Abstract
In the tsetse fly, the protozoan parasite Trypanosoma congolense is covered by a dense layer of glycosylphosphatidylinositol (GPI)-anchored molecules. These include a protease-resistant surface molecule (PRS), which is expressed by procyclic forms early in infection, and a glutamic acid- and alanine-rich protein (GARP), which appears at later stages. Since neither of these surface antigens is expressed at intermediate stages, we investigated whether a GPI-anchored protein of 50 to 58 kDa, previously detected in procyclic culture forms, might constitute the coat of these parasites. We therefore partially purified the protein from T. congolense Kilifi procyclic forms, obtained an N-terminal amino acid sequence, and identified its gene. Detailed analyses showed that the mature protein consists almost exclusively of 13 heptapeptide repeats (EPGENGT). The protein is densely N glycosylated, with up to 13 high-mannose oligosaccharides ranging from Man5GlcNAc2 to Man9GlcNAc2 linked to the peptide repeats. The lipid moiety of the glycosylphosphatidylinositol is composed of sn-1-stearoyl-2-lyso-glycerol-3-HPO4-1-(2-O-acyl)-d-myo-inositol. Heavily glycosylated proteins with similar repeats were subsequently identified in T. congolense Savannah procyclic forms. Collectively, this group of proteins was named T. congolense procyclins to reflect their relationship to the EP and GPEET procyclins of T. brucei. Using an antiserum raised against the EPGENGT repeat, we show that T. congolense procyclins are expressed continuously in the fly midgut and thus form the surface coat of cells that are negative for both PRS and GARP.
At all stages during the life cycles of African trypanosomes, the parasite surface is covered by glycosylphosphatidylinositol (GPI)-anchored molecules. In the mammalian host, bloodstream-form trypanosomes express a dense layer of variant surface glycoproteins. The periodic switching of expression between immunologically distinct antigens, which are encoded by several hundred genes, allows the parasite to escape the mammalian immune system (4, 7, 13, 30). After being taken up by the tsetse fly vector as part of a blood meal, bloodstream forms rapidly differentiate to procyclic forms and express a more restricted set of surface molecules (30, 33). In Trypanosoma brucei procyclic forms, the surface coat consists of two related types of proteins, called procyclins, which contain extensive tandem repeat units of glutamic acid and proline (EP) or five to six GPEET pentapeptide repeats (9, 32). The expression profiles of the procyclins change during development of the parasite in the tsetse fly: initially, both EP and GPEET are expressed on the surface of early procyclic forms, but after several days in the fly midgut, GPEET is repressed and is replaced by the glycosylated forms of EP (2, 42). This down-regulation of GPEET can also be observed to occur in culture (44). Recently, it was shown that, in addition to the GPI-anchored coat proteins, T. brucei procyclic forms in culture express free GPIs on their surface; these represent the major surface molecules in procyclin null mutants (43).
Despite the fact that T. congolense is of much greater importance than T. brucei as a pathogen for animal trypanosomiasis (nagana) (39), relatively little is known about the surface composition of the life cycle stages in the tsetse fly. Over a decade ago, two groups simultaneously identified the first major surface antigen in T. congolense procyclic culture forms and named it GARP for glutamate- and alanine-rich protein (5, 6). Only recently, this protein has been shown to be highly conserved among the T. congolense subgroups Savannah, Forest, and Kilifi and to be present in other trypanosomes of the subgenus Nannomonas (3). GARP, in contrast to the T. brucei procyclins, contains no amino acid repeats in the primary sequence, and yet the proteins from the two strains have been proposed to be functional equivalents because they share the properties of surface orientation, acidity, immunodominance, and stage specificity (5, 6, 21, 36). In addition, they are attached to the cell membrane via similar GPI anchors (40, 41). At the nucleic acid level, GARP and the T. brucei procyclins share a conserved stretch of 16 nucleotides in the 3′ untranslated region, the so-called 16-mer region (5, 24). This region, which is predicted to adopt similar secondary structures in the mRNAs of both trypanosome species, is known to affect procyclin RNA stability and translation in T. brucei (20, 22, 38).
T. congolense procyclic culture forms express two additional GPI-anchored surface molecules besides GARP: a protease-resistant surface molecule (PRS) with an apparent molecular mass of 24 to 34 kDa, which may be nonproteinaceous, and a protein of approximately 58 kDa in T. congolense Kilifi or 50 kDa in T. congolense Savannah (10). In common with the EP and GPEET procyclins in T. brucei, the relative expression of PRS and GARP in T. congolense changes during parasite development in the tsetse fly. PRS is strongly expressed in early procyclic forms in the fly midgut but absent from the epimastigote form in the proboscis, whereas GARP is absent or only weakly expressed in early-stage procyclic forms but abundant in epimastigotes (10). Since epimastigote forms develop in the proboscis in T. congolense but in the salivary glands in T. brucei (45), it is possible that surface molecules are involved in tropism of the parasites within their common insect host, the tsetse fly.
The identities and expression profiles of the 58- and 50-kDa proteins in T. congolense Kilifi and Savannah strains, respectively, are completely unknown. Since established procyclic forms of T. congolense in the fly midgut are negative for both PRS and GARP (10), we hypothesized that the third GPI-anchored molecule might represent the major coat protein of the parasite during this stage of the parasite life cycle. Here we show that this is indeed the case and that the 58- and 50-kDa GPI-anchored proteins from T. congolense Kilifi and Savannah procyclic forms, respectively, consist almost entirely of long heptapeptide repeats (EPGENGT), thereby closely resembling the T. brucei procyclins. Interestingly, the repeats were found to be modified by N-linked carbohydrate structures and possibly phosphodiester-linked glycans, rendering the molecules some of the most densely glycosylated proteins known to date.
MATERIALS AND METHODS
Unless otherwise specified, all reagents were of analytical grade and were purchased from Fluka (Buchs, Switzerland), Sigma (St. Louis, MO), Merck (Darmstadt, Germany), or Invitrogen (Basel, Switzerland). [1-3H]ethan-1-ol-2-amine hydrochloride ([3H]ethanolamine; 18 to 29 Ci mmol−1) was from Amersham (Zürich, Switzerland). Acrylamide solution was purchased from National Diagnostics (Hull, United Kingdom) and Tris-HCl and glycine from ICN (Tägerig, Switzerland). Octyl-Sepharose was from Sigma and trypsin from Roche Applied Science (Rotkreuz, Switzerland). Thermus aquaticus DNA polymerase was purchased from QIAGEN (Basel, Switzerland) and deoxynucleoside triphosphate mix from Invitrogen. Protran BA 85 filters were from Schleicher & Schuell (Dassel, Germany).
Antibodies.
Polyclonal rabbit antibodies against T. congolense procyclin (termed CP1) were raised against a 1:1 mixture of a synthetic peptide, AD(EPGENGT)2C, coupled N or C terminally to keyhole limpet hemocyanin (Affiniti Research Products Ltd., Exeter, United Kingdom). A polyclonal rabbit antibody against a GARP-glutathione S-transferase fusion protein (α-GARP) was generously provided by J. D. Barry (Wellcome Centre of Molecular Parasitology, Glasgow, Scotland). Monoclonal antibody (MAb) no. 491 was raised against live T. congolense Kilifi 45/1 procyclic forms and recognizes T. congolense PRS (10). Horseradish peroxidase-conjugated swine anti-rabbit antibodies were from DAKO (Glostrup, Denmark) and Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G and Alexa Fluor 568-conjugated goat anti-mouse immunoglobulin G from Molecular Probes (Eugene, OR).
Trypanosomes.
Procyclic forms of T. congolense Kilifi STIB745 and T. congolense Savannah TREU1457 were provided by the Swiss Tropical Institute (Basel, Switzerland) and cultured at 27°C in a 1:1 mixture of SM (14) and SDM-79 (8) containing 15% heat-inactivated fetal bovine serum (Gibco BRL, Basel, Switzerland).
Labeling and isolation of GPI-anchored molecules.
GPI-anchored molecules of T. congolense procyclic culture forms were radioactively labeled with [3H]ethanolamine exactly as described before (9). Subsequently, trypanosomes (1 × 109 to 3 × 109 cells) were centrifuged, washed, and sequentially extracted with chloroform:methanol (2:1, vol/vol), chloroform:methanol:water (CMW) (10:10:3, vol/vol/vol), 9% (vol/vol) butan-1-ol in water (butanol extract), 0.1% (wt/vol) Triton X-100 in 20 mM Tris-HCl, pH 7.4 (Triton extract), and 1% (wt/vol) sodium dodecyl sulfate (SDS) (SDS extract), as described before (9, 10). The pooled CMW-soluble fractions were dried and partitioned between butan-1-ol and water (43). Aliquots of the extracts were stored at −20°C.
Extracts of interest were fractionated on an octyl-Sepharose column by using a linear gradient of 25 to 40% (vol/vol) propan-1-ol in 0.1 M ammonium acetate (9). Aliquots of all fractions were counted for radioactivity, and the fractions of interest were pooled and stored at 4°C until use.
Amino acid sequencing and design of an oligonucleotide probe.
Octyl-Sepharose-purified material from T. congolense Kilifi procyclic forms was desalted using a ProSorb column and sequenced on an Applied Biosystems Procise 492-cLC sequencer. Based on the amino acid sequence information, an oligonucleotide (Tc58) with the sequence 5′-GAYGARCCNGGNGARGARGG-3′ was synthesized commercially (Microsynth GmbH, Balgach, Switzerland).
cDNA library.
Total RNA was extracted from T. congolense Kilifi procyclic culture forms by use of the hot phenol-SDS method (34). Poly(A) mRNA was isolated from total RNA by use of an Oligotex mRNA kit (QIAGEN). A cDNA library was made using ZAP Express cDNA synthesis and Gigapack III Gold cloning kits from Stratagene (La Jolla, CA) according to the manufacturer's instructions. The library was screened by hybridization using the 5′-[γ-32P]-labeled oligonucleotide Tc58: the filters were prehybridized for 5 h at 37°C in hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS, 0.1% sodium pyrophosphate, 5× Denhardt's reagent, 100 μg/ml herring sperm DNA) (37) before the labeled probe was added and hybridized overnight at 37°C. The nitrocellulose filters were washed three times for 20 min each at room temperature in 4× SSC and 0.05% SDS. Positive clones were identified by autoradiography; inserts were amplified by PCR using primers T7 and T3 (Microsynth GmbH). PCR was performed for 30 cycles under the following conditions: 94°C for 1 min, 50°C for 1 min, and 74°C for 3 min.
For the identification of the corresponding gene(s) in T. congolense Savannah, total RNA was extracted as described above. An aliquot (4.5 μg) was transcribed into cDNA using oligo(dT)12-18 primers and SuperScript II reverse transcriptase according to the manufacturer's instructions (Invitrogen). PCR was performed using primers containing a conserved region of the miniexon (spliced leader sequence) (MEcong, 5′-AGTTTCTGTACTATATTG-3′) and the conserved 16-mer region (16cong, 5′-AGAATTCTACAGGGCT-3′) for 35 cycles under the following conditions: 94°C for 1 min, 44°C for 1 min, and 72°C for 3 min. The PCR products were cloned using a Topo-TA cloning kit (Invitrogen) according to the manufacturer's instructions. All DNA sequencing was carried out commercially (Microsynth GmbH).
DNA extraction and Southern blotting.
Genomic DNA from T. congolense Kilifi was analyzed by Southern blotting following standard protocols (37). For hybridization using digoxigenin (DIG)-labeled plasmid DNA F1/1 and detection, a PCR DIG probe synthesis kit, DIG easy hyb, DIG wash and block buffer set, anti-digoxigenin-AP, and CDP-Star (Roche Applied Science) were used according to the manufacturers' instructions.
SDS-PAGE and immunoblotting.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed under reducing conditions (26) on 12% polyacrylamide gels. For fluorography, gels were soaked in Amplify (Amersham), dried, and exposed to Kodak X-Omat S films (Integra Biosciences) at −70°C. Semidry blotting of antigens from polyacrylamide gels onto polyvinylidene difluoride membranes (Immobilon P; Millipore Corp., Bedford, MA) and subsequent blocking of membranes with milk powder, followed by antibody incubation and detection of antigen binding using an enhanced chemiluminescence detection system (Pierce Chemicals Co., Rockford, IL), were done as described before (21). The CP1 antiserum was used at a dilution of 1:100. Binding of concanavalin A (ConA) to T. congolense molecules was analyzed by incubating the blotted membranes two times for 1 h each at room temperature with 0.4 μg/ml biotinylated ConA in 10 mM Tris, pH 7.4, 140 mM NaCl, 0.05% (wt/vol) Tween 20, followed by horseradish peroxidase-conjugated streptavidin (DAKO) at a dilution of 1:10,000 in the same buffer, and detection by enhanced chemiluminescence.
For silver staining, the gel was fixed for 45 min in acetic acid:ethanol:water (1:4:5, vol/vol/vol) and subsequently washed six times for 10 min each in water and incubated for 30 min in reacting solution (1 mg dithiothreitol in 200 ml water). After incubation for 30 min in silver solution (0.1% silver nitrate in water), the gel was washed in water for 60 s and then preincubated for a few seconds in 50 ml developing solution (6 g sodium carbonate and 75 μl 37% formaldehyde in 200 ml H2O) to reduce background. The developing solution was removed, and the gel was finally incubated for 1 to 2 min under gentle shaking in 150 ml developing solution. The staining reaction was stopped by the addition of solid citric acid.
MALDI-TOF MS.
An octyl-Sepharose-purified butanol extract from 2.5 × 108 T. congolense Kilifi procyclic forms was freeze-dried and treated with 25 μl ice-cold 48% aqueous hydrogen fluoride (aqHF) for 24 h at 0°C to cleave the ethanolamine-phosphate bond in the GPI anchor. After freeze-drying, the sample was deglycosylated with either peptide N4(N-acetyl-β-glucosaminyl) asparagine amidase F (PNGase F) or endo-β-N-acetylglucosaminidase H (endo-H) (see below) and processed for mass spectrometry analysis. Aliquots (0.5 μl, approximately 5 × 106 parasite equivalents) of each sample were mixed with 0.5 μl 10 mg/ml sinapinic acid in 70% acetonitrile and 0.1% trifluoroacetic acid and analyzed by negative-ion-mode matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Data collection was done in linear mode with a PerSeptive Biosystems Voyager-DE mass spectrometer. The accelerating voltage was 2,500 V, and grid voltage was set at 94%, with an extraction time delay of 700 ns. Data were collected manually at 200 shots per spectrum, with laser intensity set at 2,800.
Enzymatic deglycosylation of procyclins. (i) Deglycosylation for MALDI-TOF MS analysis.
An aliquot of aqHF-treated procyclin (approximately 2.5 × 108 parasite equivalents) was deglycosylated with 250 U of PNGase F (New England Biolabs) in 10 μl of 25 mM sodium phosphate, pH 7.5, at 37°C for 18 h. To confirm the occupancy of the glycosylation sites on T. congolense procyclin, aqHF-treated proteins (approximately 2.5 × 108 parasite equivalents) were incubated with 100 U of recombinant endo-H (New England Biolabs) in 20 μl of 50 mM sodium citrate, pH 5.5, at 37°C for 18 h. After digestion, samples were desalted using a ZipTip (containing C18 silica; Millipore Corp.) as described by the manufacturer, and an aliquot was analyzed by MALDI-TOF MS as described above.
(ii) Deglycosylation for electrospray ionization mass spectrometry (ESI MS) analysis of N-glycans.
An aliquot of octyl-Sepharose-purified procyclin (approximately 109 parasite equivalents) was exhaustively digested with 500 U of PNGase F (two additions) as described above, except that the incubation was carried out for 36 h. After digestion, the sample was boiled, dried in a Speed Vac concentrator, and resuspended in 100 μl of 5% 1-propanol and 100 mM ammonium acetate (buffer A). In order to separate the released glycans from the remaining GPI-peptide, the sample was loaded onto a mini-octyl-Sepharose (1-ml-bed-volume) column previously equilibrated with buffer A, and after several washes with the same buffer, the glycans were collected in the flowthrough of the column and processed for permethylation as described below.
(iii) Deglycosylation for SDS-PAGE.
Butanol or Triton extracts were resuspended in 10× denaturing buffer containing 5% SDS and 10% β-mercaptoethanol for 10 min at 100°C and incubated in the absence or presence of 1 μl (500 U) PNGase F for 2 h at 37°C.
Chemical O deglycosylation.
Butanol extracts containing the GPI-anchored molecules (0.5 × 108 to 1.5 × 108 cell equivalents) were dried and resuspended in 100 μl freshly prepared 40 mM trifluoroacetic acid, incubated for 25 min at 100°C, and then put on ice. Subsequently, the samples were dried under a constant flow of nitrogen in a water bath (40°C), washed twice with 100 μl H2O to eliminate the acid, dried again, and resuspended in sample buffer for analysis by SDS-PAGE and immunoblotting.
Analysis of permethylated N-glycans by ESI MS and electrospray ionization tandem mass spectrometry (ESI MS-MS).
Glycans released by PNGase F and concentrated after octyl-Sepharose chromatography (see above) were dried in a 2-ml glass vial and permethylated by the sodium hydroxide method as described elsewhere (18). The permethylated mixture was then resuspended in 20 μl of 80% acetonitrile, and an aliquot (2 μl) was mixed with 2 μl of 80% acetonitrile and 1 mM sodium acetate to give a final concentration of 80% acetonitrile and 0.5 mM sodium acetate. Samples were then analyzed in positive-ion mode with an ABI Q-Star XL instrument with tip and declustering potentials of 900 and 60 V, respectively. Daughter ion spectra were collected in product ion scanning mode using collision voltages of 35 to 90 V.
ESI MS and ESI MS-MS of phosphatidylinositol moieties.
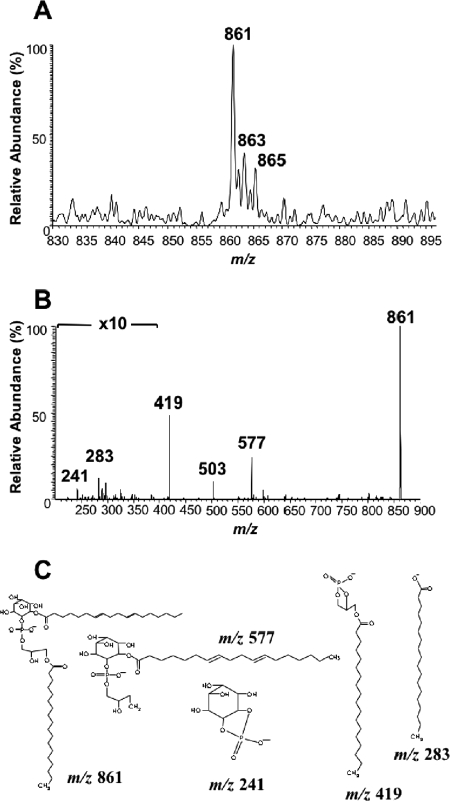
An aliquot (approximately 5 × 108 parasite equivalents) of octyl-Sepharose-purified procyclin was dried in a Speed Vac concentrator, resuspended in 100 μl of water, and washed twice with 200 μl of 1-butanol to eliminate possible phospholipid contaminants. The sample (recovered in the lower phase of the butanol-water partition) was then dried and submitted to deamination (27). The released phosphatidylinositol molecules were recovered after partition with 1-butanol and analyzed, in negative-ion mode, with a Finnigan-Thermoquest LCQ-Duo ion trap electrospray mass spectrometer. Samples were introduced at 5 μl/min. Source voltage and current were 4.52 kV and 0.24 mA, and capillary voltage and temperature were set at 19 to 36 V and 200°C. To collect the product ion spectrum of the [M-H]− pseudomolecular ion at m/z 861, the collision energy was 40 V under helium pressure. Fragmentation spectra were collected at the 200- to 1,000-m/z range at a rate of three microscans over a maximum ion injection time of 200 ms.
Infection of tsetse flies and isolation of trypanosomes.
Pupae of the tsetse fly Glossina morsitans morsitans were obtained from the International Atomic Energy Agency (Vienna, Austria). Teneral flies (newly eclosed, unfed) were fed through artificial membranes as described before (35, 44). The first blood meal consisted of 3 × 106 T. congolense Kilifi or Savannah procyclic forms per ml SM supplemented with 30% fetal bovine serum and washed horse red blood cells. The midguts and proboscises were isolated from the flies at different time points after infection and disrupted by mechanical force in phosphate-buffered saline (8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.6 mM KCl) containing 1% (wt/vol) bovine serum albumin. Large tissue pieces were removed by sedimentation, and the trypanosomes were collected from the supernatant by centrifugation at 1,300 × g for 10 min.
Immunofluorescence microscopy.
Trypanosomes were air dried onto microscope slides and fixed for 30 min in 4% formaldehyde and 0.04% glutaraldehyde in phosphate-buffered saline. MAb no. 491 (α-PRS) was used at a dilution of 1:250, α-GARP antiserum at 1:500, and CP1 antiserum at 1:100. The secondary antibodies were used at a dilution of 1:1,000. Cells were mounted with Vectashield mounting medium containing the DNA dye 4,6-diamidino-2-phenylindole (DAPI) (Reactolab SA, Servion, Switzerland).
Nucleotide sequence accession numbers.
The sequence of K3/1 was deposited in GenBank under accession number AY827559. The sequences of Sav1 and Sav2 were deposited in GenBank under accession numbers AY827557 and AY827558, respectively.
RESULTS
Characterization of a novel major GPI-anchored molecule in T. congolense procyclic culture forms.
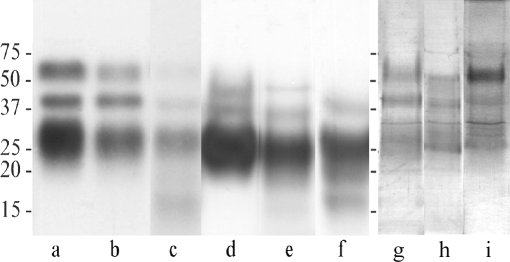
Incubation with [3H]-labeled GPI precursors is a valuable tool for the identification of GPI-anchored molecules in procyclic form trypanosomes in culture (9, 10). We found that labeling of 1 × 108 to 2.4 × 109 procyclic forms of T. congolense Kilifi and Savannah with [3H]ethanolamine resulted in the incorporation of 9.8 × 107 to 1.1 × 108 cpm (range of three independent experiments) into the delipidated protein pellets. Analysis by SDS-PAGE and fluorography revealed that the butanol and Triton extracts from T. congolense Kilifi procyclic forms contain three distinct bands with apparent molecular masses of 24 to 34 kDa, 43 kDa, and 58 kDa (Fig. 1, lanes a and b). Similarly, the butanol and Triton extracts from T. congolense Savannah procyclic forms show a major band at 24 to 34 kDa and two fainter bands at 40 kDa and 50 kDa (Fig. 1, lanes d and e). In addition, the aqueous phase of the CMW extract after partitioning between butanol and water shows a faint band at 15 to 18 kDa in T. congolense Kilifi and one at 17 to 18 kDa in T. congolense Savannah (Fig. 1, lanes c and f). The same molecules are labeled when T. congolense Kilifi and Savannah procyclic forms are incubated with [3H]myristic acid as a GPI precursor (results not shown). Bands with apparent molecular masses similar to those of the labeled molecules are also seen after staining the gel with silver (Fig. 1, lanes g and h). The labeling results are in good agreement with previous findings and reflect incorporation of [3H]ethanolamine into GPI-anchored molecules (10). The bands at 24 to 34 kDa in both trypanosome subgroups represent the previously characterized PRS (10), whereas the bands at 43 kDa in T. congolense Kilifi and 40 kDa in T. congolense Savannah correspond to GARP (10, 36, 40).
FIG. 1.
GPI-anchored molecules in T. congolense procyclic culture forms. T. congolense Kilifi (lanes a to c, g, and i) and Savannah (lanes d to f and h) procyclic forms were incubated in the presence of [3H]ethanolamine and sequentially extracted as described in Materials and Methods. The equivalent of 5 × 107 trypanosomes of the butanol (lanes a, d, g, and h) and Triton (lanes b and e) extracts and of the aqueous phases of the CMW extracts after partitioning between water and butanol (lanes c and f) were analyzed by SDS-PAGE followed by fluorography (lanes a to f) or silver staining (lanes g to i). Lane i shows the enrichment of the 58-kDa protein in the pooled fractions after octyl-Sepharose chromatography of a T. congolense Kilifi butanol extract. Apparent molecular mass markers (in kilodaltons) are indicated.
To characterize the 58-kDa [3H]ethanolamine-labeled molecule, the butanol extract from 2 × 109 T. congolense Kilifi procyclic forms was subjected to octyl-Sepharose chromatography. This procedure has been applied successfully before to purify GPI-anchored molecules from trypanosome extracts and results in preparations that are essentially devoid of other proteins (9, 10, 17). We found that the [3H]-labeled molecules elute from the column as distinct but poorly separated peaks, with the 58-kDa protein preferentially eluting in the first peak (results not shown). Since PRS is likely to be nonproteinaceous (10) and GARP cannot be sequenced by Edman degradation because its N terminus may be blocked (6), the pooled fractions that enriched the 58-kDa protein (Fig. 1, lane i) were subjected to N-terminal amino acid sequencing. Three independent sequencing reactions resulted in the consensus sequence ADEPGE(E)GTEPG (the assignment of the glutamic acid residue in parentheses was tentative), which allowed the design of a degenerate oligonucleotide (Tc58) encoding the amino acid sequence DEPGEEG.
cDNA library and sequence analysis.
A cDNA library derived from T. congolense Kilifi procyclic forms was screened in three rounds by using the labeled oligonucleotide probe Tc58. Positive clones were isolated, and inserts from two clones (F1/1 and K3/1) were amplified by PCR and sequenced. The results revealed two sequences of about 860 bp, which were identical to each other except for a short stretch of 13 nucleotides at the 5′ end and in the poly(A) addition site. The open reading frame consists of 489 nucleotides and translates into the amino acid sequence shown in Fig. 2A (the Kil1 sequence). Based on the SignalP prediction program (29), amino acids 1 to 20 represent a signal sequence for import into the endoplasmic reticulum, whereas the big-PI Predictor (16) and DGPI predictor (25) programs identify a C-terminal GPI addition sequence with a predicted anchor attachment site at Gly141 or Ser142, respectively. The stretch of amino acids identified by N-terminal sequencing of the purified material from the butanol extract matches with amino acids 44 to 55 of the deduced sequence (with the exception of position 50), suggesting that the N terminus of the protein was removed proteolytically during maturation or during extraction and isolation of the protein; alternatively, the prediction programs may not accurately identify the cleavage site of the signal peptide. Most interestingly, the remaining protein (amino acids 44 to 141) consists almost entirely of heptapeptide repeats, with 13 identical EPGENGT units. The Asn residues in the repeats represent potential N-linked glycosylation sites.
FIG. 2.
Protein sequences. (A) Deduced amino acid sequence of the 58-kDa protein from T. congolense Kilifi. Underlined amino acids represent the predicted N-terminal signal sequence and the C-terminal GPI addition signal, respectively. The stretch of amino acid residues in italics is absent from the mature protein (see text). The glycine residue at position 141 indicates the experimentally confirmed GPI anchor addition site (see text). The 13 identical EPGENGT heptapeptide repeats are indicated by alternating boldface type and lightface type (positions 46 to 136). (B) Alignment of the deduced amino acid sequence from T. congolense Kilifi (Tc58; the Kil1 sequence) with those from T. congolense Savannah (the Sav1 and Sav2 sequences) and the related sequence from the Sanger Institute T. congolense Genome Project (the Sav3 sequence). Dots reflect identical amino acids and dashes indicate gaps.
Analysis of the DNA sequences showed that the 5′ untranslated region of K3/1 contained a stretch of 13 nucleotides belonging to the miniexon of T. congolense (12). In addition, 151 nucleotides downstream of the stop codon we found a sequence (5′-TAGCCCTGTAGAATTCT-3′) with a high degree of similarity to the consensus 16-mer sequence of T. brucei procyclins and T. congolense GARP (20, 22). These two sequences were subsequently used to construct primers to amplify the gene(s) corresponding to Kil1 from a strain belonging to the T. congolense Savannah subgroup. By reverse transcriptase PCR, we obtained two sequences (the Sav1 and Sav2 sequences) showing a high degree of identity to that of Kil1 (Fig. 2B). The putative N-terminal signal sequences of Sav1 and Sav2 are identical and differ from that of Kil1 at 4 out of 20 amino acids. In addition, the two T. congolense Savannah sequences have identical GPI addition signals (positions 170 to 191) and closely resemble that of T. congolense Kilifi. The difference between the sequences of Sav1 and Sav2 lies entirely in the number of heptapeptide repeats, with 13 for Sav1 and 11 for Sav2. In addition, single amino acid changes between the Kilifi and Savannah proteins are seen within the first and third heptapeptide repeats (Fig. 2B). The compositions and numbers of repeats of the Kil1, Sav1, and Sav2 sequences and a similar sequence found in the database of the Sanger Institute T. congolense Genome Project (the Sav3 sequence [Fig. 2B]) are compiled in Table 1 for clarity. Because of their similarity to the EP and GPEET procyclins in T. brucei, we named the GPI-anchored proteins encoded by Kil1, Sav1, Sav2, and Sav3 T. congolense procyclins.
TABLE 1.
Comparison of the heptapeptide repeats in T. congolense procyclins
| Procyclin | No. of heptapeptide repeatsa
|
||||||
|---|---|---|---|---|---|---|---|
| Total | EPGENGT | EPGESGT | EPGVNGT | KPGESGT | KPGENGT | EPGANGT | |
| Kil1 | 13 | 13 | |||||
| Sav1 | 13 | 11 | 1 | 1 | |||
| Sav2 | 11 | 9 | 1 | 1 | |||
| Sav3 | 17 | 1 | 3 | 4 | 2 | 7 | |
The amino acid sequences of the heptapeptides are given. Amino acid changes compared to the heptapeptide from T. congolense Kilifi (Kil1) are underlined.
Digestion of genomic DNA from T. congolense Kilifi procyclic forms with a selection of restriction enzymes revealed several bands by Southern blot analysis, suggesting that Kil1 occurs in more than one copy (results not shown).
Expression of T. congolense procyclins in procyclic culture forms.
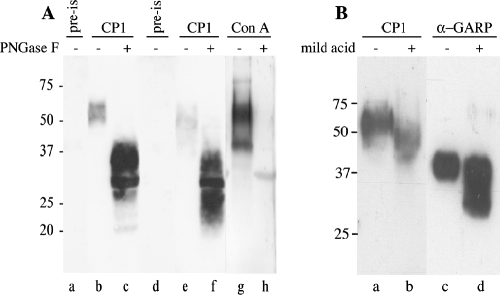
To study the expression of the proteins encoded by Kil1, Sav1, and Sav2 in T. congolense procyclic forms, polyclonal antibodies (termed CP1 antiserum) were raised against a synthetic peptide, AD(EPGENGT)2C, containing two copies of the heptapeptide repeat. Immunoblot analysis showed that CP1 antiserum recognizes bands of approximately 58 and 50 kDa in the butanol extracts from T. congolense Kilifi and Savannah procyclic culture forms, respectively (Fig. 3A, lanes b and e). The preimmune serum shows no reactivity with the extracts (Fig. 3A, lanes a and d).
FIG. 3.
Immunoblot analysis of T. congolense extracts. T. congolense Kilifi (A, lanes a to c, g, and h; B, lanes a to d) and Savannah (A, lanes d to f) procyclic forms were sequentially extracted as described in the legend for Fig. 1. (A) Aliquots (5 × 107 cell equivalents) of the butanol extracts were incubated in the absence (−) or presence (+) of PNGase F and analyzed by SDS-PAGE and immunoblotting using preimmune serum (pre-is), CP1 antiserum (CP1), or biotinylated ConA (ConA), followed by the respective peroxidase-conjugated secondary antibodies or streptavidin, to detect the antigens. (B) Aliquots (5 × 107 cell equivalents) of the butanol extracts were incubated in the absence (−) or presence (+) of mild acid (40 mM trifluoroacetic acid) and analyzed by SDS-PAGE and immunoblotting using CP1 antiserum (CP1) or α-GARP antiserum (α-GARP), followed by the respective peroxidase-conjugated secondary antibodies, to detect the antigens. Apparent molecular mass markers (in kilodaltons) are indicated.
Since the EPGENGT repeats contain potential N-glycosylation sites, which could result in the attachment of up to 13 carbohydrate side chains to the polypeptide, we treated T. congolense extracts with PNGase F and found that this procedure markedly increased the reactivity of CP1 antiserum, which then recognized bands with apparent molecular masses of 25 to 40 and 22 to 37 kDa in T. congolense Kilifi and Savannah extracts, respectively (Fig. 3A, lanes c and f). A similar decrease in apparent molecular mass was obtained when the PNGase F treatment was performed on [3H]ethanolamine-labeled butanol extracts from T. congolense procyclic forms and the samples were analyzed by SDS-PAGE followed by fluorography (results not shown). In addition, the presence of carbohydrates was analyzed by ConA binding, which showed that procyclin and GARP from T. congolense Kilifi procyclic forms were recognized by ConA and that the binding was completely abolished after PNGase F treatment (Fig. 3A, lanes g and h). To test for possible protein glycosylation via phosphate group, T. congolense Kilifi extracts were treated with mild acid (40 mM trifluoroacetic acid), which cleaves acid-labile linkages such as sugar 1-phosphate bonds (40). Immunoblot analysis using CP1 antiserum shows that this treatment resulted in a drop in molecular mass of T. congolense procyclin from 58 kDa to approximately 50 kDa (Fig. 3B, lanes a and b). A similar effect was observed when the same extract was probed with α-GARP antiserum (Fig. 3B, lanes c and d), which can be explained by the presence of oligosaccharide side chains linked to GARP via P-Thr/Ser (40).
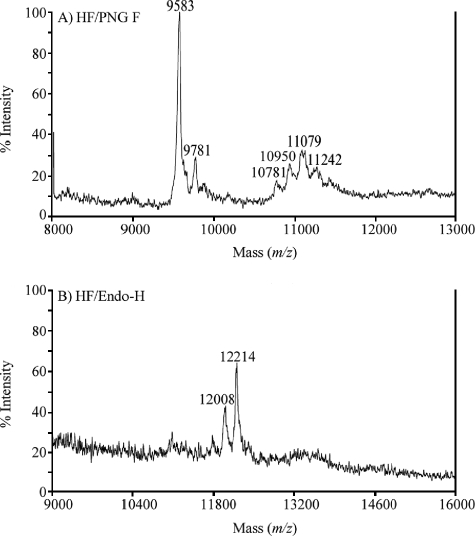
To determine the site of GPI anchor addition and corroborate the length of the peptide, octyl-Sepharose-purified butanol extract from T. congolense Kilifi procyclic forms was dephosphorylated with aqHF, followed by deglycosylation using PNGase F, and analyzed by negative-ion MALDI-TOF MS. The data revealed a major [M-H]− pseudomolecular ion at m/z 9,583, which is slightly higher than the expected nominal mass (m/z 9,573) of the deglycosylated peptide Ala44-Gly141 including the C-terminal ethanolamine (Fig. 4A). The mass difference is due to conversion of Asn to Asp after PNGase F deglycosylation, which increases the mass of the protein by 1 Da per amino acid converted, suggesting that most potential N-glycosylation sites in the native protein are occupied with an oligosaccharide chain.
FIG. 4.
Negative-ion MALDI-TOF MS analysis of T. congolense procyclin. Octyl-Sepharose-purified material from T. congolense Kilifi procyclic culture forms was analyzed by negative-ion MALDI-TOF MS after dephosphorylation with aqHF, followed by deglycosylation with PNGase F (A) or endo-H (B), as described in Materials and Methods.
To further analyze the number of N-glycans attached to procyclin, we incubated the aqHF-treated sample with endo-H, which cleaves specifically at the chitobiose core of high mannose and some hybrid oligosaccharides of N-glycosylated proteins. Since endo-H treatment truncates N-glycan chains to single N-acetylglucosamine (GlcNAc) residues, MALDI-TOF MS analysis of the treated protein allows the determination of the exact number of modified N-glycosylation sites. The results show a major [M-H]− pseudomolecular ion at m/z 12,214 after endo-H treatment, which is close to the calculated nominal mass (m/z 12,212) of the polypeptide Ala44-Gly141 with 13 N-acetylhexosamine (GlcNAc) residues (Fig. 4B). The smaller fragment at m/z 12,008 probably represents the same polypeptide with one GlcNAc residue less (calculated nominal mass of 12,009). Taken together, the MALDI-TOF MS analyses demonstrate that procyclin isolated from T. congolense Kilifi procyclic culture forms consists of at least 12 N-glycosylated heptapeptide repeats and that Gly141 is the site of GPI anchor attachment.
Characterization of the N-linked oligosaccharides.
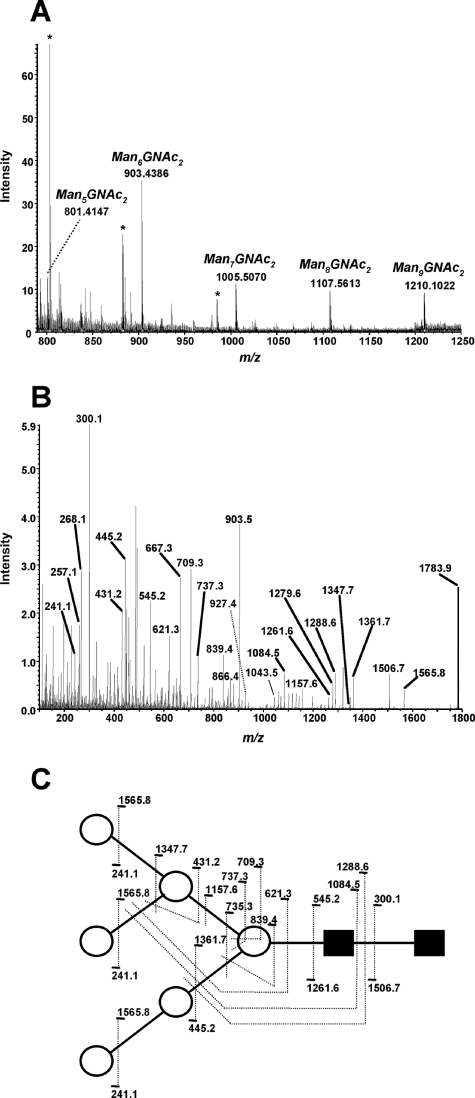
To further characterize the type of N glycosylation present in T. congolense procyclin, an aliquot of material was digested with PNGase F and the released glycans recovered in the flowthrough of a mini-octyl-Sepharose column. The oligosaccharides were then permethylated and analyzed by positive-ion-mode ESI MS. The spectrum (Fig. 5A) revealed a major doubly charged [M + 2Na]2+ ion at m/z 903.4, which after collision-induced fragmentation (Fig. 5B) was assigned as the permethylated [Hex6HexNAc2 + 2Na]2+. A detailed analysis of the product ions formed after collision (Fig. 5C) strongly suggests that the latter structure is the oligomannose Man6GlcNAc2. Furthermore, collision-induced fragmentation (not shown) of other less intense doubly charged [M + 2Na]2+ pseudomolecular ions at m/z 801.4, 1,005.5, 1,107.5, and 1,210.1 (Fig. 5A) suggests that T. congolense procyclins are modified with a series of mannosylated N-glycans ranging from Man5GlcNAc2 up to Man9GlcNAc2, which is in agreement with the strong binding of T. congolense procyclin to ConA (Fig. 3A, lane g).
FIG. 5.
Positive-ion-mode ESI MS and ESI MS-MS analysis of T. congolense procyclin permethylated N-glycans. (A) ESI MS analysis of total permethylated N-glycans. The ions at m/z 801.4, 903.4, 1,005.5, 1,107.6, and 1,210.1 correspond to the series of [M + 2Na]2+ ions of compositions Hex5HexNAc2, Hex6HexNAc2, Hex7HexNAc2, Hex8HexNAc2, and Hex9HexNAc2, respectively. The latter structures represent the whole oligomannose series ranging from Man5GlcNAc2 up to Man9GlcNAc2. Asterisks correspond to contaminants also present in control samples. (B) Product ion ESI MS-MS spectrum of the main [M + 2Na]2+ ion species at m/z 903.4. For simplification, not all [M + Na]+ sodium adducts of the B/Y, C series (15) are shown. (C) Assignment of product ions shown in panel B. The presence of the same intraring cleavage ions (i.e., m/z 709.3 and m/z 737.3) in the product ion ESI MS-MS spectra of [M + 2Na]2+ ion species at m/z 801.4 (not shown) and m/z 903.4 (Man5GlcNAc2 and Man6GlcNAc2 oligosaccharides, respectively), together with the presence of an ion at m/z 1,157.6 (panel B), which represents the loss of the trihexose fragment from the α1-6 arm of the core β-Man, strongly suggests that the extra Man residue in the suggested Man6 isomer is attached to the α1-3 arm of the core β-Man. Open circles and filled squares represent Man and GlcNAc residues, respectively.
Analysis of the GPI lipid moiety of T. congolense procyclin.
Octyl-Sepharose-purified procyclin was deaminated and the released phosphatidylinositol moieties analyzed by negative-ion ESI MS and ESI MS-MS. The spectrum (Fig. 6A) showed three major [M-H]− pseudomolecular ions at m/z 861, 863, and 865. The collision-induced daughter ion spectrum (Fig. 6B) defined the ion at m/z 861 as sn-1-(stearoyl)-2-lyso-glycerol-3-HPO4-1-(2-O-linoleoyl)-d-myo-inositol. The collision-induced daughter ion spectra (not shown) of the less abundant ions at m/z 863 and 865 indicated that they also contain sn-1-stearoyl-2-lyso-glycerol but differ in the type of fatty acid linked to the second position of the inositol ring (oleoyl and stearoyl, respectively). The assignments of the major characteristic product ions at m/z 241, 283, 419, and 577 are shown in Fig. 6C. The structures of the lipid moieties of the T. congolense procyclins are the same species previously found in the GPI anchor of T. congolense GARP but in a slightly different ratio (40).
FIG. 6.
Negative-ion ESI MS and ESI MS-MS analyses of the lipid moiety of T. congolense procyclin. (A) ESI MS analysis of the phosphatidylinositol fraction released by nitrous acid deamination. (B) Collision-induced dissociation ESI MS-MS product ion spectrum of the ion at m/z 861. (C) Assignment of the main product ions shown in panel B.
Expression of T. congolense procyclins by trypanosomes in culture and in the tsetse fly.
Analysis by immunofluorescence microscopy using CP1 antiserum demonstrated that the T. congolense procyclins are expressed together with PRS and GARP by procyclic culture forms of both T. congolense Kilifi (Fig. 7A) and T. congolense Savannah (data not shown). In contrast, bloodstream forms of T. congolense Kilifi STIB745 showed no expression of procyclin (result not shown).
FIG. 7.
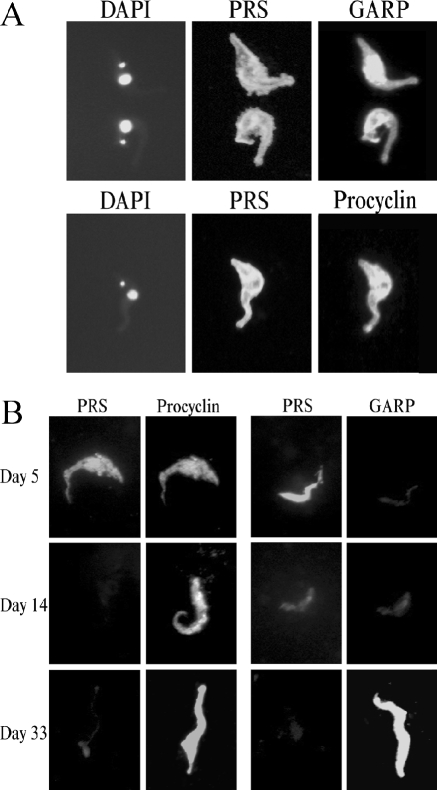
Immunofluorescence microscopy of T. congolense Kilifi procyclic forms. (A) T. congolense procyclic culture forms were air dried onto microscope slides and fixed with formaldehyde and glutaraldehyde. Surface antigens were detected using MAb no. 491 against PRS, α-GARP antiserum against GARP, and CP1 antiserum against T. congolense procyclin, in combination with the appropriate fluorescently labeled secondary antibodies. DNA was stained using DAPI. (B) After infection of tsetse flies with T. congolense Kilifi procyclic culture forms, parasites were collected from the midguts at the times indicated. Antigens were visualized as described for panel A.
To study the expression of T. congolense procyclins on trypanosomes during fly infection, two independent experiments involving 130 tsetse flies each were carried out. T. congolense Kilifi or Savannah culture forms were fed to teneral flies as part of their first blood meal, and the midguts and proboscises of the flies were isolated and analyzed for the presence of trypanosomes at various time points. We found that 2 days postinfection 100% of the flies were heavily infected, whereas after 5 days 40% of the flies showed heavy and 60% intermediate infections; the total number of infected flies subsequently decreased to 50% at 14 days postinfection and to 33% at 33 days postinfection. Parasites in the proboscis were extremely rare and could be found only in flies infected with T. congolense Savannah.
Immunofluorescence microscopy using antibodies against PRS, GARP, and T. congolense procyclin showed that the expression of the antigens changes during midgut infection (Fig. 7B and 8). T. congolense Kilifi and Savannah procyclic forms isolated from the midgut 5 days after infection are strongly positive for PRS and procyclin and negative for GARP (Fig. 7B, top panels). During the course of a midgut infection, the expression of PRS progressively decreases and is barely recognized by MAb no. 491 (α-PRS) at days 14 and 33. In contrast, GARP is hardly detectable in early procyclic forms in the midgut but increases during infection (Fig. 7B, middle and bottom panels, and 8). Interestingly, parasites are positive for procyclin(s) at all stages in the midgut (Fig. 7B and 8). The occasional parasite isolated from the proboscis of a fly infected with T. congolense Savannah also stained positive for procyclin (data not shown). Together, our results show that PRS and GARP are markers for early and late procyclic midgut forms, respectively, whereas T. congolense procyclin is present during the entire time course of a midgut infection. These developmental changes in antigen expression, i.e., the down-regulation of PRS and the up-regulation of GARP, occurred more slowly in the T. congolense Savannah strain than in the T. congolense Kilifi strain used in this study (Fig. 8).
FIG. 8.
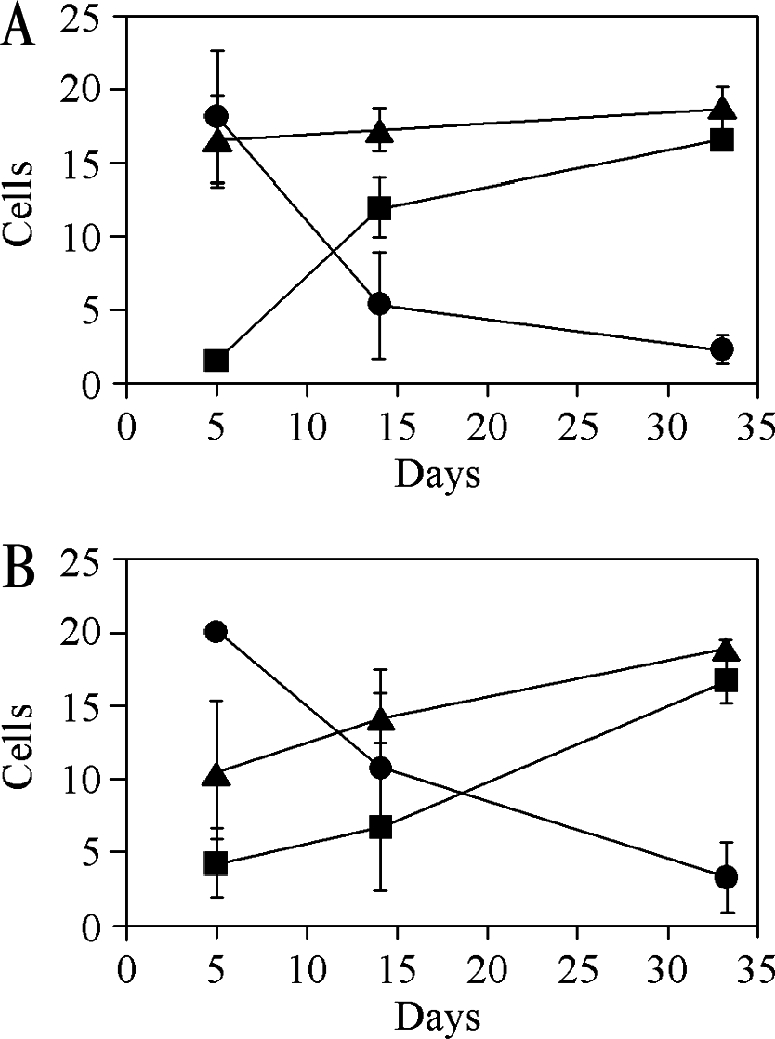
Changes in surface antigen expression during fly infection. Tsetse flies were infected with T. congolense Kilifi (A) or T. congolense Savannah (B) procyclic forms as described in the legend for Fig. 7, and parasites positive for PRS (•), GARP (▪), and T. congolense procyclin (▴) were counted on microscope slides prepared from trypanosomes isolated from fly midguts after the indicated times. The data points represent the means ± standard deviations after counting at least 20 parasites on three to four different microscope slides from two independent infections.
DISCUSSION
For many years it was believed that EP procyclin and GARP constituted the so-called invariant surface coats of T. brucei and T. congolense procyclic forms, respectively (30, 33). This concept had to be adjusted when additional GPI-anchored molecules, GPEET procyclin in T. brucei (9, 35, 41) and PRS in T. congolense (10), were discovered and when it was found that the composition of the surface coat changes during parasite development in the tsetse fly (2, 10, 42, 44). In the present work, we describe the identification of novel abundant GPI-anchored proteins in T. congolense procyclic forms that share a number of characteristics with T. brucei EP and GPEET and, thus, were named T. congolense procyclins. It is unclear why these proteins have not been detected in the past (5, 6, 40). Possible explanations include their extensive posttranslational modification, variation in the repeats between isolates/subspecies of T. congolense, altered expression levels in different strains or culture media, and the use of different protocols for protein extraction and analysis.
In common with EP and GPEET from T. brucei, T. congolense procyclins are small, highly acidic proteins consisting almost exclusively of repetitive peptide sequences. In T. congolense Kilifi, we have so far identified a single DNA sequence encoding a protein with 13 identical EPGENGT heptapeptides, whereas two genes encoding proteins with 11 to 13 heptapeptides were found in T. congolense Savannah; two of these repeats show single amino acid substitutions compared to the Kilifi peptide repeat. Despite the variability in the numbers of repeats and in the heptapeptide sequences between the procyclins of the two strains we have analyzed (Kil1, Sav1, and Sav2) and a procyclin from the database (Sav3), the heptapeptide unit seems to be a feature that is shared by all T. congolense procyclins. Interestingly, four amino acids in the T. congolense repeats (EPGT) are also present in the EP and GPEET repeats in T. brucei. Southern blot analysis suggests that there is more than one copy of the procyclin gene in T. congolense Kilifi; multiple gene copies located in two distinct loci have also been reported previously for GARP (31). The 3′ untranslated regions of the T. congolense procyclin transcripts contain a 16-mer sequence that is highly similar to the corresponding sequence in the GARP and T. brucei EP and GPEET procyclin mRNAs (20, 22, 38). The conserved sequences in T. congolense procyclin mRNAs are also located ∼100 bases upstream of the poly(A) tails and are predicted to adopt the same secondary structure (46). It remains to be established, however, whether they have the same functions in regulating gene expression.
Various programs that predict posttranslational modifications indicate that the T. congolense procyclins are all GPI anchored and heavily glycosylated. Our results using radiolabeled GPI precursor molecules and PNGase F treatment demonstrate that this is indeed the case. The procyclins in both T. congolense strains could be labeled by incubating parasites in culture with [3H]ethanolamine or [3H]myristic acid as the GPI precursor (10). In addition, part of the label was recovered in a faint band in the 15- to 18-kDa range after SDS-PAGE of the CMW extract. A similar labeling pattern has been observed previously for T. brucei (43) and probably reflects labeling of free GPI anchors in procyclic forms.
The T. congolense procyclins migrate by SDS-PAGE with a much higher apparent molecular mass than predicted, i.e., 50 to 58 kDa instead of 8.1 to 9.5 kDa, based on their amino acid sequences (the mass values vary depending on the polypeptide sequence used for calculation). A similar observation has been made before for T. congolense GARP (5, 6, 10) and the T. brucei EP and GPEET procyclins (9, 11, 19) and is, in part, due to the attachment of the proteins to complex GPI anchors (17, 40, 41). In addition, the T. congolense procyclins undergo extensive N glycosylation on the polypeptide chains, as demonstrated by the substantial reduction of their molecular masses after treatment with PNGase F. Interestingly, the results from the mass spectrometry analysis indicate that the majority of potential N-glycosylation sites in the heptapeptide repeats of T. congolense Kilifi procyclin are modified. Furthermore, removal of the N-linked carbohydrates resulted in increased antibody binding to procyclins in immunoblots; this result was not unexpected since the anti-procyclin antiserum used in this study was raised against an unglycosylated EPGENGT peptide repeat. The ConA blotting experiments suggest that, based on the binding specificity of this lectin (23), the N-glycans are of an oligomannose nature. In fact, ESI MS analysis of permethylated N-glycans clearly showed that the T. congolense procyclin polypeptides can be modified with a series of high-Man-type oligosaccharides ranging from Man5GlcNAc2 to Man9GlcNAc2. The heavy glycosylation on the polypeptide chain seems to distinguish the T. congolense surface proteins from the T. brucei procyclins, which contain, at most, a single homogenous Man5GlcNAc2 glycan (1). In addition, the Thr residues in the heptapeptide repeats may be modified with oligosaccharide chains linked via phosphodiester bonds (as judged by the susceptibility of T. congolense procyclins to mild-acid treatment), which would also contribute to an increase in the total mass of the molecule. The latter modification is absent in mammalian cells and has been reported only for certain Dictyostelium sp. and protozoal glycoproteins, including T. congolense GARP, which contains very large side chains rich in Man and Gal residues (40). The elucidation of the detailed structure of the mild-acid-sensitive modification will require additional work. Taken together, the T. congolense procyclins are among the most densely glycosylated parasite surface molecules ever reported, with at least 10 oligomannose N-glycans (depending on the parasite strain), possibly additional phosphodiester-linked glycans, and a GPI anchor with potential complex GPI modifications (Fig. 9). The overall structural features of the T. congolense procyclins are similar (except for the GPI modification) to those described for NETNES, a highly mannosylated surface protein expressed in T. cruzi epimastigote forms (27).
FIG. 9.
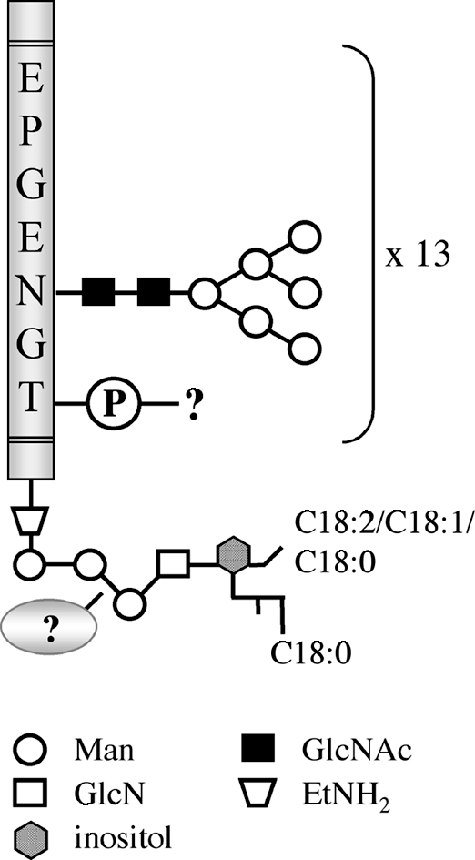
Partial chemical structure of T. congolense Kilifi procyclins. The schematic representation shows the proposed N-glycan structures and possible phosphoglycan modifications linked to the EPGENGT repeats of the mature protein (Ala44-Gly141; gray bar) and the partial structure of the GPI anchor with the identified fatty acyl chains. For clarity, the phosphate groups in the GPI anchor are not shown.
N-terminal amino acid sequencing and analysis by MALDI-TOF MS revealed that the N terminus of procyclin from T. congolense Kilifi procyclic culture forms is shorter than predicted. A similar observation has been made previously for T. brucei GPEET: amino acid sequencing (9) and mass spectrometry analysis (2, 28) showed that the N terminus of GPEET in T. brucei procyclic culture forms is truncated by 7 to 11 amino acids compared to the prediction made using SignalP (29). In addition, during infection in the tsetse fly midgut, the N-terminal domains of GPEET and EP are further cleaved proteolytically, leaving little other than the protease-resistant amino acid repeats (2). It is possible that a similar process may also occur during infection of tsetse flies with T. congolense.
Our previous work with T. congolense showed that early procyclic forms in the tsetse fly midgut strongly express PRS, whereas GARP appears only at a later time during fly infection (10). Remarkably, a large number of parasites in between these two stages were negative for both antigens. We now show that the parasite surface during that phase is covered with T. congolense procyclins, which are expressed continuously during the course of a midgut infection after tsetse flies are infected with procyclic forms. In this respect, T. congolense procyclins resemble T. brucei EP, which is expressed throughout the course of a midgut infection (44). At present, we cannot determine which surface molecule appears first during parasite differentiation in the insect host since tsetse flies were infected with procyclic culture forms that already expressed all three GPI-anchored molecules. We were unable to perform fly experiments using bloodstream forms since the T. congolense Kilifi stock used in this study did not establish an infection in mice. In addition, it should be noted that T. congolense bloodstream forms that have been adapted to rodents are often poorly infectious for tsetse flies (J. D. Barry, personal communication). Nevertheless, our studies suggest that PRS represents a marker for early T. congolense procyclic forms whereas GARP is a marker for late-stage parasites in the midgut and the proboscis and procyclin is expressed continuously during midgut infection. Finally, a BLAST search of the database of the Sanger Institute T. vivax Genome Project for proteins similar to the T. brucei and T. congolense procyclins did not reveal any candidate sequences. This may not be particularly surprising, however, if the role of the procyclins is to help the parasites survive in the fly midgut, since the development of T. vivax in the tsetse fly is restricted to the proboscis.
Acknowledgments
This work was supported by Swiss National Science Foundation grants 3100-103695 (to P.B.) and 3100-063987 (to I.R.). A.A.-S. is supported by a Wellcome Trust Research Career Development Fellowship and I.C.A. by a BBRC/Biology/UTEP grant (NIH no. 5G12RR008124).
We thank Mike Ferguson for the generous use of the Q-Star instrument, S. Jungi for her help with preparing the figures, and B. J. Armstrong and M. Bütikofer for support.
REFERENCES
- 1.Acosta-Serrano, A., R. N. Cole, A. Mehlert, M. G. Lee, M. A. Ferguson, and P. T. Englund. 1999. The procyclin repertoire of Trypanosoma brucei. Identification and structural characterization of the Glu-Pro-rich polypeptides. J. Biol. Chem. 274:29763-29771. [DOI] [PubMed] [Google Scholar]
- 2.Acosta-Serrano, A., E. Vassella, M. Liniger, C. Kunz Renggli, R. Brun, I. Roditi, and P. T. Englund. 2001. The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc. Natl. Acad. Sci. USA 98:1513-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asbeck, K., U. Kurath, I. Roditi, and W. Gibson. 2004. Trypanosoma (Nannomonas) simiae and T. (N.) godfreyi have genes encoding glutamic acid and alanine-rich proteins. Mol. Biochem. Parasitol. 134:159-162. [DOI] [PubMed] [Google Scholar]
- 4.Barry, D., and M. Carrington. 2004. Antigenic variation, p. 25-37. In I. Maudlin, P. H. Holmes, and M. A. Miles (ed.), The trypanosomiases. CABI Publishing, Wallingford, United Kingdom.
- 5.Bayne, R. A., E. A. Kilbride, F. A. Lainson, L. Tetley, and J. D. Barry. 1993. A major surface antigen of procyclic stage Trypanosoma congolense. Mol. Biochem. Parasitol. 61:295-310. [DOI] [PubMed] [Google Scholar]
- 6.Beecroft, R. P., I. Roditi, and T. W. Pearson. 1993. Identification and characterization of an acidic major surface glycoprotein from procyclic stage Trypanosoma congolense. Mol. Biochem. Parasitol. 61:285-294. [DOI] [PubMed] [Google Scholar]
- 7.Borst, P. 2002. Antigenic variation and allelic exclusion. Cell 109:5-8. [DOI] [PubMed] [Google Scholar]
- 8.Brun, R., and M. Schönenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 9.Bütikofer, P., S. Ruepp, M. Boschung, and I. Roditi. 1997. ‘GPEET’ procyclin is the major surface protein of procyclic culture forms of Trypanosoma brucei brucei strain 427. Biochem. J. 326:415-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bütikofer, P., E. Vassella, M. Boschung, C. Kunz Renggli, R. Brun, T. W. Pearson, and I. Roditi. 2002. Glycosylphosphatidylinositol-anchored surface molecules of Trypanosoma congolense insect forms are developmentally regulated in the tsetse fly. Mol. Biochem. Parasitol. 119:7-16. [DOI] [PubMed] [Google Scholar]
- 11.Clayton, C. E., and M. R. Mowatt. 1989. The procyclic acidic repetitive proteins of Trypanosoma brucei. Purification and post-translational modification. J. Biol. Chem. 264:15088-15093. [PubMed] [Google Scholar]
- 12.Cook, G. A., and J. E. Donelson. 1987. Mini-exon gene repeats of Trypanosoma (Nannomonas) congolense have internal repeats of 190 base pairs. Mol. Biochem. Parasitol. 25:113-122. [DOI] [PubMed] [Google Scholar]
- 13.Cross, G. A. 1996. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays 18:283-291. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham, I. 1977. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J. Protozool. 24:325-329. [DOI] [PubMed] [Google Scholar]
- 15.Domon, B., and C. E. Costello. 1988. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 5:397-409. [Google Scholar]
- 16.Eisenhaber, B., P. Bork, and F. Eisenhaber. 1998. Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 11:1155-1161. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson, M. A., P. Murray, H. Rutherford, and M. J. McConville. 1993. A simple purification of procyclic acidic repetitive protein and demonstration of a sialylated glycosyl-phosphatidylinositol membrane anchor. Biochem. J. 291:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson, M. A. J. 1993. GPI membrane anchors: isolation and analysis, p. 349-383. In M. Fukuda and A. Kobata (ed.), Glycobiology: a practical approach. IRL Press, Oxford, United Kingdom.
- 19.Field, M. C., A. K. Menon, and G. A. Cross. 1991. A glycosylphosphatidylinositol protein anchor from procyclic stage Trypanosoma brucei: lipid structure and biosynthesis. EMBO J. 10:2731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furger, A., N. Schürch, U. Kurath, and I. Roditi. 1997. Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol. Cell. Biol. 17:4372-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hehl, A., T. W. Pearson, J. D. Barry, R. Braun, and I. Roditi. 1995. Expression of GARP, a major surface glycoprotein of Trypanosoma congolense, on the surface of Trypanosoma brucei: characterization and use as a selectable marker. Mol. Biochem. Parasitol. 70:45-58. [DOI] [PubMed] [Google Scholar]
- 22.Hehl, A., E. Vassella, R. Braun, and I. Roditi. 1994. A conserved stem-loop structure in the 3′ untranslated region of procyclin mRNAs regulates expression in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 91:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwa, K. Y., A. Acosta-Serrano, K. H. Khoo, T. Pearson, and P. T. Englund. 1999. Protein glycosylation mutants of procyclic Trypanosoma brucei: defects in the asparagine-glycosylation pathway. Glycobiology 9:181-190. [DOI] [PubMed] [Google Scholar]
- 24.König, E., H. Delius, M. Carrington, R. O. Williams, and I. Roditi. 1989. Duplication and transcription of procyclin genes in Trypanosoma brucei. Nucleic Acids Res. 17:8727-8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kronegg, J., and D. Buloz. 1999. Detection/prediction of GPI cleavage site (GPI-anchor) in a protein (DGPI). [Online.] http://129.194.185.165/dgpi/.
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.MacRae, J. I., A. Acosta-Serrano, N. A. Morrice, A. Mehlert, and M. A. Ferguson. 2005. Structural characterization of NETNES, a novel glycoconjugate in Trypanosoma cruzi epimastigotes. J. Biol. Chem. 280:12201-12211. [DOI] [PubMed] [Google Scholar]
- 28.Mehlert, A., A. Treumann, and M. A. Ferguson. 1999. Trypanosoma brucei GPEET-PARP is phosphorylated on six out of seven threonine residues. Mol. Biochem. Parasitol. 98:291-296. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 30.Pays, E., and D. P. Nolan. 1998. Expression and function of surface proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:3-36. [DOI] [PubMed] [Google Scholar]
- 31.Rangarajan, D., T. I. Harvey, and J. D. Barry. 2000. Characterisation of the loci encoding the glutamic acid and alanine rich protein of Trypanosoma congolense. Mol. Biochem. Parasitol. 105:281-290. [DOI] [PubMed] [Google Scholar]
- 32.Roditi, I., and C. Clayton. 1999. An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol. Biochem. Parasitol. 103:99-100. [DOI] [PubMed] [Google Scholar]
- 33.Roditi, I., A. Furger, S. Ruepp, N. Schürch, and P. Bütikofer. 1998. Unravelling the procyclin coat of Trypanosoma brucei. Mol. Biochem. Parasitol. 91:117-130. [DOI] [PubMed] [Google Scholar]
- 34.Roditi, I., H. Schwarz, T. W. Pearson, R. P. Beecroft, M. K. Liu, J. P. Richardson, H. J. Bühring, J. Pleiss, R. Bülow, R. O. Williams, et al. 1989. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J. Cell Biol. 108:737-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruepp, S., A. Furger, U. Kurath, C. Kunz Renggli, A. Hemphill, R. Brun, and I. Roditi. 1997. Survival of Trypanosoma brucei in the tsetse fly is enhanced by the expression of specific forms of procyclin. J. Cell Biol. 137:1369-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruepp, S., U. Kurath, C. Kunz Renggli, R. Brun, and I. Roditi. 1999. Glutamic acid/alanine-rich protein from Trypanosoma congolense is the functional equivalent of ‘EP’ procyclin from Trypanosoma brucei. Mol. Biochem. Parasitol. 98:151-156. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schürch, N., A. Furger, U. Kurath, and I. Roditi. 1997. Contributions of the procyclin 3′ untranslated region and coding region to the regulation of expression in bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 89:109-121. [DOI] [PubMed] [Google Scholar]
- 39.Tabel, H., R. S. Kaushik, and J. E. Uzonna. 2000. Susceptibility and resistance to Trypanosoma congolense infections. Microbes Infect. 2:1619-1629. [DOI] [PubMed] [Google Scholar]
- 40.Thomson, L. M., D. J. Lamont, A. Mehlert, J. D. Barry, and M. A. Ferguson. 2002. Partial structure of glutamic acid and alanine-rich protein, a major surface glycoprotein of the insect stages of Trypanosoma congolense. J. Biol. Chem. 277:48899-48904. [DOI] [PubMed] [Google Scholar]
- 41.Treumann, A., N. Zitzmann, A. Hülsmeier, A. R. Prescott, A. Almond, J. Sheehan, and M. A. Ferguson. 1997. Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J. Mol. Biol. 269:529-547. [DOI] [PubMed] [Google Scholar]
- 42.Vassella, E., A. Acosta-Serrano, E. Studer, S. H. Lee, P. T. Englund, and I. Roditi. 2001. Multiple procyclin isoforms are expressed differentially during the development of insect forms of Trypanosoma brucei. J. Mol. Biol. 312:597-607. [DOI] [PubMed] [Google Scholar]
- 43.Vassella, E., P. Bütikofer, M. Engstler, J. Jelk, and I. Roditi. 2003. Procyclin null mutants of Trypanosoma brucei express free glycosylphosphatidylinositols on their surface. Mol. Biol. Cell 14:1308-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vassella, E., J. Van Den Abbeele, P. Bütikofer, C. Kunz Renggli, A. Furger, R. Brun, and I. Roditi. 2000. A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 14:615-626. [PMC free article] [PubMed] [Google Scholar]
- 45.Vickerman, K., L. Tetley, K. A. Hendry, and C. M. Turner. 1988. Biology of African trypanosomes in the tsetse fly. Biol. Cell 64:109-119. [DOI] [PubMed] [Google Scholar]
- 46.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]