were used. Cells were grown at 25°C to a cell density of 1 × 107 to 2 × 107 cells/ml with the cultures split in two. To one half, YES medium containing 2 M KCl was added to a final concentration of 1 M KCl, and the cells were frozen in liquid nitrogen after 15 or 60 min. The other half of the cultures were used as controls and immediately frozen in liquid nitrogen. In the case of the gcn5− strain, the cells were subjected to KCl for 60 min. RNA was collected as previously described (31). For each experiment, about 25 μg of total RNA was subjected to reversed transcription (catalog no. 11904-018; Invitrogen) and labeled with Cy3- or Cy5-labeled dCTP (Amersham). Reverse transcription primers (Eurogentec SA,) include an S. pombe-specific primer mix plus oligo(dT)18-21 and T20VN anchor primers. Labeled probes were hybridized to DNA microarrays containing S. pombe gene probes spotted in duplicate (31) (Eurogentec SA, Seraing, Belgium). Hybridization was performed as described previously (31) except that the hybridization volume was 72 μl. Slides were scanned using a Scan Express laser scanner and quantified with Imagene 4.2 software. Data were analyzed and normalized by the Lowess per-spot per-chip method using Genespring software (Silicon Genetics). Genes with changed expression were identified by selecting genes for which the mean ratio (sample to reference) reproducibly exceeded a chosen threshold value. Reproducibility was assessed using Student's t test to assess the significance of the difference of the mean ratio from a ratio of 1, expected for nonregulated genes. GeneSpring software was also used to calculate overlaps between gene groups using Fisher's exact test, for which P values were corrected for multiple testing (Bonferroni). Overlap between two groups of genes with a P value higher than 0.05 was not considered significant.
Promoter sequence analysis.
RSAT software (http://rsat.ulb.ac.be/) was used to analyze promoter sequences. We used the Pattern Discovery tool to look for overrepresented sequences of six to eight nucleotides in the 800-bp upstream region of Gcn5-dependent KCl-induced genes. The Pattern Matching tool was then used to look at the occurrences of these sequence motifs as well as known DNA sequence motifs in different groups of genes selected according to their change in expression in response to KCl stress in the wild-type and gcn5− mutant strains. As controls, nine groups of 30 genes were selected at random; in addition, the 800-bp upstream sequence of each S. pombe gene was also used.
Gene ontology terms.
GoMiner (http://discover.nci.nih.gov/gominer/) was used to find enriched gene ontology (GO) terms of groups of genes based on their expression profiles during the KCl stress response. The different groups of genes were compared to all genes with known GO terms. Only GO terms that were significantly enriched (P ≤ 0.05) were included in the results.
Microarray data accession number.
All microarray data are available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE5227.
RESULTS
Gcn5 is not generally required for cell growth and proliferation.
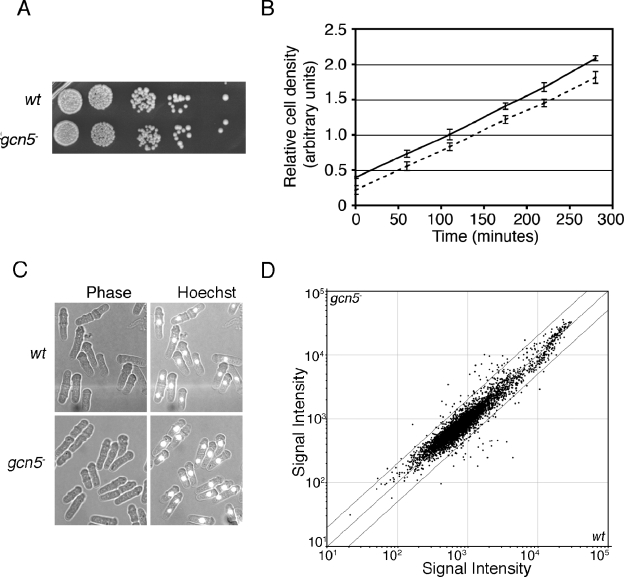
To investigate the role of Gcn5 in fission yeast, the gene encoding the S. pombe homolog of Gcn5 in S. cerevisiae (SPAC1952.05) was replaced by a kanamycin resistance marker gene. Figure 1A shows that deletion of the gcn5 gene is not associated with a detectable deficiency in the rate of colony formation during growth on rich medium plates at 30°C. Furthermore, there was only a very slight difference between the doubling time of the gcn5− mutant and the wild type during exponential growth in rich liquid medium at 30°C (Fig. 1B). Morphologically, gcn5− cells are very similar to wild-type cells (Fig. 1C). Thus, under these conditions, it is possible that Gcn5 is not required for the expression of any genes. Alternatively, Gcn5 could be required for the regulation of many genes that are not critical for cell growth, proliferation, and morphology. To examine the consequences of gcn5 deletion at the molecular level, we used DNA microarrays (31) to measure changes in genome-wide gene expression in a gcn5− mutant strain. Figure 1D shows that there are very few major differences in gene expression between gcn5− and wild-type cells. After normalization and quality control, we found only 9 of 4,825 genes to be reduced in expression by twofold or more in the gcn5− mutant. Interestingly, the expression of 32 genes increased by twofold or more in the mutant strain, even though Gcn5 has normally been associated with gene activation. The genes affected by Gcn5 represent a diverse group of functions, as shown in Table 1. We conclude that relatively few genes require Gcn5 activity for their expression under standard growth conditions.
FIG. 1.
Gcn5 is not required for normal growth. wt, wild type. (A) Fivefold dilutions of wild-type and gcn5− strains were spotted on rich medium (YEA) and the plates incubated at 30°C for 3 days. (B) Growth rate of log-phase wild-type (filled points) and gcn5− (clear points) cells in liquid culture (mean relative densities of three cultures ± standard deviations are plotted). (C) There is no morphological difference between wild-type and gcn5− cells in log phase. Cells were stained with Hoechst stain to visualize nuclear DNA. (D) gcn5 deletion causes small effects on gene expression. A scatter plot shows the mean signal intensity values of three independent microarray experiments. The flanking diagonal lines indicate a threshold value of a twofold change in expression (see Table S1 in the supplemental material for gene names, mean changes in induction [n-fold], and P values of the 260 genes with a reproducible change of >1.5-fold).
TABLE 1.
Genes with changed expression in the gcn5− mutant
| Gene namea | Description of gene product | Mean fold change | P value | |||
|---|---|---|---|---|---|---|
| Reduced expression in gcn5− mutant | ||||||
| SPAC1002.17c | Uracil phosphoribosyltransferase activity (predicted) | 0.295 | 0.0032 | |||
| dfr1 | Dihydrofolate reductase | 0.385 | 0.0010 | |||
| pex7 | Peroxisomal targeting signal receptor | 0.432 | 0.0005 | |||
| SPAC5H10.03 | Phosphoglycerate mutase family | 0.435 | 0.0001 | |||
| SPAC12G12.06c | RNA 3′-terminal phosphate cyclase (predicted) | 0.444 | 0.0124 | |||
| SPBC839.18c | Oxidoreductase (predicted) | 0.466 | 0.0020 | |||
| SPAC1039.02 | Calcineurin-like phosphoesterase (predicted) | 0.474 | 0.0005 | |||
| SPBC1683.05 | Thiamine transporter (predicted) | 0.486 | 0.0151 | |||
| obr1 | Ubiquitinated histone-like protein Uhp1 | 0.487 | 0.0003 | |||
| Increased expression in gcn5− mutant | ||||||
| mei2 | RNA-binding protein | 5.584 | 0.0055 | |||
| ght5 | Hexose transporter | 4.686 | 0.0159 | |||
| SPCC1739.08c | Short chain dehydrogenase | 4.605 | 0.0100 | |||
| SPACUNK4.10 | 2-Hydroxyacid dehydrogenase (predicted) | 4.592 | 0.0003 | |||
| SPBC1105.01 | HEAT repeat (ISS) | 3.680 | 0.0357 | |||
| fio1 | Iron transport multicopper oxidase | 2.874 | 0.0112 | |||
| plb1 | Phospholipase B homolog Plb1 | 2.834 | 0.0133 | |||
| SPBC20F10.02c | Conserved eukaryotic protein | 2.743 | 0.0090 | |||
| pho1 | Acid phosphatase activity | 2.561 | 0.0009 | |||
| SPBC359.06 | Class II aldolase and adducin N-terminal domain | 2.558 | 0.0455 | |||
| ght4 | Hexose transporter | 2.556 | 0.0488 | |||
| ppa2 | Serine/threonine protein phosphatase (catalytic subunit) | 2.530 | 0.0142 | |||
| SPAC6F12.04 | COPI coated-vesicle-associated protein (predicted) | 2.487 | 0.0039 | |||
| SPBC1709.14 | Peptide N-glycanase (predicted) | 2.357 | 0.0245 | |||
| SPAC26H5.09c | Oxidoreductase (predicted) | 2.267 | 0.0032 | |||
| inv1 | Beta-fructofuranosidase | 2.265 | 0.0050 | |||
| rst2 | Transcription factor | 2.191 | 0.0242 | |||
| nnf1 | Kinetochore protein Nnf1 | 2.149 | 0.0234 | |||
| hsp9 | Heat shock protein | 2.132 | 0.0212 | |||
| SPAC1565.07c | TATA-binding protein-interacting protein (predicted) | 2.055 | 0.0045 | |||
| SPBC887.09c | Leucine-rich repeat protein | 2.029 | 0.0002 | |||
| SPAC30.03c | Translin-like protein | 2.011 | 0.0318 | |||
Genes with a ≥2-fold change in expression in the gcn5− mutant compared to the wild type (P ≤ 0.05). The following genes, lacking descriptive terms, also had increased expression in the gcn5− mutant: SPAC4F8.08, SPAC27D7.10c, SPAC27D7.11c, SPAC1952.04c, SPBC1861.10, SPCC1393.07c, SPCC63.15, wtf13, and wtf7.
Gcn5 is required for adaptation to specific environmental conditions.
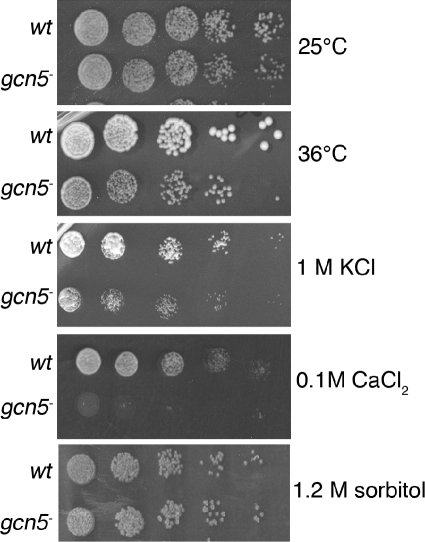
Gcn5 has been suggested to play a specific role in the activation of highly regulated genes in S. cerevisiae in response to environmental stress (see the introduction). We therefore studied the growth characteristics of gcn5− mutants under a range of environmental conditions. As shown in Fig. 2, the gcn5− strain has a reduced growth rate at 36°C and is sensitive to KCl and CaCl2 stress. The salt stress phenotype is not due to a general defect in the osmotic-stress response because the gcn5− mutant is not sensitive to sorbitol. Rather, the Gcn5 defect appears to cause a specific sensitivity to KCl and CaCl2. In comparison, we found no sensitivity to NaCl, MnCl2, or FeCl3 (data not shown) at concentrations that inhibit the growth of mutants defective in the Cta4 ion transporter, which is thought to be involved cation homeostasis (9). Importantly, the conditions (36°C, 1 M KCl, 0.1 M CaCl2) that inhibit colony development of gcn5− mutants do not reduce the viability of the mutant cells in relation to the wild type. Cells exposed to inhibitory doses of KCl, CaCl2, or to 36°C for 24, 48, or 72 h form colonies as efficiently as the wild type when plated on normal medium (data not shown). We conclude that Gcn5 has a role in some stress responses but that it is not required for others.
FIG. 2.
Gcn5 is required for growth under specific environmental stress conditions. The gcn5− mutant spotted on YEA plates at 36°C or YEA plates supplemented with 1 M KCl or 100 mM CaCl2 displayed reduced growth relative to the wild type (wt), whereas gcn5− mutant cells spotted on YEA plates supplemented with 1.2 M sorbitol showed no difference in growth from wild-type cells. Plates were incubated for 3 to 5 days at 25°C unless otherwise indicated.
Characterization of the response to KCl stress.
We have previously studied the differential role of the Tup11 and Tup12 corepressors in the KCl adaptation response (10), and therefore we decided to use this system as a model for further studies of Gcn5 function. Our approach was to identify Gcn5-regulated genes involved in KCl adaptation. In a first step, we used DNA microarrays to identify genes regulated during adaptation to KCl stress in wild-type cells. Figure 3 compares gene expression levels after 15-min (Fig. 3A) and 60-min (Fig. 3B) exposures to KCl to expression levels in untreated cells. The numbers of genes for which changes in expression level were reproducibly greater than twofold and threefold are tabulated in Table 2. There is a clear difference between the gene expression changes at 15 min and 60 min after KCl addition. Induction of gene expression after 15 min was of lower amplitude than induction after 60 min, and interestingly, relatively few genes were induced at both time points. In contrast, relatively large reductions in expression were observed for many genes after 15 min, and the expression levels of many of these genes were still reduced after 60 min of KCl treatment. A previous study of gene expression changes under a range of different stresses, though not under salt-induced stress, led to the identification of a group of core environmental stress response (CESR) genes (6). The genes induced after 60-min KCl treatment show a highly significant overlap with the CESR genes (P = 8.18e−50). No significant overlap was observed for genes induced after a 15-min KCl treatment or for genes that are down-regulated by KCl. Many CESR genes are dependent on the Sty1/Atf1 MAP kinase signal transduction pathway for their regulation (6). As expected, there is a significant overlap between the group of 54 KCl-induced genes that are also CESR genes and the Sty1/Atf1-dependent genes (16/54 genes, P = 4.45e−12), but there is no significant overlap between the other classes of KCl-regulated genes and Sty1/Atf1-dependent genes. We conclude that the response to KCl-induced stress involves different types of changes in gene expression patterns. The group of KCl-induced genes contains many genes that are induced early, transiently, and to a limited degree. These genes are subsequently replaced by another set of genes, many of which are Sty1/Atf1-dependent CESR genes, which are induced later and with greater amplitude. The group of genes that are down-regulated by KCl contains many genes whose expression is rapidly and persistently reduced to a relatively large extent.
FIG. 3.
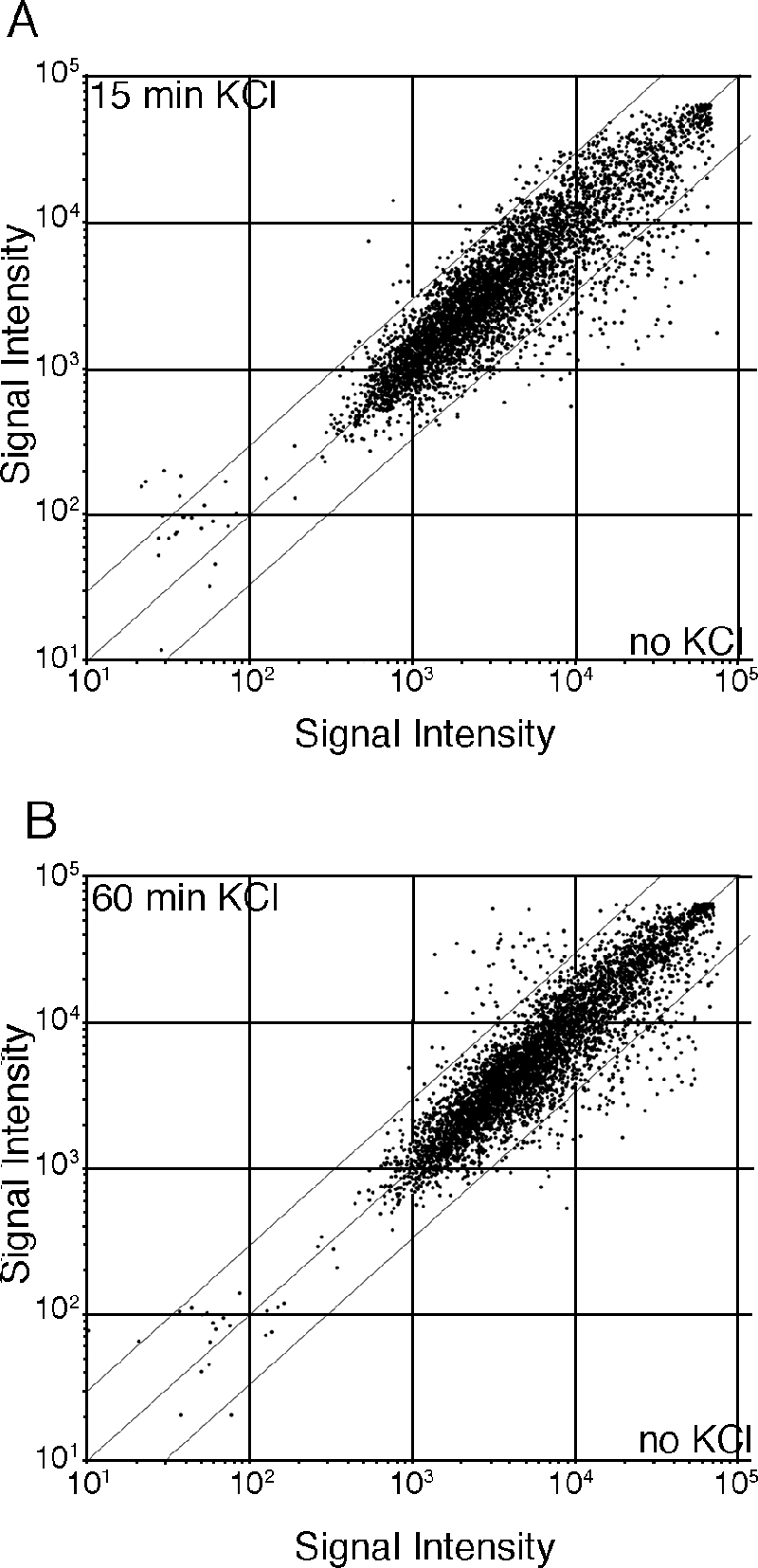
Characterization of gene expression changes during the response to 1 M KCl. (A) Gene expression changes after 15-min KCl treatment are characterized by robust down-regulation of genes. A scatter plot of DNA microarray signal intensities after 15 min of KCl stress plotted against signal intensities of nonstressed control cells is shown. The flanking diagonal lines indicate a threefold change in expression. (B) Gene expression changes after 60 min of KCl treatment are characterized by robust up- and down-regulation of genes. A scatter plot of DNA microarray signal intensities after 60 min of KCl stress plotted against signal intensities of nonstressed control cells is shown. Annotations are as shown in panel A.
TABLE 2.
Number of genes with altered expression during KCl adaptation
| Change in expression after indicated time (min) | No. of genes with indicated mean fold changea
|
||||
|---|---|---|---|---|---|
| ≥2-fold
|
≥3-fold
|
||||
| Total | Overlapb | Total | Overlapb | CESRc | |
| Up | |||||
| 15 | 350 | 75 | 57 | 4 | 1 |
| 60 | 364 | 141 | 54d | ||
| Down | |||||
| 15 | 285 | 100 | 105 | 25 | 1 |
| 60 | 204 | 49 | 4 | ||
See Tables S2 and S3 in the supplemental material for complete lists of gene names, mean changes in expression (n-fold), and P values.
Number of genes changed at both 15 and 60 min.
Number of genes that have also been designated as CESR genes; a total of 140 induced CESR genes and 104 repressed CESR genes were designated (6).
Significant overlap (P = 8.18e−50). The size of the other overlaps with CESR genes are not statistically significant (P > 0.05).
The correspondence of KCl response genes with known stress genes can be regarded as one line of evidence that validates our results. An independent approach is to determine whether the biological processes represented by the KCl-responsive genes belong to functional classes that might be expected. Although the majority of response genes are associated with at least one gene ontology term, most genes are not found in the different groups of response genes at a frequency greater than that expected by chance. Table 3 shows the subset of gene ontology (biological process) terms that are significantly enriched by the sets of genes whose expression changed up or down at 15 min or 60 min. The enriched genes induced at 15 min are involved RNA polymerase II transcription and ribosome biogenesis, while down-regulated enriched genes at this time point are involved in energy generation and nitrogen compound metabolism. By 60 min, the induced group includes enriched genes with a broad range of functions, including metabolism, cell cycle, transport, and response to various types of stimuli. Genes that are significantly enriched in the group that is down-regulated at 60 min are mainly associated with transport but also with cytokinesis and cell septation. The functions enriched by these sets of genes are generally consistent with current views of the stress response, whereby cells stop growing (manifested by reduced expression of genes involved in energy generation and cell division) while they initiate new gene expression programs to deal with stress conditions (manifested by early induction of gene expression and protein synthesis components followed by later induction of diverse groups of genes and up- and down-regulation of genes associated with transport).
TABLE 3.
Biological processes specifically associated with KCl response genes
| GO term annotationa | No. of genes | Frequency (%)b |
|---|---|---|
| Genes with increased expression after 15 min | ||
| Cytoplasm organization and biogenesis (184) | 4 | 13 |
| Ribosome biogenesis (148) | 4 | 13 |
| rRNA processing (122) | 4 | 13 |
| Transcription from RNA Pol II promoter (24) | 2 | 7 |
| Genes with reduced expression after 15 min | ||
| ATP synthesis coupled electron transport (17) | 2 | 3 |
| Electron transport (117) | 4 | 5 |
| Energy derivation by oxidation of organic compounds (101) | 5 | 6 |
| Generation of precursor metabolites and energy (249) | 9 | 12 |
| Nitrogen compound metabolism (249) | 3 | 4 |
| Genes with increased expression after 60 min | ||
| Carbohydrate metabolism (184) | 11 | 14 |
| Cation transport (101) | 5 | 6 |
| Cell communication (258) | 9 | 11 |
| Generation of precursor metabolites and energy (249) | 7 | 9 |
| M phase (309) | 9 | 11 |
| Meiosis (143) | 8 | 10 |
| Negative regulation of cellular metabolism (86) | 5 | 6 |
| Nicotinamide metabolism (24) | 3 | 4 |
| Nucleotide metabolism (107) | 5 | 6 |
| Oxidoreduction coenzyme metabolism (39) | 4 | 5 |
| Positive regulation of biological process (44) | 2 | 3 |
| Protein-mitochondrial targeting (42) | 3 | 4 |
| Regulation of enzyme activity (46) | 2 | 3 |
| Regulation of transcription by glucose (10) | 2 | 3 |
| Response to abiotic stimulus (102) | 5 | 6 |
| Response to nutrients (16) | 3 | 4 |
| Response to stimulus (388) | 13 | 16 |
| Signal transduction (232) | 8 | 10 |
| Genes with reduced expression after 60 min | ||
| Carbohydrate transport (24) | 2 | 5 |
| Cell separation during cytokinesis (22) | 2 | 5 |
| Cytokinesis (125) | 5 | 13 |
| Monosaccharide transport (7) | 2 | 5 |
| Transport (747) | 12 | 30 |
Biological-process GO terms specifically associated with genes with a ≥3-fold reproducible change in expression compared to the expression in untreated cells (P ≤ 0.05). The number of S. pombe genes annotated with each term is shown in parentheses. Pol, polymerase.
Percentage of GO-process annotated genes in each expression group that are annotated with each specific GO term. The numbers of GO process-annotated genes analyzed for each expression group were 30 and 77 and 80, and 40 for the increased and decreased expression groups at 15 and 60 min, respectively.
Identification of Gcn5-dependent genes during adaptation to KCl stress.
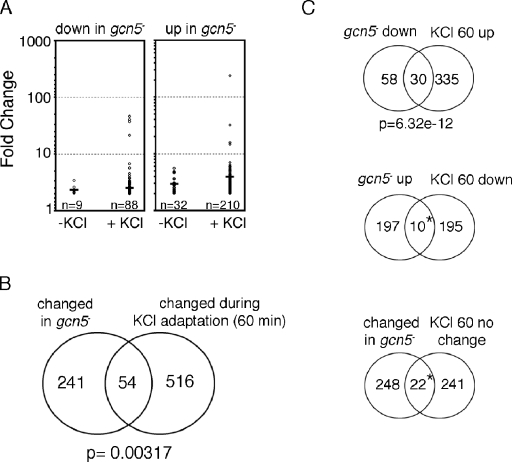
Next, we used the DNA microarray approach to identify genes whose expression is dependent on Gcn5 during KCl adaptation. We focused on genes dependent on Gcn5 after 60 min of KCl treatment due to the larger number of induced genes as well as the greater extent of their induction at this time point. As predicted by the KCl-sensitive phenotype of the gcn5− mutant, there were many more Gcn5-dependent genes under these conditions than were detected in the absence of KCl (Fig. 4A). A subset of these genes also showed a much greater degree of dependence on Gcn5 on KCl than was seen for Gcn5-dependent genes in the absence of KCl (Fig. 4A).
FIG. 4.
Gcn5 is required for regulation of a subset of genes involved in the KCl stress response. (A) Scatter plot showing an increase in the number of Gcn5-dependent genes and the extent of their dependence during KCl stress. n, number of genes with a mean change in induction of ≥2-fold (P ≤ 0.05). The mean change in induction (n-fold) for all genes in each class is indicated (dark line). (B) There is a significant overlap between genes involved in adaptation to KCl and genes with changed expression in the gcn5− mutant during KCl stress. A P of 0.00317 is the probability with which the observed overlap of 54 genes would be expected by chance. (C) KCl-induced genes show a significant overlap (P = 6.32 × 10−12) with genes that are dependent on Gcn5 during KCl adaptation. The overlaps (*) between genes with increased expression in the gcn5− mutant and KCl-repressed genes and between Gcn5-dependent genes and genes that are not involved in KCl adaptation were not considered significant (P > 0.05).
Figure 4B shows that the 295 genes, which show significantly changed expression by at least twofold in the gcn5− mutant after a 60-min KCl treatment, overlap with the set of 570 genes that are regulated at least twofold during KCl adaptation (60 min). The size of the overlap between the two data sets is much greater than that expected by chance (P = 0.00317), indicating that the patterns of KCl-induced gene expression and their dependence on Gcn5 reflect the KCl-sensitive growth phenotype. As shown in Fig. 4C, the overlap between KCl-regulated genes and Gcn5-dependent genes is highly significant for KCl-induced genes that require Gcn5 (30 genes) and not significant for KCl-repressed genes that require Gcn5 (10 genes). For a control, we checked the Gcn5 dependence of a group of genes that are reproducibly not regulated by KCl, for which the overlap of 22 genes that are Gcn5 dependent is not significant. We conclude that there is a significant subset of KCl adaptation genes that are Gcn5 dependent but that there are larger groups of KCl adaptation genes and Gcn5-dependent genes that are not associated with each other.
In order to get an overall view of Gcn5 dependence during KCl treatment in relation to the transcriptional response to KCl, we compared the change in expression of KCl adaptation genes (60 min) with the effect of gcn5 deletion on expression (KCl, 60 min) for genes that were either induced or repressed by KCl stress. For both sets of genes, the relationship between expression change in response to KCl and the degree of Gcn5 dependence (positive or negative) was essentially random (data not shown). Thus, there is no correlation between the extent of KCl induction and Gcn5 dependence. We conclude that Gcn5 dependence is not correlated to the extent to which genes respond to KCl-mediated stress. Therefore, there must be an alternative explanation that accounts for why some genes require Gcn5 for their regulation while others do not.
Gcn5 dependence is associated with a subset of KCl adaptation processes.
To further investigate the reason for the Gcn5 dependence of some genes, we used gene ontology biological process terms to determine whether Gcn5-dependent genes during KCl adaptation are associated with any of the biological functions that change in response to KCl treatment (Table 3). Table 4 shows that genes with reduced expression in the gcn5− strain are significantly enriched and associated with some of the same functions of enriched genes in the set that was induced by KCl after 60 min. Most significantly, these functions include carbohydrate metabolism and nicotinamide metabolism, whose genes were further enriched in the set of Gcn5-dependent genes, even in relation to the enrichment level observed during KCl adaptation. However, other terms that are closely related in the gene ontology, such as vitamin metabolism, generation of precursor metabolites and energy, alcohol metabolism, main pathways of carbohydrate metabolism, and glycolysis, had genes that were also significantly enriched. While several of these terms are also associated with KCl adaptation genes, many classes of adaptation genes are not enriched in the Gcn5-dependent group (e.g., cation transport, cell communication, M phase, meiosis, response to stimulus, and signal transduction). Furthermore, the genes associated with ontology terms were significantly enriched in the gene set with increased expression in the gcn5− mutant (Table 4) and do not overlap with those for the genes that are down-regulated after KCl treatment for 60 min (Table 3). This is consistent with the low significance of the overlap between these sets of genes (Fig. 4). We conclude that a subset of functions associated with KCl-induced genes may have evolved together with Gcn5 dependence, while other adaptation responses may have evolved independently of Gcn5.
TABLE 4.
Biological processes specifically associated with Gcn5-dependent genes during KCl adaptation
| GO term annotationa | No. of genes | Frequency (%)b |
|---|---|---|
| Genes with reduced expression in gcn5− mutant | ||
| Alcohol metabolism (106) | 5 | 10 |
| Carbohydrate metabolism (184)c | 7 | 14 |
| Generation of precursor metabolites and energy (249) | 5 | 10 |
| Glycolysis (23) | 2 | 4 |
| Main pathways of carbohydrate metabolism (59) | 3 | 6 |
| Nicotinamide metabolism (24)c | 2 | 4 |
| Protein folding (99) | 4 | 8 |
| Vitamin metabolism (50) | 3 | 6 |
| Genes with increased expression in gcn5− mutant | ||
| Aerobic respiration (51) | 6 | 4 |
| Alcohol biosynthesis (12) | 2 | 1 |
| Alcohol metabolism (106) | 9 | 6 |
| Aspartate family amino acid metabolism (47) | 6 | 4 |
| Carbohydrate metabolism (184) | 14 | 9 |
| Cellular macromolecule catabolism (278) | 12 | 8 |
| Cellular catabolism (366) | 17 | 11 |
| Conjugation with cellular fusion (82) | 7 | 5 |
| Energy derivation by oxidation of organic compounds (107) | 15 | 10 |
| Generation of precursor metabolites and energy (245) | 19 | 12 |
| Glucose catabolism (32) | 6 | 4 |
| Glycerophospholipid catabolism (6) | 2 | 1 |
| Glycolysis (23) | 5 | 3 |
| Hydrogen transport (37) | 4 | 3 |
| Main pathways of carbohydrate metabolism (59) | 11 | 7 |
| Methionine metabolism (25) | 5 | 3 |
| Nucleosome assembly (13) | 2 | 1 |
| Sulfur metabolism (52) | 6 | 4 |
| Threonine metabolism (8) | 2 | 1 |
| Transcription from RNA Pol II promoter (116) | 8 | 5 |
| Transcription initiation from RNA Pol II promoter (32) | 3 | 2 |
Biological-process GO terms specifically associated with genes with a ≥2-fold reproducible change in expression in the gcn5− mutant during KCl adaptation compared to the expression in the wild type (P ≤ 0.05). The number of S. pombe genes annotated with each term is shown in parentheses. For the list of gene names and the corresponding mean changes in expression (n-fold) and P values, see Table S4 in the supplemental material. Pol, polymerase.
Percentage of GO-process annotated genes in each expression group that are annotated with each specific GO term. The numbers of GO process-annotated genes analyzed for each expression group were 51 for the decreased-expression group and 153 for the increased-expression group.
GO terms further enriched in the group of Gcn5-dependent KCl-induced genes in relation to the enrichment level observed during KCl adaptation (Table 3).
Putative cis-acting sequences enriched upstream of Gcn5-dependent KCl-induced genes.
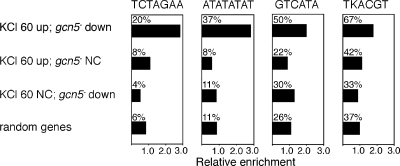
The correlation of Gcn5 dependence with a subset of functions associated with KCl-induced genes suggested that such genes might contain cis-acting regulatory sequences in their upstream regions. To identify putative cis-acting sequences, we looked for sequence motifs that are overrepresented in the region 800 bp upstream of the initiator ATG of Gcn5-dependent KCl-induced genes. For controls, we used groups of genes that are induced by KCl independently of Gcn5 and that are Gcn5 dependent but not KCl induced as well as groups of randomly chosen genes. We identified three motifs, ATATATAT, GTCATA, and TCTAGAA, that that are specifically overrepresented in the test group. Figure 5 shows the relative enrichment of these motifs in the test group in relation to their genomic frequency. Figure 5 also shows the relative frequency of the cis-acting replication element motif (TKACGT), which has previously been observed in CESR genes (6) and is also specifically enriched in Gcn5-dependent KCl-induced genes.
FIG. 5.
Identification of DNA sequence motifs that are specifically overrepresented in Gcn5-dependent KCl-induced genes. The bar charts show the frequency of four indicated DNA sequence motifs upstream (−800 bp from ATG) of genes within different classes relative to the genomic frequency of the motifs. The classes contain (i) 30 genes that are induced ≥2-fold after KCl treatment for 60 min, (ii) 52 genes that are induced ≥2-fold by KCl at 60 min but are not Gcn5 dependent (gcn5− NC; change in induction [n-fold] ≤ 1.1-fold), (iii) 27 genes that show reduced expression in the gcn5− mutant after 60 min of KCl treatment (≥2-fold) but are not KCl induced (KCl NC; change in induction [n-fold] ≤ 1.3-fold), and (iv) 9 × 30 randomly selected genes. The proportion (%) of genes in each class that contain each motif is indicated above the bars.
DISCUSSION
Our results suggest that relatively few genes in S. pombe require Gcn5 for their expression during growth on rich medium. This may be because Gcn5 functions at very few genes or because its function can be provided by other proteins under these conditions. However, under specific stress conditions, such as exposure to KCl and CaCl2, Gcn5 plays an essential role in reprogramming the expression of a much larger group of genes. Some aspects of KCl adaptation, such as potassium transport, have been studied previously (5). Under conditions of high external potassium, intracellular potassium levels are primarily regulated at the level of potassium efflux. Cta3 is the best-characterized potassium efflux transporter in fission yeast. As confirmed by our data, transcription of the cta3+ gene is strongly induced by KCl-mediated stress. In S. cerevisiae, potassium ions are pumped out of the cell by the Kha1 and Sod2 transporters. S. pombe contains homologues of both proteins, but neither of them was induced by KCl in our experiments. This is consistent with characterization of the S. pombe Sod2 protein as a specific transporter of sodium ions, unlike the S. cerevisiae Sod2 that transports both sodium and potassium. Kha1 in S. pombe has not been characterized. Another process that is regulated by KCl stress is protein synthesis (7). Upon exposure to KCl, there is a rapid but transient decrease in protein synthesis. Our observation that genes involved in ribosome function are rapidly induced by KCl is consistent with the requirement for restoration of protein synthesis capacity that follows the transient decrease.
It has been suggested that, in S. cerevisiae, Gcn5 function is characterized by the highly inducible nature of the genes it regulates such that a large proportion of stress-induced genes are associated with Gcn5 function (20). In S. pombe, Gcn5 is required for adaptation to some stress conditions but not others. Furthermore, during the KCl stress response, only a minority of the induced genes are Gcn5 dependent, and we have not observed any correlation between the extent of gene induction and Gcn5 dependence. Consistently, it has been suggested that very small changes in nucleosome positioning in individual promoters can have a large effect on the requirement for histone acetylation during gene activation (24).
Interestingly, we have recently shown that the stress sensitivity profile of gcn5− mutants (KCl and CaCl2) is shared by strains that lack the Tup12 corepressor (10). Tup12, together with its homologue Tup11, are involved in the regulation of KCl-induced genes, such as cta3 (14), and they have recently been shown to function as specificity factors for the stress response in S. pombe (18). In strains lacking both Tup11 and Tup12, the cta3 gene, which is normally specifically derepressed by KCl, is derepressed by a broadened range of stress conditions (18). Tup11 and Tup12 have also been shown to repress chromatin remodeling of the ade6-M26 locus (19), where Gcn5 is required for H3 acetylation and chromatin remodeling during meiosis (32). In addition, the S. cerevisiae GAL1 gene, which is repressed by Tup1-Ssn6, requires the SAGA complex during induction (26). Thus, one possibility that we will investigate in future work is that Gcn5 has a specific role in the derepression of at least some of the genes that are repressed by Tup11 and Tup12 corepressors.
Over half of the genes that are induced in a Gcn5-dependent fashion by KCl after 60 min have been identified previously as CESR genes that are induced by many environmental stresses (6). The overlapping group of genes includes zym1 (encoding metallothionein), mex6 (encoding an mRNA export factor), gut2 (encoding glycerol-3-phosphate dehydrogenase), and pyp2 (encoding a serine/threonine protein phosphatase). Metallothionein is a zinc-binding protein that is thought to transfer zinc to newly synthesized zinc finger transcription factors, and thus, its function, together with those of Mex6 and glycerol-3-phosphate dehydrogenase, can easily be understood in the context of a cell adapting its proteome and metabolism to external stress. Many CESR genes have been shown to respond to the Sty1/Atf1 stress response system (6). Phosphorylation by the Sty1 MAP kinase is thought to activate the Atf1 transcription factor, which subsequently activates many CESR genes. Pyp2 has been shown to inactivate Sty1 in response to stress (25) and may thereby constitute a negative feedback mechanism that regulates the stress response.
As expected from the overlap with CESR genes, we found that the Atf1-binding site (TKACGT) is significantly overrepresented in the upstream region Gcn5-dependent KCl-induced genes. Interestingly, the same site is not significantly overrepresented in KCl-induced genes that are Gcn5 independent or in Gcn5-dependent genes that are not induced by KCl. We found several other sequence motifs that are significantly overrepresented, specifically in the upstream regions of Gcn5-dependent KCl-induced genes. The TCTAGAA motif is contained within the binding site for the S. cerevisiae heat shock factor Hsf1 (33) and is thus a likely binding site for the S. pombe heat shock factor. The occurrence of this motif in CESR genes might be expected, since heat shock is one of the stresses by which they are induced. Indeed, five out of six Gcn5-dependent KCl-induced genes containing the motif have been shown to be induced by heat shock (≥2-fold) (6). The other motifs are more obscure. The central hexamer within the ATATATAT motif is a high-affinity binding site for the TBP (15). However, the first and last bases of the motif are not part of the sequence required for TBP binding, and therefore, these sites are not like ordinary TATA boxes. Additionally, ordinary TATA boxes would not be expected to be specifically associated with this set of genes. Since the distance between the TATA box and transcription site is around 30 bp in S. pombe (23), it is possible to calculate the predicted length of the 5′ untranscribed regions (UTRs) for genes containing the ATATATAT motif if the motif were to represent a specialized type of TATA box. In this case, the mean 5′ UTR length would be about 290 nucleotides, which is significantly longer than the mean length of 170 nucleotides for the 390 5′ UTRs that have been mapped (P = 0.056). Taken together, our data suggest that the ATATATAT motifs are not TATA boxes. Proteins that might bind to the GTCATA motif have not to our knowledge been identified.
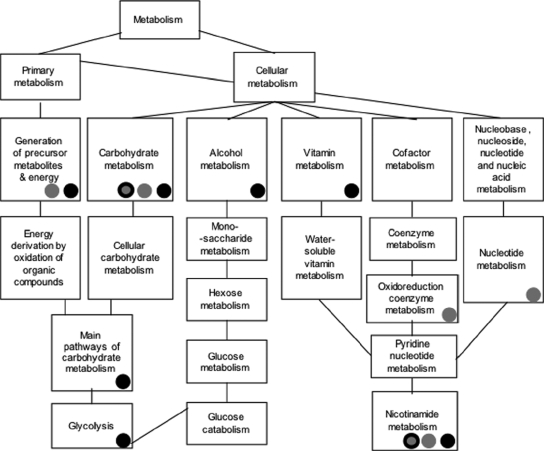
We have found that a set of related biological-process gene ontology functions are specifically enriched in the group of Gcn5-dependent KCl-induced genes. As shown in Fig. 6, the ontology terms are associated with carbohydrate metabolism, glycolysis, and nicotinamide metabolism. The latter term includes metabolism of nicotinamide adenine dinucleotide, which is an essential cofactor in glycolysis. The only enriched ontology term that is not closely connected to the others is protein folding. Interestingly, the group of related gene ontology terms overlaps with a similar group of genes that are enriched by genes that are induced by KCl treatment for 60 min. As shown in Fig. 6, some terms are enriched by both groups, and two terms are significantly further enriched by the Gcn5-dependent group in relation to their enrichment level by the KCl-induced group. Thus, the group of carbohydrate metabolism-associated terms that is associated with Gcn5-dependent genes is independently associated with KCl-induced genes. Genes induced by KCl after 60 min are also associated with other gene ontology terms, such as cation transport, mitosis, meiosis, and cell communication. Therefore, only a subset of related functions associated with KCl induction are dependent upon Gcn5.
FIG. 6.
Gcn5-dependent KCl-induced genes are involved in similar biological processes. Relationships between GO processes enriched in the groups of KCl-induced genes (gray) and Gcn5-dependent genes (black) (Tables 3 and 4) are shown. Two GO processes are further enriched by Gcn5-dependent, KCl-induced genes (gray and black) (Table 3).
We conclude that Gcn5 is required for the regulation of a subset of genes, the expression of which changes in response to specific stress conditions. Gcn5 dependence appears to be a characteristic of genes associated with a subset of functions that changes in response to stress as well as genes enriched in a certain type of promoter motifs.
Supplementary Material
Acknowledgments
DNA microarray slides were scanned at the KI-CHIP core facility at the Karolinska Institute, which is supported by the Wallenberg Foundation. This research was funded by project grants from the Swedish Research Council. A.P.H.W. is a senior investigator for the Swedish Research Council.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 2.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 3.Bhaumik, S. R., T. Raha, D. P. Aiello, and M. R. Green. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 5.Calero, F., and J. Ramos. 2003. K+ fluxes in Schizosaccharomyces pombe. FEMS Yeast Res. 4:1-6. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bahler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14:214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunand-Sauthier, I., C. A. Walker, J. Narasimhan, A. K. Pearce, R. C. Wek, and T. C. Humphrey. 2005. Stress-activated protein kinase pathway functions to support protein synthesis and translational adaptation in response to environmental stress in fission yeast. Eukaryot. Cell 4:1785-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekwall, K. 2005. Genome-wide analysis of HDAC function. Trends Genet. 21:608-615. [DOI] [PubMed] [Google Scholar]
- 9.Facanha, A. L., H. Appelgren, M. Tabish, L. Okorokov, and K. Ekwall. 2002. The endoplasmic reticulum cation P-type ATPase Cta4p is required for control of cell shape and microtubule dynamics. J. Cell Biol. 157:1029-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fagerstrom-Billai, F., and A. P. Wright. 2005. Functional comparison of the Tup11 and Tup12 transcriptional corepressors in fission yeast. Mol. Cell. Biol. 25:716-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira, M. E., S. Hermann, P. Prochasson, J. L. Workman, K. D. Berndt, and A. P. Wright. 2005. Mechanism of transcription factor recruitment by acidic activators. J. Biol. Chem. 280:21779-21784. [DOI] [PubMed] [Google Scholar]
- 12.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 13.Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner, and J. L. Workman. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895-5900. [DOI] [PubMed] [Google Scholar]
- 14.Greenall, A., A. P. Hadcroft, P. Malakasi, N. Jones, B. A. Morgan, C. S. Hoffman, and S. K. Whitehall. 2002. Role of fission yeast Tup1-like repressors and Prr1 transcription factor in response to salt stress. Mol. Biol. Cell 13:2977-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, S., S. Buratowski, P. A. Sharp, and L. Guarente. 1989. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc. Natl. Acad. Sci. USA 86:5718-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermann, S., K. D. Berndt, and A. P. Wright. 2001. How transcriptional activators bind target proteins. J. Biol. Chem. 276:40127-40132. [DOI] [PubMed] [Google Scholar]
- 17.Hertel, C. B., G. Langst, W. Horz, and P. Korber. 2005. Nucleosome stability at the yeast PHO5 and PHO8 promoters correlates with differential cofactor requirements for chromatin opening. Mol. Cell. Biol. 25:10755-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirota, K., T. Hasemi, T. Yamada, K. I. Mizuno, C. S. Hoffman, T. Shibata, and K. Ohta. 2004. Fission yeast global repressors regulate the specificity of chromatin alteration in response to distinct environmental stresses. Nucleic Acids Res. 32:855-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirota, K., C. S. Hoffman, T. Shibata, and K. Ohta. 2003. Fission yeast Tup1-like repressors repress chromatin remodeling at the fbp1+ promoter and the ade6-M26 recombination hotspot. Genetics 165:505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 21.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269-272. [DOI] [PubMed] [Google Scholar]
- 22.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y., P. M. Flanagan, H. Tschochner, and R. D. Kornberg. 1994. RNA polymerase II initiation factor interactions and transcription start site selection. Science 263:805-807. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Campa, C., P. Politis, J. L. Moreau, N. Kent, J. Goodall, J. Mellor, and C. R. Goding. 2004. Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol. Cell 15:69-81. [DOI] [PubMed] [Google Scholar]
- 25.Millar, J. B., V. Buck, and M. G. Wilkinson. 1995. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 9:2117-2130. [DOI] [PubMed] [Google Scholar]
- 26.Papamichos-Chronakis, M., T. Petrakis, E. Ktistaki, I. Topalidou, and D. Tzamarias. 2002. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 9:1297-1305. [DOI] [PubMed] [Google Scholar]
- 27.Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy, A. Rolfe, J. L. Workman, D. K. Gifford, and R. A. Young. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16:199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 30.Wallberg, A. E., K. E. Neely, J. A. Gustafsson, J. L. Workman, A. P. Wright, and P. A. Grant. 1999. Histone acetyltransferase complexes can mediate transcriptional activation by the major glucocorticoid receptor activation domain. Mol. Cell. Biol. 19:5952-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue, Y., S. A. Haas, L. Brino, A. Gusnanto, M. Reimers, D. Talibi, M. Vingron, K. Ekwall, and A. P. Wright. 2004. A DNA microarray for fission yeast: minimal changes in global gene expression after temperature shift. Yeast 21:25-39. [DOI] [PubMed] [Google Scholar]
- 32.Yamada, T., K. I. Mizuno, K. Hirota, N. Kon, W. P. Wahls, E. Hartsuiker, H. Murofushi, T. Shibata, and K. Ohta. 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 23:1792-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, W. M., W. Gahl, and D. Hamer. 1991. Role of heat shock transcription factor in yeast metallothionein gene expression. Mol. Cell. Biol. 11:3676-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.