Abstract
Autophagy is a well-known degradation system, induced by nutrient starvation, in which cytoplasmic components and organelles are digested via vacuoles/lysosomes. Recently, it was reported that autophagy is involved in the turnover of cellular components, development, differentiation, immune responses, protection against pathogens, and cell death. In this study, we isolated the ATG8 gene homologue Aoatg8 from the filamentous fungus Aspergillus oryzae and visualized autophagy by the expression of DsRed2-AoAtg8 and enhanced green fluorescent protein-AoAtg8 fusion proteins in this fungus. While the fusion proteins were localized in dot structures which are preautophagosomal structure-like structures under normal growth conditions, starvation or rapamycin treatment caused their accumulation in vacuoles. DsRed2 expressed in the cytoplasm was also taken up into vacuoles under starvation conditions or during the differentiation of conidiophores and conidial germination. Deletion mutants of Aoatg8 did not form aerial hyphae and conidia, and DsRed2 was not localized in vacuoles under starvation conditions, indicating that Aoatg8 is essential for autophagy. Furthermore, Aoatg8 conditional mutants showed delayed conidial germination in the absence of nitrogen sources. These results suggest that autophagy functions in both the differentiation of aerial hyphae and in conidial germination in A. oryzae.
Autophagy is a protein degradation system that is conserved in eukaryotic cells and used to recycle macromolecules and aid cell survival under nutritional starvation conditions (12, 13). When autophagy is induced, bulk cytoplasm and/or organelles are sequestered within double-membrane vesicles termed autophagosomes. The outer membranes of the autophagosomes then fuse to the vacuolar/lysosomal membrane and deliver single-membrane vesicles, called autophagic bodies, into the lumina of the vacuoles/lysosomes. The subsequent breakdown of the vesicle membranes allows the degradation of the contents of the autophagic bodies by vacuolar hydrolases. ATG8 is an autophagy-related gene found in Saccharomyces cerevisiae that plays an important role in the formation of autophagosomes (9). Atg8 is localized in the membranes of preautophagosomal structures (PAS), autophagosomes, and autophagic bodies and has therefore been used as a marker of these organelles (33).
In addition to helping cells to survive starvation, autophagy is involved in stress-induced differentiation and development. In S. cerevisiae diploid cells, atg mutations block starvation-induced sporulation (34). In Dictyostelium discoideum, starvation, overcrowding, and high temperature induce the formation of fruiting bodies and atg mutations block these multicellular developmental processes (25). Additionally, in Caenorhabditis elegans, atg mutations result in abnormal dauer development, which is also induced by starvation, overcrowding, high temperature and so on (17). Recent studies have suggested that autophagy might participate in diseases such as cancer, liver disease, muscular disorder, and neurodegeneration (31) and protect from infection by bacterial pathogens (e.g., group A Streptococcus and Shigella) (20, 22).
In filamentous fungi, the processes of autophagy have been observed and studied for Podospora anserina (26, 27). In this species, autophagy is induced during cell death by incompatibility, which occurs when cells of different genotypes fuse. Null mutants of the idi-7/PaATG8 gene (the orthologue of S. cerevisiae ATG8) form fewer aerial hyphae and no protoperithecia (27). Furthermore, a null mutant of PaATG1 (the orthologue of S. cerevisiae ATG1) shows the same defects as those of the Δidi-7/PaATG8 mutants (26). However, studies on autophagy and the identification of the genes involved have not been reported in deuteromycetes. The deuteromycete filamentous fungus Aspergillus oryzae is an important microorganism in Japanese fermentative industries because it plays a role in the production of sake, miso, and soy sauce (11). More recently, A. oryzae has been described as an excellent host for the production of homologous and heterologous enzymes (28). In the present study, we isolated the ATG8 gene homologue Aoatg8 from A. oryzae and generated strains expressing the DsRed2-AoAtg8 and enhanced green fluorescent protein (EFGP)-AoAtg8 fusion proteins to visualize autophagy in A. oryzae. Moreover, we constructed a mutant strain in which the Aoatg8 gene was disrupted and described the involvement of autophagy in the formation of aerial hyphae, conidiation, and germination in A. oryzae.
MATERIALS AND METHODS
Strains and growth media.
The A. oryzae strains used in this study are listed in Table 1. A. oryzae RIB40 is a wild-type strain that was used as a DNA donor. A. oryzae niaD300 (niaD−) and A. oryzae strains expressing carboxypeptidase Y (CPY)-EGFP, which is the vacuolar marker (niaD−) (23), named NSCE1, were used as host strains to visualize autophagy. A. oryzae NSR13 (niaD−, sC−, and adeA−) (6) was used to disrupt the Aoatg8 gene. The NSR13AA strain, NSR13 transformed with adeA (6), was used as a control for the phenotypic assay. Czapek Dox (CD) medium (0.3% NaNO3, 0.2% KCl, 0.1% KH2PO2, 0.05% MgSO4 · 7H2O, 0.002% FeSO4 · 7H2O, and 2% glucose [pH 5.5]) was used as a selective medium and for microscopic observations. Methionine (0.0015%) was added to select for positive clones when the ΔAoatg8-1-1 strain was transformed with plasmids harboring the niaD gene as a selection marker. M medium [0.2% NH4Cl, 0.1% (NH4)2SO4, 0.05% KCl, 0.05% NaCl, 0.1% KH2PO4, 0.05% MgSO4 · 7H2O, 0.002% FeSO4 · 7H2O, and 2% glucose (pH 5.5)] supplemented with 0.15% methionine was used as a selective medium for disrupting the Aoatg8 gene. CD medium lacking sodium nitrate (CD−N) or CD medium containing 200 ng/ml rapamycin (CD+R) was used for inducing autophagy. The transformation of A. oryzae was carried out using the standard method (7).
TABLE 1.
Strains of A. oryzae
| Strain | Genotype | Source or reference |
|---|---|---|
| RIB40 | Wild type | |
| niaD300 | niaD | 18 |
| NSCE1 | niaD− and sC−::(PamyB-cpyA-egfp sC) | 23 |
| NSR13 | niaD−sC−adeA− | 6 |
| NSR13AA | niaD−sC−adeA−::adeA | 6 |
| GEGA8 | niaD−::(PAoatg8-egfp-Aoatg8 niaD) | This study |
| GDRA8 | niaD−::(PAoatg8-DsRed2-Aoatg8 niaD) | This study |
| CEDA8 | niaD−::(PAoatg8-DsRed2-Aoatg8 niaD) sC−::(PamyB-cpyA-egfp sC) | This study |
| CEDR1 | niaD−::(PamyB-DsRed2 niaD) sC−::(PamyB-cpyA-egfp sC) | This study |
| ΔAoatg8-1-1 | niaD−sC−adeA− ΔAoatg8::adeA | This study |
| ΔAoatg8-1-1DR | niaD−::(PamyB-DsRed2 niaD) sC−adeA− ΔAoatg8::adeA | This study |
| CSC1-2-1 | niaD−::(PthiA-Aoatg8 niaD) sC−::sC adeA− ΔAoatg8::adeA | This study |
Construction of A. oryzae strains for visualizing autophagy.
To clone the promoter, gene, and terminator regions of Aoatg8 with the Multisite Gateway cloning system (Invitrogen Co., Tokyo, Japan), four primers were designed: attB4-PAoatg8-F (5′-GGGGACAACTTTGTATAGAAAAGTTGGATAGGATCCTTAGGGTCTG-3′), attB1-PAoatg8-R (5′-GGGGACTGCTTTTTTGTACAAACTTGATTGATGGATCGAATCAGTTAATGG-3′), attB2-Aoatg8-F (5′-GGGGACAGCTTTCTTGTACAAAGTGGTTATGCGCTCCAAGTTCAA G-3′), and attB3-Aoatg8-R (5′-GGGGACAACTTTGTATAATAAAGTTGTTTTCACTACTTATTTTCAATTACC-3′). These primers were based on the A. oryzae EST database (http://www.nrib.go.jp/ken/EST/db/index.html). The underlined sequences are the attB recombination sites for the Multisite Gateway. Using the genomic DNA of A. oryzae RIB40 as a template, a 1,722-bp Aoatg8 promoter region was amplified by PCR using the primers attB4-PAoatg8-F and attB1-PAoatg8-R and an 890-bp fragment (gene, 535 bp; terminator, 355 bp) was amplified using the primers attB2-Aoatg8-F and attB3-Aoatg8-R. The amplified attB-flanked promoter and gene plus terminator were introduced into pDNORP4-P1R and pDNORP2R-P3, respectively, using the Gateway BP clonase reaction mix (Invitrogen) and sequenced. The resultant entry clone plasmids were named pg5′PAoatg8 and pg3′Aoatg8, respectively. Gateway LR reactions (Gateway LR clonase reaction mix; Invitrogen) were carried out with these and an entry clone plasmid containing the egfp gene (constructed in our laboratory) plus the destination vector pgDN containing the A. oryzae niaD gene as a selection marker (16) in order to generate pgEGA8. The plasmid pgDRA8 was constructed by performing the Gateway LR reaction with the following four plasmids: pg5′PAoatg8, pg3′Aoatg8, an entry clone plasmid containing the DsRed2 gene (constructed in our laboratory), and pgDN. The pgEGA8 (or pgDRA8) plasmid was introduced into A. oryzae niaD300. The transformants GEGA8 (expressing EGFP-AoAtg8) and GDRA8 (expressing DsRed2-AoAtg8) were used to visualize autophagy. pgDRA8 was also introduced into A. oryzae NSCE1. The transformed strain, which expresses CPY-EGFP and DsRed2-AoAtg8, was named CEDA8. Furthermore, to visualize autophagy by the uptake of cytoplasm, the expression plasmid pUNDR was constructed as follows. The DsRed2 gene was removed from pDsred2 with XbaI (TaKaRa Bio, Inc., Tokyo, Japan). The resulting fragment was blunt ended and inserted into the SmaI site of pUNA, which contains the amyB promoter and niaD. The plasmid pUNDR, consisting of DsRed2 expressed under the control of the amyB promoter and the selective marker niaD, was introduced into A. oryzae NSCE1. The strain expressing CPY-EGFP and DsRed2 was named CEDR1.
Conidia from the CEDR1, CEDR8, GEGA8, and GDRA8 strains were cultured on a coverslip with 100 μl CD medium for 24 h at 30°C. The medium was replaced with fresh CD medium (control), CD−N or CD+R (for the induction of autophagy), and the cells were further incubated for 4 to 24 h at 30°C. The strains were then observed under a fluorescence microscope (BX52; Olympus Co., Tokyo, Japan). To visualize the autophagosomes and autophagic isolation membranes, the conidia of GEGA8 were cultured on a glass base dish (Asahi Techno Glass Co., Funabashi, Japan) under the above-mentioned conditions and observed with an IX71 confocal laser scanning microscope (Olympus). To observe the aerial hyphae and conidiophores, the conidia of CEDR1 were inoculated on minimal agar medium (CD) on glass slides and then coverslips were placed on top of the culture setup. After incubation for 3 days at 30°C, aerial hyphae and conidiophores elongating from the medium into the air were observed with a fluorescence microscope (23). The aerial hyphae and conidiophores of CEDR1 were observed by differential interference and fluorescence microscopy.
Construction of the Aoatg8 disruption mutant.
The plasmid pgΔAoatg8 was constructed to disrupt the Aoatg8 gene using the Multisite Gateway cloning system. The downstream region of the Aoatg8 gene (1.7 kb) was amplified by PCR using the primers attB2-downAoatg8-F (5′-GGGGACAGCTTTCTTGTACAAAGTGGGCTCTGATAAGCAGTTCTCC-3′) and attB3-downAoatg8-R (5′-GGGGACAACTTTGTATAATAAAGTTGATTGCGAGCAGCAGTCCA-3′), the sequences of which were based on the A. oryzae EST database. The underlined sequences are the Multisite Gateway attB recombination sites. The fragment was introduced into pDNORP2R-P3 (Invitrogen) using the Gateway BP clonase reaction, and the resulting plasmid was named pg3′downAoatg8. The Gateway LR reaction (Gateway LR clonase reaction mix; Invitrogen) was carried out with the entry clone plasmids pg3′downAoatg8 and pg5′PAoatg8 (mentioned above), the entry clone plasmid containing the A. oryzae adeA gene as a selective marker (constructed in our laboratory), and the destination vector pDESTR4-R3. The resultant plasmid was named pgΔAoatg8. Using the plasmid pgΔAoatg8 as a template, the sequence containing the deletion cassette was amplified by PCR with the primers attB4-PAoatg8-F and attB3-downAoatg8-R. The amplified deletion fragment contained the promoter region of Aoatg8 (1.7 kb), the adeA gene (2.0 kb), and the downstream region of Aoatg8 (1.7 kb) and was transformed into A. oryzae NSR13. The disruption of the Aoatg8 gene was confirmed by Southern blotting using a 1.2-kb fragment as a probe, which was generated by PCR with the primers pro4-F (5′-TATAGACCCGATATCACCGG-3′) and attB1-PAoatg8-R (5′-GGGGACTGCTTTTTTGTACAAACTTGATTGATGGATCGAATCAGTTAATGG-3′). To visualize autophagy in the strain carrying ΔAoatg8, ΔAoatg8-1-1, the DsRed2 protein-expressing plasmid pUNDR was introduced into this strain, which was named ΔAoatg8-1-1DR. For phenotypic analysis of ΔAoatg8-1-1, the hyphae were point inoculated on M medium with the addition of 0.15% methionine, dextrin-polypeptone-yeast extract (DPY), and potato dextrose (PD) (Nissui, Tokyo, Japan) agar media and incubated for 4 days at 30°C. To test whether autophagy was defective in the disruptant, it was grown on CD medium supplemented with 0.0015% methionine and then shifted to the autophagy-inducing conditions (described above). NSR13AA was used as a control.
Complementation of the Aoatg8 disruption mutant.
To complement the Aoatg8 disruption mutant, the plasmid pgPthiAAoatg8 was constructed using the Multisite Gateway cloning system as mentioned above. Three entry clone plasmids harboring the promoter region of thiA, the sequence containing the gene and terminator of Aoatg8 and niaD, respectively, were used for the Gateway LR reaction (Invitrogen). pgPthiAAoatg8 and pBAsC containing the A. oryzae sC gene were then introduced into ΔAoatg8-1-1. The strain obtained, CSC1-2-1, expresses AoAtg8 under the control of the thiA promoter; its conidia were subsequently used for germination analysis.
Conidial germination.
Conidia of CSC1-2-1 were inoculated onto a coverslip with 100 μl CD−N medium with or without thiamine (200 nM) and incubated at 30°C. The conidia were observed under a microscope for germ tube emergence.
RESULTS
Isolation of the A. oryzae Aoatg8 gene.
S. cerevisiae Atg8 is an essential protein in the autophagic pathway, and orthologues of Atg8 in other organisms have been used as specific markers of autophagy (8, 19, 27, 35). In order to isolate the A. oryzae ATG8 gene homologue, we searched the A. oryzae EST and genome databases using the BLAST algorithm. The ATG8 homologue in A. oryzae, Aoatg8, contains two introns and three exons and encodes a polypeptide of 118 amino acids with a calculated molecular mass of 14 kDa. AoAtg8 (DDBJ accession number AB246664) displays similarity to Atg8 of S. cerevisiae (79% identity) (14), AtAtg8a of Arabidopsis thaliana (NCBI accession number AAM70188) (73%), and LC3 of Homo sapiens (NCBI accession number NP_115093) (29%) and shows even higher similarity to its orthologues from the ascomycete fungi Aspergillus nidulans (NCBI accession number EAA62312) (98%) and P. anserina (98%) (27) and the basidiomycete fungus Ustilago maydis (NCBI accession number EAL64271) (86%).
Visualization of autophagy in A. oryzae.
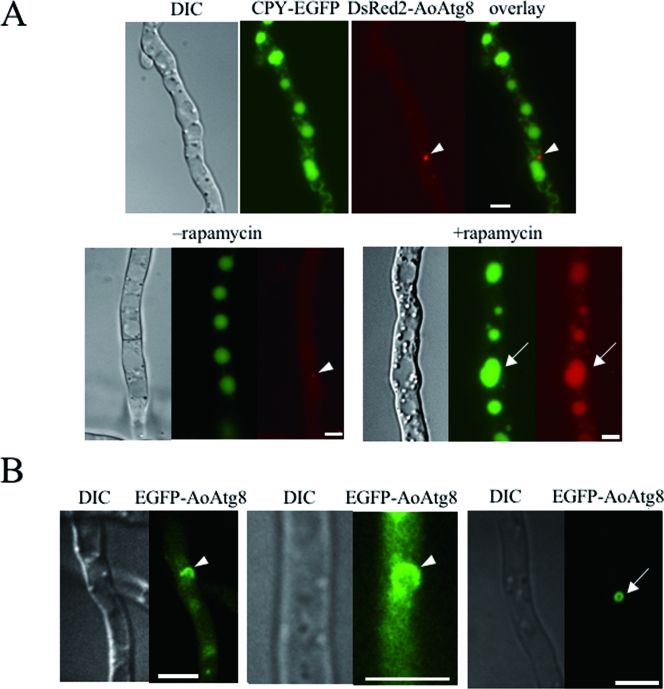
To visualize autophagy in A. oryzae, we constructed the CEDA8 strain coexpressing vacuolar carboxypeptidase Y (CPY)-EGFP and DsRed2-AoAtg8 fusion proteins. After growing for 24 h at 30°C in the CD medium, CEDA8 was cultured in the nitrogen-deprived medium (CD−N) or CD medium containing rapamycin (CD+R) to induce autophagy. During the growth in CD, DsRed2-AoAtg8 was localized to the PAS-like structures found in the vicinity of vacuoles (Fig. 1A). DsRed2-AoAtg8 was not localized in vacuoles when autophagy was not induced, although CPY-EGFP was observed in the vacuoles (Fig. 1A, lower left panels). When autophagy was induced, DsRed2-AoAtg8 was translocated to vacuoles (Fig. 1A, lower right panels). EGFP-AoAtg8 expressed in the GEGA8 strain showed behavior similar to that of DsRed2-AoAtg8 in CEDA8 (data not shown).
FIG. 1.
Localization of DsRed2-AoAtg8 and EGFP-AoAtg8 under starvation conditions. (A) The CEDA8 strain expressing CPY-EGFP and DsRed2-AoAtg8 was grown on CD medium (upper panels) and then shifted to CD medium with (+rapamycin) or without (−rapamycin) rapamycin (lower panels). After incubation for 4 h, hyphae were observed by DIC and fluorescence microscopy. The arrowheads and arrows indicate PAS-like structures and vacuoles, respectively. (B) The GEGA8 strain expressing EGFP-AoAtg8 was grown on CD medium for 24 h and then shifted to CD−N medium. After incubation for 2 h, hyphae were observed by DIC and confocal laser scanning microscopy. The arrowheads and the arrow indicate a cup-shaped and ring-like structure, respectively. Scale bars, 5 μm.
In order to find autophagic isolation membranes and autophagosomes in A. oryzae, EGFP-AoAtg8 was observed using a confocal laser scanning microscope. GEGA8 was cultured for 24 h at 30°C in CD medium on glass base dishes. When cells grown in CD were shifted to CD−N and cultured for an additional 2 h, ring-like and cup-shaped structures reminiscent of autophagosomes and isolation membranes, respectively, were observed (Fig. 1B). Moreover, some spherical structures were seen in the center of the ring-like and cup-shaped structures in differential interference contrast (DIC) images. In P. anserina, lipid bodies are accumulated during cell death by incompatibility (26). The spherical structures accumulated under starvation conditions or during treatment with rapamycin in CEDA8 (Fig. 1A, lower right panels) were also stained by Nile red (data not shown), indicating that the structures are most likely lipid droplets.
Induction of autophagy during the formation of aerial hyphae, conidiation, and conidial germination.
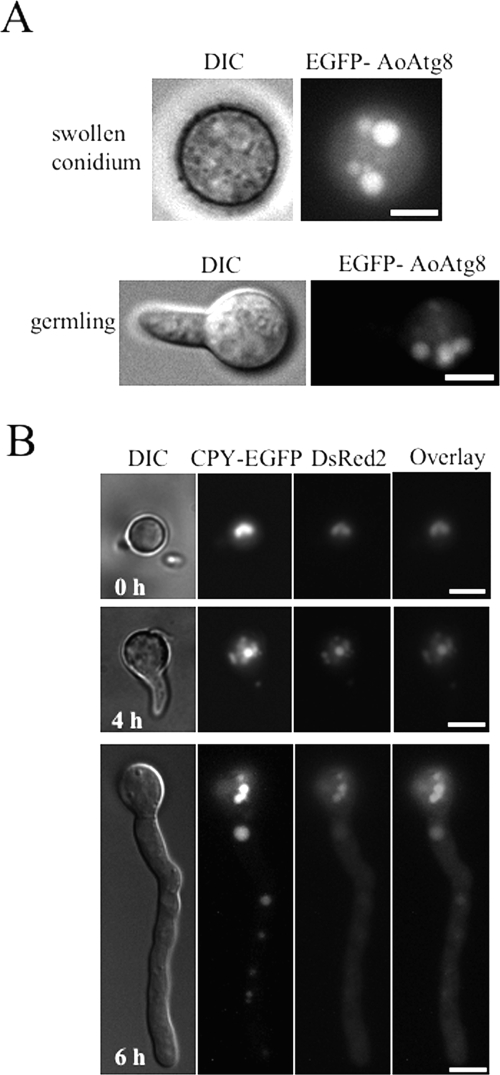
Recently, it has been suggested that autophagy is involved in development and differentiation in various eukaryotes (15). To observe autophagy in the development and differentiation of A. oryzae, conidia of GEGA8 expressing EGFP-AoAtg8 were cultured under noninduced conditions of autophagy (the presence of nitrogen sources and the absence of rapamycin). Interestingly, in swollen conidia, germlings, and germ tubes, EGFP-AoAtg8 was localized in vacuoles, even under nutrient-rich growth conditions (Fig. 2).
FIG. 2.
Localization of EGFP-AoAtg8 and DsRed2 during germination under normal growth conditions. (A) Conidia of the GEGA8 strain expressing EGFP-AoAtg8 were suspended in CD medium. After incubation for 7 h, swollen conidia and germlings were observed by DIC and fluorescence microscopy. (B) Conidia of the CEDR1 strain expressing CPY-EGFP and DsRed2 were suspended in CD medium. After incubation for the indicated time periods, the conidia and hyphae were observed by DIC and fluorescence microscopy. Scale bars, 5 μm.
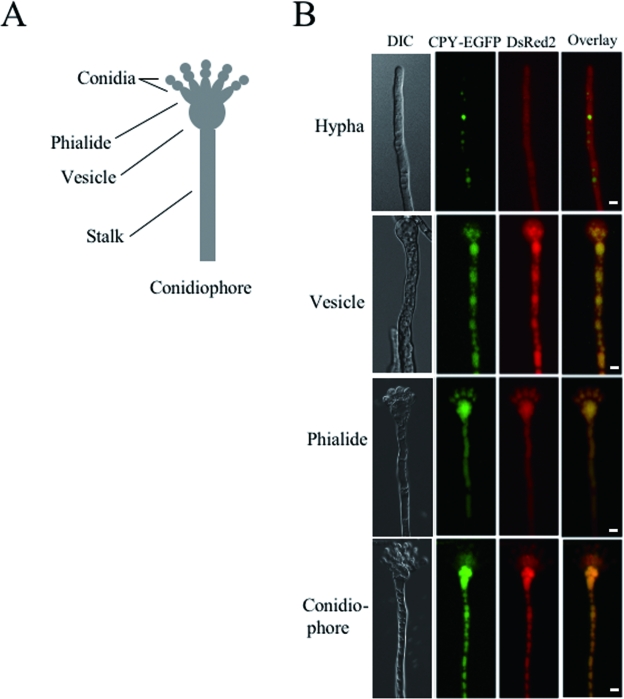
When autophagy is induced, the isolation membranes sequester cytoplasm and organelles nonselectively (12, 13). Therefore, we predicted that autophagy could be observed by detecting fluorescent proteins inside vacuoles, which had been incorporated from the cytoplasm. We obtained the CEDR1 strain, which constitutively expresses CPY-EGFP in vacuoles and DsRed2 protein in the cytoplasm. DsRed2 in CEDR1 was observed in vacuoles 12 h after the shift to the starvation condition (see Fig. 4D, left panels). In A. oryzae, conidiation occurs through the characteristic morphogenesis of differentiated cells. Aerial hyphae are formed from foot cells, and some differentiate into conidiophores, which are composed of vesicles, phialides, and conidia (Fig. 3A). To investigate whether autophagy is induced in differentiated cells of the aerial hyphae, CEDR1 was grown on minimal agar medium (CD) on glass slides for 3 days at 30°C and observed with a fluorescence microscope. In conidiophores growing on CD medium, DsRed2 was localized to vacuoles in stalk, conidiophore vesicles, and phialides, whereas it was localized to cytoplasm in aerial hyphae that have not developed into conidiophores (Fig. 3B). These results suggest that autophagy is involved in conidiation and the formation of conidiophores.
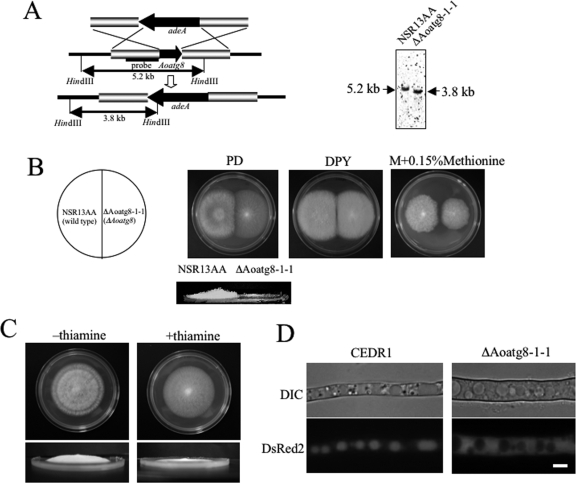
FIG. 4.
Disruption of A. oryzae Aoatg8 gene. (A) Schema for the integration of the adeA gene (left panel). Southern blot analysis was performed with the genomic DNA of the NSR13AA (wild type) and ΔAoatg8-1-1 strains (right panel). The 1.2-kb fragment carrying a portion of the promoter region of Aoatg8 was used as a probe in hybridization. The HindIII fragments in ΔAoatg8-1-1 and the wild type were 3.8 kb and 5.2 kb, respectively. (B) NSR13AA and ΔAoatg8-1-1 strains were grown on PD, DPY, and M medium with 0.15% methionine (M plus 0.15% methionine) agar plates for 4 days at 30°C. The lower panel shows the side view of the NSR13AA and ΔAoatg8-1-1 strains grown on a PD plate. (C) The CSC1-2-1 strain expressing AoAtg8 under the control of the thiA promoter was grown on PD agar plates with (+thiamine) or without (−thiamine) thiamine for 4 days at 30°C. The lower panels show the side view of the strain. (D) The CEDR1 strain expressing CPY-EGFP and DsRed2 (left panels) and ΔAoatg8-1-1DR expressing DsRed2 (right panels) were grown on CD medium for 24 h. The medium was replaced with CD−N and the cells were further incubated for 12 h at 30°C. The cells were then observed by DIC (upper panels) and fluorescence microscopy (lower panels). Scale bar, 5 μm.
FIG. 3.
Localization of DsRed2 in conidiophores. (A) Schematic figure of the A. oryzae conidiophore. (B) The CEDR1 strain expressing CPY-EGFP and DsRed2 was grown on CD medium containing 1.5% agar for 3 days. Aerial hyphae and conidiophores were observed by DIC and fluorescence microscopy. Scale bar, 5 μm.
Phenotypes of Aoatg8 gene disruption mutants in A. oryzae.
In order to examine the function of autophagy in A. oryzae, we constructed strains disrupted for the Aoatg8 gene by replacement with the selective marker adeA (Fig. 4A). Disruption was confirmed by PCR and Southern blot analyses (Fig. 4A). Hyphae of the disruptant, ΔAoatg8-1-1, were grown on PD, DPY, and M plus 0.15% methionine agar media for 4 days at 30°C. ΔAoatg8-1-1 formed no aerial hyphae and conidia and showed slower growth on synthetic medium than the wild-type strain did (Fig. 4B). Conidiation is usually enhanced when A. oryzae is grown on PD medium; therefore, differences in the formation of conidia and aerial hyphae were clearly identified between ΔAoatg8-1-1 and the control strain (Fig. 4B, lower panel). These phenotypes were complemented by the introduction of a plasmid for the conditional expression of AoAtg8 under the control of the thiA promoter into ΔAoatg8-1-1. The thiA promoter is controlled by the addition of thiamine: in the absence of thiamine, the expression of a gene controlled by the thiA promoter is induced, whereas in the presence of 100 nM thiamine, it is repressed (32). This strain, CSC1-2-1, was grown on a PD medium plate with or without thiamine. In the absence of thiamine, the formation of aerial hyphae and conidiation occurred; however, in the presence of thiamine, CSC1-2-1 displayed a phenotype similar to that of ΔAoatg8-1-1 (Fig. 4C). These results suggest that Aoatg8 is involved in the formation of aerial hyphae. Moreover, conidia of the wild-type strain germinated on CD−N medium and then formed a mycelial colony. Similarly, CSC1-2-1 elongated the hyphae on CD−N medium containing thiamine but the differentiation of aerial hyphae and conidiation was inhibited and its mycelial colony was larger than that of the wild-type strain (data not shown).
Next, we tested whether ΔAoatg8-1-1 displays defects in autophagy. To visualize autophagy in ΔAoatg8-1-1, we constructed ΔAoatg8-1-1DR expressing DsRed2 in the cytoplasm. ΔAoatg8-1-1DR showed a phenotype similar to that of ΔAoatg8-1-1. During autophagy, DsRed2 expressed in the cytoplasm in CEDR1 was incorporated into autophagosomes and transported to vacuoles (Fig. 4D). By contrast, in ΔAoatg8-1-1DR, DsRed2 was homogeneously distributed in the cytoplasm under starvation conditions. This observation suggests that ΔAoatg8-1-1DR is defective in autophagy and AoAtg8 is essential for autophagy in A. oryzae.
Conidial germination of the Aoatg8 conditional mutant.
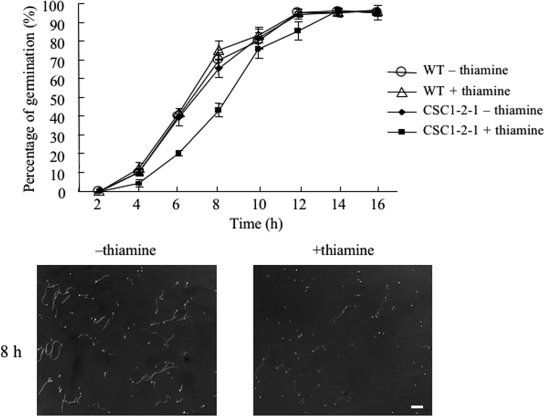
Analysis of the phenotype of ΔAoatg8-1-1 suggested that autophagy is involved in the formation of conidia and aerial hyphae. Moreover, the localization of EGFP-AoAtg8 during germination implied that autophagy is involved in conidial germination (Fig. 2 and 3). To investigate the role of autophagy in conidial germination, conidia were harvested from the CSC1-2-1 strain grown on a CD medium plate with 50 nM thiamine because the expression of AoAtg8 was decreased as much as possible. Conidia of CSC1-2-1 were cultured in liquid CD−N medium with or without thiamine, and the number of germ tubes formed was counted under a microscope (Fig. 5). The conidia started to germinate 4 h after inoculation. In the absence of thiamine, 65% of the conidia formed germ tubes by 8 h (Fig. 5, lower left panel). By contrast, when conidia were inoculated in CD−N containing thiamine, conidial germination was delayed: 40% of the conidia formed germ tubes by 8 h (Fig. 5, lower right panel). However, more than 90% of the conidia formed germ tubes 16 h after inoculation. Thus, conidial viability was not affected by the defect in autophagy. These data suggest that autophagy functions partially during the early stage of conidial germination.
FIG. 5.
Conidial germination of the CSC1-2-1 strain. Kinetics of germ tube outgrowth in the wild type and the CSC1-2-1 strain are shown. Conidia of these strains were inoculated onto a coverslip with 100 μl CD−N medium, with or without thiamine, and incubated at 30°C. The conidia were observed under a microscope for germ tube emergence at the indicated time points. All values are the means and standard deviations of three independent experiments. Photographs show conidial germination of the CSC1-2-1 strain after 8 h of incubation on CD−N (left panel) or CD−N with thiamine (right panel). Scale bars, 50 μm.
DISCUSSION
Autophagy is a degradation process using vacuolar enzymes and primarily functions as a survival strategy during nutrient starvation and for the turnover of nonselective cytoplasmic contents. We isolated the A. oryzae ATG8 homologue Aoatg8 as a molecular marker and visualized autophagy in A. oryzae using fluorescent proteins. AoAtg8 is composed of a polypeptide of 118 amino acids, and its sequence is highly conserved among other eukaryotes, including humans and plants. In S. cerevisiae, the carboxy (C)-terminal glycine of Atg8 conjugates with phosphatidylethanolamine mediated by a ubiquitination-like system (5). This lipidation is essential for autophagy (10). In A. oryzae, AoAtg8 also has a conserved glycine residue at the C terminus, suggesting the evolutionary conservation of this conjugation system.
We visualized autophagy by expressing EGFP-AoAtg8 and DsRed2-AoAtg8 fusion proteins. Furthermore, we constructed complemented strains expressing EGFP-AoAtg8 in the ΔAoatg8 mutants. In these strains, the phenotypes of the ΔAoatg8 mutants were restored, and EGFP-AoAtg8 was localized in vacuoles under starvation conditions and during conidial germination (data not shown). Therefore, EGFP-AoAtg8 was functional and behaved in a manner similar to that of the native AoAtg8 protein. Our observations suggest that a similar system of autophagy takes place in the deuteromycete A. oryzae. In addition, we found that DsRed2 and EGFP-AoAtg8 were taken up into vacuoles in conidiophores, swollen conidia, and germlings under growth conditions. Thus, we propose that autophagy occurs during asexual differentiation and conidial germination in A. oryzae.
Under starvation conditions, DsRed2 expressed in the cytoplasm was taken up into vacuoles, indicating the induction of autophagy. Therefore, autophagy can be detected by using this visualization method much more conveniently than by using an electron microscope. AoAtg8 was localized in the vacuoles within 4 h, whereas DsRed2 fluorescence in the vacuoles was observed 12 h after the shift to starvation conditions. The reason for this time difference might be the increased time needed for the fluorescence intensity inside the vacuoles to exceed that in the cytoplasm in CEDR1 during autophagy.
In P. anserina, Δidi-7/ΔPaATG8 mutants show defects in the differentiation of aerial hyphae and the protoperithecia, which is a female reproductive organ (27). By contrast, A. oryzae has no sexual reproduction, and conidia are the sole reproductive organs. Here, although the differentiation of aerial hyphae and conidiation in the ΔAoatg8 mutants were defective, the mutants could grow on minimal medium without any nitrogen source and the mycelial colony was large compared with that of the control strain. This might have been due to a defect in the differentiation of aerial hyphae; vegetative hyphae searched for nutrients around the colony because the mutants could not develop aerial hyphae or conidiophores, which were induced by nutrient starvation. Therefore, unlike P. anserina, the defect in conidiation in A. oryzae ΔAoatg8-1-1 prohibited it from producing progeny. Thus, the effect of the deletion of Aoatg8 was more severe in A. oryzae in extreme environments (such as limited nutrients) than in P. anserina.
In A. nidulans, brlA mutants develop abnormal conidiophore stalks that are 20 to 30 times longer than those of the wild type and give the colony a “bristle” phenotype (1). Moreover, chitin synthase chsA disruptants in A. nidulans form normal aerial hyphae but cannot form conidiophores (3). In the current paper, we show that a strain defective in autophagy (ΔAoatg8-1-1) failed to form aerial hyphae and that DsRed2 expressed in the cytoplasm in CEDR1 was taken up into the vacuole during conidiophore formation. Therefore, the defect in conidiation might have been due to defects in aerial hyphae or conidiophore stalk formation, rather than in conidiophore-specific cells, such as vesicles, phialides and metulae, suggesting that autophagy is involved in the development of both aerial hyphae and conidiophores.
Thus, the formation of aerial hyphae in filamentous fungi is regulated by various factors. Although the involvement of autophagy during the differentiation of aerial hyphae and conidiophores is not yet clearly understood, autophagy might take part in the reconstitution of intracellular components during aerial hypha and conidiophore formation. Furthermore, it is thought that cells that are not in contact with the medium might acquire nutrients through the recycling of intracellular components by autophagy.
In Neurospora crassa, conidial germination requires a carbon source and a salt (30). In A. nidulans, conidial germination occurs in the presence of a carbon source alone (21, 24). We examined germination in A. oryzae and observed that conidial germination of the wild type was induced on CD medium lacking a nitrogen source (CD−N). This suggests that the nitrogen source might be stored in the conidia for germination. Furthermore, we showed that on CD−N, conidial germination of CSC1-2-1 was delayed, suggesting that autophagy was at least partly involved in supplying a nitrogen source at an early stage of conidial germination. This result agrees with the localization of AoAtg8 and DsRed2 during conidial germination. In A. nidulans and Aspergillus fumigatus, the Ras signaling pathway, which plays an important role in the control of cell growth and response to nutrients, is involved in germination and autophagy (2, 4, 24). Furthermore, in S. cerevisiae, a relationship has been reported between the Ras signaling pathway and the Tor kinase (central negative regulator of autophagy) (29), which supports our data. Thus, the involvement of autophagy in conidial germination has been demonstrated for the first time in filamentous fungi.
How autophagy plays a role in differentiation and development is of interest and remains an important question. Because yeast is unicellular, an analysis of the relationship between autophagy and differentiation is difficult. Thus, we predict that investigations of the multicellular organism A. oryzae will be more useful than those involving yeast in elucidating an overview of autophagy.
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research (B) (no. 15380058) to K. Kitamoto from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and by a program for the promotion of basic research activities for innovative biosciences of the Bio-Oriented Technology Research Advancement Institution.
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J.-H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budovskaya, Y. V., J. S. Stephan, F. Reggiori, D. J. Klionsky, and P. K. Herman. 2004. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279:20663-20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culp, D. W., C. L. Dodge, Y. Miao, L. Li, D. Sag-Ozkal, and P. T. Borgia. 2000. The chsA gene from Aspergillus nidulans is necessary for maximal conidiation. FEMS Microbiol. Lett. 182:349-353. [DOI] [PubMed] [Google Scholar]
- 4.Fortwendel, J. R., J. C. Panepinto, A. E. Seitz, D. S. Askew, and J. C. Rhodes. 2004. Aspergillus fumigatus rasA and rasB regulate the timing and morphology of asexual development. Fungal Genet. Biol. 41:129-139. [DOI] [PubMed] [Google Scholar]
- 5.Ichimura, Y., T. Kirisako, T. Takao, Y. Satomi, Y. Shimonishi, N. Ishihara, N. Mizushima, I. Tanida, E. Kominami, M. Ohsumi, T. Noda, and Y. Ohsumi. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408:488-492. [DOI] [PubMed] [Google Scholar]
- 6.Jin, F. J., J. Maruyama, P. R. Juvvadi, M. Arioka, and K. Kitamoto. 2004. Adenine auxotrophic mutants of Aspergillus oryzae: development of a novel transformation system with triple auxotrophic hosts. Biosci. Biotechnol. Biochem. 68:656-662. [DOI] [PubMed] [Google Scholar]
- 7.Jin, F. J., J. Maruyama, P. R. Juvvadi, M. Arioka, and K. Kitamoto. 2004. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol. Lett. 239:79-85. [DOI] [PubMed] [Google Scholar]
- 8.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirisako, T., M. Baba, N. Ishihara, K. Miyazawa, M. Ohsumi, T. Yoshimori, T. Noda, and Y. Ohsumi. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirisako, T., Y. Ichimura, H. Okada, Y. Kabeya, N. Mizushima, T. Yoshimori, M. Ohsumi, T. Takao, T. Noda, and Y. Ohsumi. 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151:263-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitamoto, K., K. Gomi, K. Goto, and S. Hara. 1991. Genetic transfer applied to traditional sake brewing. Biotechnol. Genet. Eng. Rev. 9:89-125. [PubMed] [Google Scholar]
- 12.Klionsky, D. J., and Y. Ohsumi. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:1-32. [DOI] [PubMed] [Google Scholar]
- 13.Klionsky, D. J., and S. D. Emr. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290:1717-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang, T., E. Schaeffeler, D. Bernreuther, M. Bredschneider, D. H. Wolf, and M. Thumm. 1998. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 17:3597-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine, B., and D. J. Klionsky. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6:463-477. [DOI] [PubMed] [Google Scholar]
- 16.Mabashi, Y., T. Kikuma, J. Maruyama, M. Arioka, and K. Kitamoto. Biosci. Biotechnol. Biochem., in press. [DOI] [PubMed]
- 17.Meléndez, A., Z. Tallóczy, M. Seaman, E.-L. Eskelinen, D. H. Hall, and B. Levine. 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301:1387-1391. [DOI] [PubMed] [Google Scholar]
- 18.Minetoki, T., Y. Nunokawa, K. Gomi, K. Kitamoto, C. Kumagai, and G. Tamura. 1996. Deletion analysis of promoter elements of the Aspergillus oryzae agdA gene encoding alpha-glucosidase. Curr. Genet. 30:432-438. [DOI] [PubMed] [Google Scholar]
- 19.Monastyrska, I., M. van der Heide, A. M. Krikken, J. A. Kiel, I. J. van der Klei, and M. Veenhuis. 2005. Atg8 is essential for macropexophagy in Hansenula polymorpha. Traffic 6:66-74. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa, I., A. Amano, N. Mizushima, A. Yamamoto, H. Yamaguchi, T. Kamimoto, A. Nara, J. Funao, M. Nakata, K. Tsuda, S. Hamada, and T. Yoshimori. 2004. Autophagy defends cells against invading group A Streptococcus. Science 306:1037-1040. [DOI] [PubMed] [Google Scholar]
- 21.Ni, M., S. Rierson, J.-A. Seo, and J.-H. Yu. 2005. The pkaB gene encoding the secondary protein kinase A catalytic subunit has a synthetic lethal interaction with pkaA and plays overlapping and opposite roles in Aspergillus nidulans. Eukaryot. Cell 4:1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa, M., T. Yoshimori, T. Suzuki, H. Sagara, N. Mizushima, and C. Sasakawa. 2005. Escape of intracellular Shigella from autophagy. Science 307:727-731. [DOI] [PubMed] [Google Scholar]
- 23.Ohneda, M., M. Arioka, H. Nakajima, and K. Kitamoto. 2002. Visualization of vacuoles in Aspergillus oryzae by expression of CPY-EGFP. Fungal Genet. Biol. 37:29-38. [DOI] [PubMed] [Google Scholar]
- 24.Osherov, N., and G. May. 2000. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 155:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otto, G. P., M. Y. Wu, N. Kazgan, O. R. Anderson, and R. H. Kessin. 2003. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J. Biol. Chem. 278:17636-17645. [DOI] [PubMed] [Google Scholar]
- 26.Pinan-Lucarré, B., A. Balguerie, and C. Clavé. 2005. Accelerated cell death in Podospora autophagy mutants. Eukaryot. Cell 4:1765-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinan-Lucarré, B., M. Paoletti, K. Dementhon, B. Coulary-Salin, and C. Clavé. 2003. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol. Microbiol. 47:321-333. [DOI] [PubMed] [Google Scholar]
- 28.Punt, P. J., N. van Biezen, A. Conesa, A. Albers, J. Mangnus, and C. van den Hondel. 2002. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20:200-206. [DOI] [PubMed] [Google Scholar]
- 29.Schmelzle, T., T. Beck, D. E. Martin, and M. N. Hall. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24:338-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmit, J. C., and S. Brody. 1976. Biochemical genetics of Neurospora crassa conidial germination. Bacteriol. Rev. 40:1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shintani, T., and D. J. Klionsky. 2004. Autophagy in health and disease: a double-edged sword. Science 306:990-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoji, J. Y., J. Maruyama, M. Arioka, and K. Kitamoto. 2004. Development of Aspergillus oryzae thiA promoter as a tool for molecular biological studies. FEMS Microbiol. Lett. 244:41-46. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, K., T. Kirisako, Y. Kamada, N. Mizushima, T. Noda, and Y. Ohsumi. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20:5971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169-174. [DOI] [PubMed] [Google Scholar]
- 35.Yoshimoto, K., H. Hanaoka, S. Sato, T. Kato, S. Tabata, T. Noda, and Y. Ohsumi. 2004. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16:2967-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]