Abstract
The filamentous fungus Neurospora crassa is able to utilize a wide variety of carbon sources. Here, we examine the involvement of a predicted G-protein-coupled receptor (GPCR), GPR-4, during growth and development in the presence of different carbon sources in N. crassa. Δgpr-4 mutants have reduced mass accumulation compared to the wild type when cultured on high levels of glycerol, mannitol, or arabinose. The defect is most severe on glycerol and is cell density dependent. The genetic and physical relationship between GPR-4 and the three N. crassa Gα subunits (GNA-1, GNA-2, and GNA-3) was explored. All three Gα mutants are defective in mass accumulation when cultured on glycerol. However, the phenotypes of Δgna-1 and Δgpr-4 Δgna-1 mutants are identical, introduction of a constitutively activated gna-1 allele suppresses the defects of the Δgpr-4 mutation, and the carboxy terminus of GPR-4 interacts most strongly with GNA-1 in the yeast two-hybrid assay. Although steady-state cyclic AMP (cAMP) levels are normal in Δgpr-4 strains, exogenous cAMP partially remediates the dry mass defects of Δgpr-4 mutants on glycerol medium and Δgpr-4 strains lack the transient increase in cAMP levels observed in the wild type after addition of glucose to glycerol-grown liquid cultures. Our results support the hypothesis that GPR-4 is coupled to GNA-1 in a cAMP signaling pathway that regulates the response to carbon source in N. crassa. GPR-4-related GPCRs are present in the genomes of several filamentous ascomycete fungal pathogens, raising the possibility that a similar pathway regulates carbon sensing in these organisms.
The multicellular fungus Neurospora crassa has been used as a model organism for several decades for the study of genetics, biochemistry, and molecular biology (reviewed in reference 8). Like other filamentous fungi, N. crassa can grow in diverse environments and thus is confronted with a wide variety of environmental stimuli (8). One of the major systems used by N. crassa to sense extracellular changes involves heterotrimeric G proteins. In eukaryotic cells, heterotrimeric G protein signaling pathways are used to sense and respond to environmental stimuli (32, 43). Heterotrimeric G proteins are composed of a Gα subunit and tightly associated Gβ and Gγ subunits (32, 49). A class of plasma membrane proteins known as G-protein-coupled receptors (GPCRs) communicates changes in the environment to heterotrimeric G proteins (49, 74). GPCRs contain seven transmembrane helices that are connected by intracellular and extracellular loops, with the carboxy terminus extending into the cytoplasm (17, 32, 53). Ligand binding to the GPCR activates the G protein by inducing the exchange of GTP for GDP on the Gα and subsequent dissociation of Gα-GTP from the Gβγ dimer, allowing Gα and/or Gβγ to interact with downstream effector proteins (32).
Three Gα proteins have been identified in N. crassa, GNA-1, GNA-2, and GNA-3 (33, 69). GNA-1 was the first heterotrimeric G protein subunit identified in filamentous fungi (69). GNA-1 and GNA-3 play major roles in the regulation of growth and development of N. crassa through cAMP-dependent and -independent pathways (28, 31). With regard to cAMP-dependent functions, GNA-1 regulates the activity of adenylyl cyclase (CR-1) (36), while GNA-3 controls adenylyl cyclase protein levels (29, 31, 33). In contrast to GNA-1 and GNA-3, GNA-2 plays a lesser role, as effects of the gna-2 mutation were only observed in genetic backgrounds also lacking gna-1 or gna-3 (3, 31). N. crassa has one Gβ (GNB-1) and one Gγ (GNG-1) subunit, which function as a dimer during signaling and are important for the stability of all three Gα proteins (38, 80).
There are at least 10 predicted seven-transmembrane helix GPCRs in the N. crassa genome that fall into five distinct groups (8, 21). Of these 10 GPCRs, 2 are pheromone receptors (34), 3 are similar to predicted GPCRs from Arabidopsis thaliana, Caenorhabitis elegans, and Dictyostelium discoideum (8, 21), 2 are microbial opsins (5, 6), and 2 are related to putative nitrogen sensors in Schizosaccharomyces pombe (8).
The fifth group of N. crassa GPCRs contains a single member, GPR-4 (G-protein-coupled receptor 4). GPR-4 is similar to a group of putative carbon-sensing GPCRs from yeasts, including Saccharomyces cerevisiae Gpr1p, S. pombe Git3, and Candida albicans Gpr1 (25, 37, 48, 70, 73, 78, 81, 82). In the case of S. cerevisiae Gpr1p, glucose and sucrose have been implicated as agonist ligands, while mannose acts as an antagonist (42, 56). Gpr1p interacts with Gpa2p, a Gα subunit that regulates pseudohyphal differentiation, invasive growth, and meiosis in S. cerevisiae (1, 37, 65, 67, 78, 81, 82). Induction of the Gpr1p pathway leads to elevated intracellular cyclic AMP (cAMP) concentration and activation of the Tpk2p cAMP-dependent protein kinase (PKA) catalytic subunit, with subsequent increased expression of genes required for filamentation (4). In S. pombe, the GPCR Git3 is coupled to the Gα subunit Gpa2 (25, 73). Git3 and Gpa2 are required for a glucose-triggered increase in cAMP levels which in turn activates PKA (10, 25, 50, 73). In C. albicans, Gpr1 and Gpa2 have been reported to regulate filamentous growth in a cAMP- and PKA-dependent manner (46, 47, 48, 60, 70).
In this study, we present characterization of N. crassa GPR-4. We create Δgpr-4 mutants and determine effects due to loss of this gene on growth, development, and cAMP metabolism. We also examine epistatic relationships between gpr-4 and the three Gα subunits. Our results demonstrate that GPR-4 physically interacts with the Gα GNA-1 to regulate carbon source-dependent growth and development through a pathway that at least in part involves regulation of cAMP metabolism. We also report the existence of at least two carbon sensory pathways in N. crassa that require the action of heterotrimeric G proteins.
MATERIALS AND METHODS
Strains, media, and general molecular procedures.
N. crassa strains used in this study are listed in Table 1. Vogel's minimal medium (designated VM-sucrose) (71) was used for vegetative growth, while synthetic crossing medium (SCM) (75) was used to induce development of female reproductive structures. Sucrose was replaced by other carbon sources in VM medium where indicated (i.e., VM-glycerol). The concentration of the carbon source was 100 mM unless otherwise noted. Sorbose-containing medium (16) was used to facilitate colony formation on plates. If required, hygromycin B (Calbiochem, La Jolla, CA) was added to media at a concentration of 200 μg/ml. Conidia from 5- to 7-day-old cultures were used as the inoculum for new cultures. Yeast strains were propagated on synthetic dextrose (SD) medium containing the appropriate dropout mixture supplement (US Biological, Swampscott, MA). Plasmids were maintained in Escherichia coli strain DH5α (24).
TABLE 1.
N. crassa strains
| Strain | Relevant genotype | Comment(s) | Source or reference |
|---|---|---|---|
| 74A-OR23-1A (74A) | Wild-type matA | FGSCa 987 | FGSC |
| 74a-OR8-1a (74a) | Wild-type mata | FGSC 988 | FGSC |
| 7-32 | Δgpr-4::hph+matA | Δgpr-4 homokaryon | This study |
| 7-33 | Δgpr-4::hph+mata | Δgpr-4 homokaryon | This study |
| 35-6 | Δgpr-4::hph+ mata | Δgpr-4 homokaryon | This study |
| 38-2 | Δgpr-4::hph+ mata | Δgpr-4 homokaryon | This study |
| LA25 | pan-2 matA | pan-2 mutant | R. L. Weiss, UCLA |
| Rmi2 | Δgpr-4::hph+gpr-4+::his-3+mata | Complemented Δgpr-4 | This study |
| FGSC 6103 | his-3 matA | his-3 targeting strain | FGSC |
| FGSC 4008 | cr-1 matA | Allele B123 | FGSC |
| 7-33his3A | Δgpr-4::hph+his-3 matA | FGSC 6103 × 7-33 progeny | This study |
| 35-6pan2-7 | Δgpr-4::hph+ pan-2 mata | 35-6 × LA25 progeny | This study |
| 35-6pan2-8 | Δgpr-4::hph+ pan-2 matA | 35-6 × LA25 progeny | This study |
| 1B8 | Δgna-1::hph+ matA | gna-1 mutant | 29 |
| a1r54 | Δgna-1::hph+ Δgpr-4::hph+matA | 1B8 × 7-33 progeny | This study |
| a1r2 | Δgna-1::hph+ Δgpr-4::hph+mata | 1B8 × 7-33 progeny | This study |
| a1r2h7 | Δgna-1::hph+ Δgpr-4::hph+his-3 | FGSC 6103 × a1r2 progeny | This study |
| a29-1 | Δgna-2::pyrG+matA | gna-2 mutant | 3 |
| a2r2 | Δgna-2::pyrG+ Δgpr-4::hph+matA | A29-1 × 7-33 progeny | This study |
| 31c2 | Δgna-3::hph+matA | gna-3 mutant | 33 |
| 2a1 | Δgna-3::hph+ matA | gna-3 mutant | 33 |
| a3r1 | Δgna-3::hph+ Δgpr-4::hph+matA | 2a1 × 7-33 progeny | This study |
| Δ1gna-1Q204L | Δgna-1::hph+ gna-1Q204L::his-3+matA | gna-1Q204L allele | H. Kim and K. A. Borkovich, unpublished data |
| a1rQ204L-25 | Δgpr-4::hph+ Δgna-1::hph+gna-1Q204L::his-3+ mata | Δgpr-4 gna-1Q204L allele | This study |
| a1rR178C | Δgpr-4::hph+ Δgna-1::hph+ gna-1R178C::his-3+ mata | Δgpr-4 gna-1R178C allele | This study |
FGSC, Fungal Genetics Stock Center, Kansas City, Mo.
Recombinant DNA procedures, such as plasmid construction, E. coli transformation, and Southern blots, etc., were performed according to standard protocols (59). All PCR products (cloned into the pGEM-T vector; Promega Corp., Madison, WI) and recombinant vectors were verified by sequencing (Institute for Integrative Genome Biology, University of California, Riverside).
RT-PCR analysis.
The tissues used for RNA extraction and isolation of total RNA were as previously described (38, 80). The reverse transcriptase PCR (RT-PCR) was used to assess message levels for various genes. Reactions were performed using 1 μg of total RNA and specific primers with the Access RT-PCR system (Promega Corp.), as recommended by the manufacturer. Reactions were conducted using conditions previously demonstrated to yield quantitative/semiquantitative data for mRNA levels (33). Products were subjected to Southern analysis (59) using specific probes (Table 2).
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′-3′) |
|---|---|
| FpRTPCRgpr-4 | TCTTGACTCTGTCGACCTTTTACTG |
| RpRTPCRIIgpr-4 | GTAAGATGGTCATCAGGAGATGGA |
| pGPR-4UF | GCTCTAGACAATGAAGTTGCAAGTG |
| pGPR-4UR | CGGATCCGTATGACTTGAAACTCTA |
| pGPR-4DF | GGAAGCTTCGACTCGAGTCTCGG |
| pGPR-4DR | GGAGCTCTTGCACTTTACTCCTTAC |
| FpNexon2gpr-4 | TCAAGTCGGTGTGGTTTGTCATTC |
| RpNexon2gpr-4 | TTTCCAGGGTTGCTCTCTCAGAGTA |
| 1.5kbFORWARD | CTACCCTTCTGGGACAACACAATG |
| 1.5kbREVERSE | ACTGCATCTCCCTCGTCATCATC |
| GPR-4RESCUE-FP | GCTGGAATTCATTCTGGCATATGG |
| GPR-4RESCUE-RP | TGACTAGTCTGATGAATAGACTAC |
| Fgpr-4cDNAHPLC | GGAAGACGTTTCGGCATGAACTGATAGTGTTGCTTATTCAGAGTG |
| Rgpr-4cDNAHPLC | CACTCTGAATAAGCAACACTATCAGTTCATGCCGAAACGTCTTCC |
gpr-4 intron verification and expression studies.
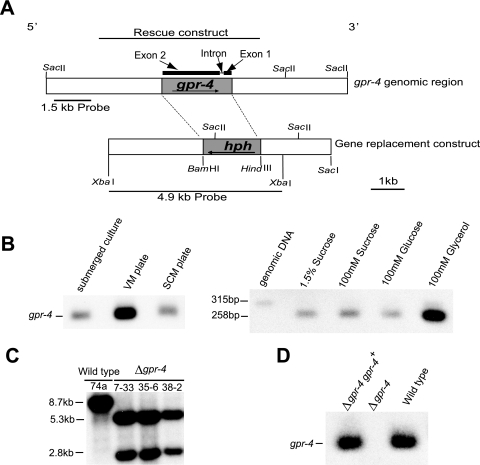
gpr-4 corresponds to predicted protein NCU06312.1 in the N. crassa genome database. The gene structure predicted by the automated gene caller contains two exons and one intron (http://www.broad.mit.edu/annotation/genome/neurospora/home.html). For intron verification, RT-PCR analysis (Access RT-PCR; Promega) was performed on total RNA isolated from conidia, 8- and 16-h submerged cultures, and VM and SCM plates using primers FpRTPCRgpr-4 and RpRTPCRIIgpr-4 (Table 2) which flank the intron (see Fig. 2A). A genomic control was provided by amplification of a 315-bp PCR product using the same primers, with cosmid pMOcosX X1 B8 as the template (contains the gpr-4 gene) (http://www.broad.mit.edu /annotation/genome/neurospora/home.html). The RT-PCRs were electrophoresed on agarose gels and subjected to Southern analysis using the amplified 315-bp genomic fragment as a probe (see Fig. 2A). Analysis of gpr-4 message levels in various tissues in wild-type Δgpr-4 and Δgpr-4 gpr-4+ strains was accomplished by RT-PCR-Southern analysis using the same set of primers and probe.
FIG. 2.
gpr-4 intron verification, expression profile and construction of Δgpr-4 mutant and complemented strains. (A) Structure of the N. crassa gpr-4 genomic region and of Δgpr-4 gene replacement and complementation vectors. The predicted gpr-4 gene structure, containing two exons and one intron (57 bp), is depicted above the bar representing the gpr-4 genomic region. The shaded areas in the genomic region and gene replacement construct depict the ORFs of gpr-4 and hph, respectively. Probes used for Southern analysis (1.5 and 4.9 kb) are shown by the bold lines. The bold line above the predicted gene structure depicts the insert in the construct used for complementation. (B) Intron verification and expression profile of gpr-4. Left panel: expression of gpr-4 during the life cycle. Shaken submerged cultures and VM and SCM plate cultures (all containing 1.5% sucrose as a carbon source) were prepared using wild-type strain 74a. Total RNA was isolated and subjected to quantitative RT-PCR analysis as described in Materials and Methods. Right panel: intron verification and gpr-4 expression level in the presence of different carbon sources. Total RNA was isolated from shaken submerged cultures of wild-type strain 74A containing the indicated sugars and subjected to quantitative RT-PCR analysis (yields a 258-bp fragment). Genomic DNA was amplified to serve as an intron-containing control (315 bp). The size difference between the genomic and cDNA fragments corresponds to that predicted by the automated gene caller (57 bp), thus verifying the presence of the intron. (C) Southern blot analysis. Genomic DNA was isolated from wild-type (74a) and Δgpr-4 homokaryotic strains (7-33, 35-6, and 38-2) and digested with SacII. The 4.9-kb XbaI fragment of gpr-4 that contains the gpr-4 ORF as well as 2.9-kb 5′ and 1-kb 3′ flanking DNA (panel A) was used as a probe. SacII cuts within hph, but not gpr-4, and the hygromycin-resistant progeny all contain the expected 5.3-kb and 2.8-kb fragments and no extra ectopic copies of the gene replacement construct. (D) Restoration of gpr-4 expression in complemented strains. Total RNA was isolated from shaken submerged cultures (containing 1.5% sucrose as a carbon source) and subjected to quantitative RT-PCR analysis as described in Materials and Methods. Strains are 74A (wild type), 7-32 (Δgpr-4), and Rmi2 (Δgpr-4 gpr-4+ complemented strain).
Western analysis.
For analysis of G protein subunit levels, a fraction enriched for plasma membranes was isolated as described previously (69) from 3-day VM and VM-glycerol plates. For detection of adenylyl cyclase (CR-1), whole-cell extracts were prepared from VM-sucrose or VM-glycerol 16-h shaken submerged cultures as previously described with some modifications (33). Tissue was collected and ground in liquid nitrogen, mixed with extraction buffer, shaken for 20 min at 4°C, and then centrifuged at 1,000 × g for 15 min at 4°C. The protein amount was quantified using the Bradford protein assay (Bio-Rad, Hercules, CA). Samples containing 30 μg of total protein were subjected to Western analysis (33). Primary antibodies against GNA-1, GNA-2, GNA-3, GNB-1, and CR-1 were used at 1:1,000, 1:500, 1:1,000, 1:1,000, and 1:5,000 dilutions, respectively (3, 27, 33, 80). A goat anti-rabbit immunoglobulin G horseradish peroxidase conjugate (Bio-Rad) was used as the secondary antibody at a 1:7,500 or 1:10,000 dilution. Detection was performed using the enhanced chemiluminescence method (Amersham Pharmacia Biotech, Little Chalfont, England), as described by the manufacturer. A duplicate gel was electrophoresed and stained with Coomassie brilliant blue to verify equal loading of protein samples, as described previously (33, 59).
Construction of N. crassa strains.
The gpr-4 gene was replaced with a hph gene cassette as follows. The gpr-4 gene replacement construct pLL1 was made by ligation of four DNA fragments: vector pGEM7Zf(+) (Promega Corp.) digested with XbaI and SacI, the HindIII-BamHI fragment from pCSN44 (containing the hph gene and Aspergillus nidulans promoter trpC) (64), and PCR products corresponding to the 2.9-kb XbaI-BamHI fragment of 5′ flanking DNA extending from the gpr-4 open reading frame (ORF) and the 2.0-kb HindIII-SacI fragment of 3′ flanking DNA extending from the gpr-4 ORF. The two flanks were amplified from cosmid pMOcosX X1 B8 using primer pairs pGPR-4UF/pGPR-4UR and pGPR-4DF/pGPR-4DR, respectively (Table 2; see Fig. 2A). To obtain the gpr-4 deletion strain, 1 μg of plasmid pLL1 was electroporated into 10-day-old conidia of N. crassa wild-type strain 74a, with selection on sorbose medium containing hygromycin B. Genomic DNA was extracted from the hygromycin B-resistant transformants using the Puregene kit according to the manufacturer's protocol (Gentra Systems, Minneapolis, MN). DNA was digested with SacII and subjected to Southern analysis using two different probes. The first was a 1.5-kb fragment corresponding to a region upstream of the gpr-4 ORF (see Fig. 2A) that was amplified using primers 1.5kbFORWARD and 1.5kbREVERSE. The second probe was a 4.9-kb XbaI fragment (see Fig. 2A) containing hph and portions of the gpr-4 5′ and 3′ flanking DNA that was excised from pLL1. Heterokaryotic Δgpr-4 strains without ectopic integrations were crossed to wild-type strain 74A (Table 1). The progeny were plated on sorbose medium containing hygromycin B. The homokaryotic status of hygromycin-resistant progeny was verified by Southern analysis using the 4.9-kb XbaI fragment from pLL1 described above as a probe.
A complemented Δgpr-4 strain was constructed by targeting the wild-type gpr-4 allele to the his-3 locus. A Δgpr-4 his-3 recipient strain (7-33his3A) was created by crossing Δgpr-4 strain 7-33 to strain FGSC 6103, with selection on hygromycin-containing medium followed by screening for his-3 auxotrophy (Table 1). For the rescue construct, a 4.7-kb wild-type genomic DNA fragment including the entire gpr-4 ORF and 2.0-kb upstream flank and 0.7-kb downstream flank (see Fig. 2A) was amplified using primers GPR-4RESCUE-FP and GPR-4RESCUE-RP and cloned into pGEM-T (Promega) to yield plasmid pLL7. The 4.7-kb fragment from pLL7 was then inserted into the his-3-targeting plasmid pHK40 (35), modified from pRAUW122) (2), to create plasmid pLL8. Strain 7-33his3A was electroporated with pLL8, and transformants were selected on histidine-free sorbose medium supplemented with hygromycin B. Heterokaryons containing the wild-type gpr-4 allele integrated at the his-3 locus were identified by Southern analysis. Genomic DNA was digested with HindIII, and a 3-kb HindIII-EcoRI fragment obtained from pHK40 corresponding to the his-3 left flank was used as the probe (data not shown). Homokaryons with homologous recombination at the his-3 locus (Δgpr-4::hph+ and gpr-4+::his-3+ strains) were obtained by microconidial isolation (18) and verified by Southern and RT-PCR analysis as described above.
To obtain strains for a forced heterokaryon test, Δgpr-4 strain 35-6 was crossed to the pan-2 strain, and progeny were plated on medium containing hygromycin B, followed by spot testing to identify Δgpr-4 pan-2 double mutants. To probe the relationship between GPR-4 and Gα proteins in N. crassa, a series of gpr-4 Gα double mutants were constructed using sexual crosses between single mutants (Table 1). Progeny were screened on medium containing hygromycin B. The presence of the Δgpr-4, Δgna-1, Δgna-2, and Δgna-3 mutations in progeny was verified by Southern analyses as described above (for gpr-4) or as reported previously (for gna-1, gna-2, and gna-3) (3, 33).
The Δgpr-4 Δgna-1 his-3 recipient strain to be used for transformation with gna-1-activated allele vectors was constructed by crossing Δgpr-4 Δgna-1 mutant a1r2 with his-3 strain FGSC 6103. Ascospore progeny were screened by plating on hygromycin B-containing medium, followed by testing for histidine auxotrophy and Southern analysis using probes for Δgpr-4 and Δgna-1, as described above. Vectors pQY15 and pQY21, containing two different predicted GTPase-deficient, constitutively activated gna-1 alleles (R178C and Q204L), have been described previously (79). pQY21 and pQY15 were electroporated into strain a1r2h7 (Table 1) and transformants plated on medium lacking histidine. Transformants were screened for homologous recombination of the his-3 targeting vector using Southern analysis (79). Homokaryotic Δgpr-4 Δgna-1 his-3::gna-1Q204L or Δgna-1 his-3::gna-1R178C strains were purified and verified using the microconidiation procedure, followed by Southern analysis as described above.
Phenotypic analysis.
Conidia from 5-day-old flask cultures were used as the inoculum, and a minimum of three independent experiments were performed for each analysis. Generally, the centers of cellophane-overlaid plates were inoculated using 1 μl of a conidial suspension (1 × 109 conidia/ml), followed by incubation in the dark at 30°C for the time specified in the figure legend. Measurement of dry mass was as described previously (79), with modifications. Briefly, cultures were scraped from cellophane-overlaid plates and then transferred to preweighed plastic weighing dishes. Collected material was dried for 2 days in a 60°C oven and then cooled to room temperature before weighing. Apical extension rates, microscopic observations, assessment of aerial hypha formation in standing liquid cultures, fertility analysis, sorbose resistance, submerged culture conidiation, H2O2 resistance, and thermotolerance were determined as previously described (27, 33, 79, 80). A SZX9 stereomicroscope with an ACH 1× objective lens or a BX41 fluorescence microscope, both outfitted with a C-4040 digital camera (Olympus America), were used for general microscopic observations.
Yeast two-hybrid assay.
A gpr-4 clone free of intron sequences was created using the QuikChange site-directed mutagenesis kit by following the manufacturer's instructions (Stratagene, La Jolla, CA). The intron of gpr-4 was deleted using plasmid pLL7 (containing the entire gpr-4 ORF; described above) as the template for primers Fgpr-4CDNAHPLC and Rgpr-4cDNAHPLC, and the resulting cDNA clone was designated pLL9. A fragment corresponding to the carboxy terminus of GPR-4 (GPR-4CT; amino acids 527 to 654) was amplified from pLL9 using primers Fgpr-4CT and Rgpr-4CT and subcloned into pGEM-T to form pLL10. pLL10 was digested with EcoRI and PstI to release the carboxy terminus of gpr-4, which was then inserted in frame into pGBKT7 (TRP1 Kanr; Clontech Laboratories, Mountain View, CA) to yield plasmid pLL12. pLL12 was then transformed into yeast strain Y187 (genotype MATα ura3-52 his3-200 ade2-101 trp1-901 leu2-3,112 gal4Δ met− gal80Δ URA3::GAL1UAS-GAL1TATA-lacZ; Clontech) using the lithium acetate method (Yeast Protocols Handbook; Clontech). Construction of plasmids in which the ORFs of gna-1, gna-2, and gna-3 were inserted in frame into pGAD424 (LEU2 Ampr; Clontech) and their transformation into yeast strain AH109 (genotype MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ; Clontech) will be described elsewhere (H. Kim, S. J. Martinez, and K. A. Borkovich, unpublished observations).
The yeast two-hybrid assay was performed according to the manufacturer's recommendations (BD Matchmaker Library Construction & Screening Kits User Manual and Yeast Protocols Handbook; Clontech). Matings were set up between yeast strains containing pLL12 (or controls) and those with a Gα vector (or controls). The mating mixtures were plated on SD minus leucine and tryptophan (selects for the presence of both pGAD424 and pGBKT7) and SD minus adenine, histidine, leucine, and tryptophan (selects for both plasmids and expression of the ADE2 and HIS3 reporter genes). Expression of the lacZ reporter was measured as β-galactosidase activity according to the manufacturer's instructions (Clontech) using a filter assay (Optitran BA-S 85 NC membrane; Schleicher & Schuell Bioscience, Keene, NH), with colonies propagated on SD minus leucine and tryptophan. No β-galactosidase activity could be detected in two hybrid assays performed using vectors with the gpr-4 carboxy terminus inserted into pGAD424 and the Gα genes cloned into pGBKT7; the reason for this is not known. The activity of the ADE2 and HIS3 reporters was quantitated using a growth assay. Yeast strains were cultured in SD liquid medium lacking leucine and tryptophan at 30°C for 1 day with shaking at 200 rpm. The number of cells in the culture was quantitated and serial dilutions prepared. A 2.5-μl aliquot of each concentration of cells (107, 106, or 105 cells/ml) was spotted onto SD plates lacking adenine, histidine, leucine, and tryptophan containing 5 mM 3-aminotriazole (to increase the stringency of the HIS3 reporter screen). The plate was incubated at 30°C for 3 days.
cAMP assays.
Tissues used for measurement of intracellular steady-state cAMP levels were obtained from VM-glucose and VM-glycerol plate cultures incubated at 30°C for 3 days in constant darkness. Mycelia were ground in liquid nitrogen as previously described (29). For analysis of the transient increase in cAMP after glucose addition to glycerol cultures, conidia were inoculated at a final concentration of 1 × 106 conidia/ml into 10 ml of VM-glycerol medium in 125-ml Erlenmeyer flasks. There were at least three flasks for each time point/strain. Cultures were incubated at 30°C for 16 h with shaking at 200 rpm, at which time one set of flasks was collected (time = 0). Glucose was then added to the remaining flasks at a final concentration of 100 mM, and samples collected at 30, 60, and 180 s. Cultures were collected using a 2.3-cm metal vacuum filter apparatus with Whatman 2.3-cm filters. The filters were immediately transferred to 2-ml microcentrifuge tubes containing 1 ml of 10% trichloroacetic acid and vortexed briefly, frozen in liquid nitrogen, and then thawed at 4°C with shaking. Samples from two individual flasks were combined during cAMP extraction when needed.
cAMP was extracted from tissue samples as previously described (29). cAMP levels were quantified using a protein binding assay according to the manufacturer's instructions (Amersham Pharmacia Biotech, Piscataway, N. J.). The protein concentration was determined using the BCA assay (Pierce, Rockford, IL) as described previously (29).
RESULTS
gpr-4 encodes a putative G-protein-coupled receptor that is highly expressed in glycerol-grown cultures.
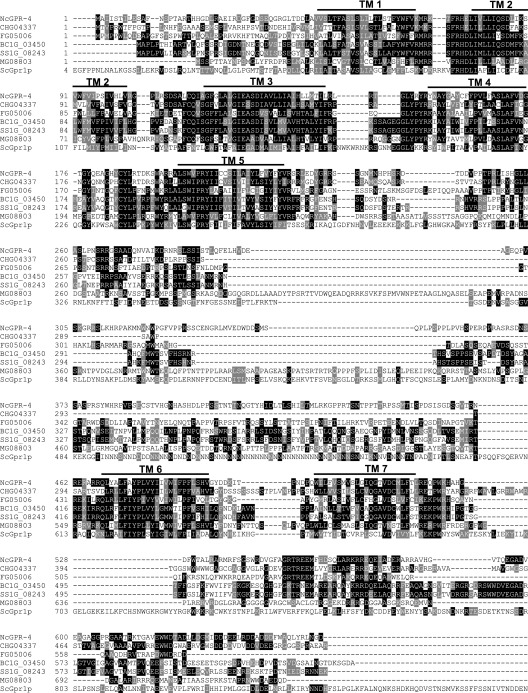
The gpr-4 gene was identified during the initial annotation of the N. crassa genome sequence as NCU06312.1 (21). The predicted GPR-4 protein contains seven transmembrane domains (TMs), characteristic of G-protein-coupled receptors (Fig. 1). GPR-4 is homologous to three putative carbon sensory GPCRs found in yeasts: Gpr1p from Saccharomyces cerevisiae (ScGpr1p; E = 3e−15), Git3 from Schizosaccharomyces pombe (SpGit3; E = 9e−3) and Gpr1 from Candida albicans (CaGpr1; E = 2e−17) (Fig. 1; data not shown) (37, 46, 48, 70, 73, 78, 82). N. crassa GPR-4, ScGpr1p, CaGpr1, and SpGit3 are similar in the transmembrane domains (especially TM1 to TM5) and connecting loops, particularly in the second and third intracellular and second extracellular loops, which are important for coupling to heterotrimeric G proteins and ligand binding, respectively (78, 81). In addition, the third intracellular loops of GPR-4 and ScGpr1p are relatively large (247 and 283 amino acids) (Fig. 1) (37, 73, 78, 82).
FIG. 1.
Alignment of GPR-4 (NcGPR-4) with homologous putative G-protein-coupled receptors from ascomycete fungi. The predicted amino acid sequences were aligned using ClustalW with shading by Boxshade (http://www.ch.embnet.org). The predicted transmembrane regions (TM1 to 7) are indicated by numbered lines above the sequences. Abbreviations: NcGPR-4, Neurospora crassa GPR-4 (accession no. NCU06312.1); ScGpr1p, Saccharomyces cerevisiae Gpr1p (accession no. JC5808); Mg08803, Magnaporthe grisea 70-15 hypothetical protein MG08803.4 (accession no. EAA51281); Fg05006, Gibberella zeae PH-1 (anamorph Fusarium graminearum) hypothetical protein FG05006.1 (accession no. XP_385182); CHG04337, Chaetomium globosum hypothetical protein CHG04337.1; SS1G_08243, Sclerotina sclerotiorum hypothetical protein SS1G_08243.1; BC1G_03450, Botrytis cinerea hypothetical protein BC1G_03450.1. An intron in FG05006.1 was misannotated, and the missing amino acid sequence LIMLLIYS was inserted between amino acids 79 and 80 in the original predicted protein sequence (http://www.broad.mit.edu/annotation/fgi/). Amino acids 121 to 137 were removed from the original predicted protein sequences of BC1G_03450.1 and SS1G_08243.1 (http://www.broad.mit.edu/annotation/fgi/), as these were misannotated and are actually part of an intron region (between amino acids 120 and 121 in this figure).
N. crassa GPR-4 is also similar to six predicted but uncharacterized proteins containing seven TMs from other ascomycete filamentous fungi. These are CHG04337 (E = 2e−65) from the white rot pathogen Chaetomium globosum, SS1G_08243.1 from the necrotrophic pathogen Sclerotinia sclerotiorum, BC1G_03450.1 (E = 4e−41) from the gray mold pathogen Botrytis cinerea, FG05006.1 (E = 3e−41) from the pathogenic filamentous ascomycete Gibberella zeae PH-1 (anamorph, Fusarium graminearum), and MG08803.4 (E = 8e−34) from the rice BLAST fungus Magnaporthe grisea (Fig. 1). These proteins have large third intracellular loops (83 to 348 amino acids) and carboxy-terminal tails (84 to 155 amino acids) and are most similar to GPR-4 in the 7-TM helix regions and connecting loops (especially in the second and third intracellular and second extracellular loops).
The gpr-4 gene structure predicted by the automated gene caller (http://www.broad.mit.edu/annotation/genome/neurospora/home.html) was verified using RT-PCR with gpr-4-specific primers that amplify the region containing the intron (Fig. 2B). The gpr-4 message is of relatively low abundance and could not be detected using Northern analysis (data not shown). To elucidate the expression profile of gpr-4 throughout growth and development, RT-PCR was performed. We began with analysis of total RNA from conidia, 8- and 16-h shaken submerged cultures, and vegetative (VM) and sexually differentiated (SCM) plate cultures, all with 1.5% sucrose as the carbon source. gpr-4 is expressed to detectable levels in three tissues (16-h submerged cultures and VM and SCM plates), with the highest level of gpr-4 mRNA present in VM plate cultures (Fig. 2B, left panel). gpr-4 message could also be detected in conidia and 8-h submerged cultures, although the relative levels of expression were more variable (data not shown). We extended our studies to tissues grown in 1.5% (43.8 mM) and 100 mM sucrose, 100 mM glucose, and 100 mM glycerol, a relatively poor carbon source (Fig. 2B, right panel). The results demonstrate that gpr-4 transcript levels are similar in glucose and both concentrations of sucrose but are highest in glycerol-grown cultures.
Deletion of gpr-4 by targeted gene replacement and isolation of a Δgpr-4 gpr-4+-complemented strain.
A Δgpr-4 mutant was created by electroporation of a wild-type strain with a construct in which the gpr-4 ORF region was replaced with the hygromycin B resistance marker gene hph (Fig. 2A) (see Materials and Methods). Heterokaryotic primary transformants were identified by Southern analysis (data not shown) and then crossed to a wild-type strain of the opposite mating type. Homokaryotic Δgpr-4 mutants were obtained by selection of the progeny on hygromycin-containing medium and verified by Southern analysis (Fig. 2C).
A complemented Δgpr-4 strain (Δgpr-4::hph+ gpr-4+::his-3+) was constructed by targeting a construct containing the wild-type gpr-4 allele to the his-3 locus as described in Materials and Methods and was verified by Southern analysis (data not shown). The gpr-4 mRNA could be detected in Δgpr-4 gpr-4+ and wild-type strains but not in Δgpr-4 mutants (Fig. 2D; data not shown), thus demonstrating restoration of gpr-4 expression in the complemented strains.
Δgpr-4 mutants accumulate less mass than the wild type on poor carbon sources.
Extensive phenotypic analysis was performed on the Δgpr-4 mutants. Δgpr-4 strains are fertile as males or females, and ascospores produced from crosses involving Δgpr-4 mutants germinated normally. Δgpr-4 mutants did not exhibit defects during asexual growth and development (colony morphology, aerial hypha height, conidiation, and dry mass) on minimal medium containing sucrose, fructose, or glucose at 30°C (Fig. 3A; data not shown). The apical extension rates of basal hyphae from Δgpr-4 mutants were normal on preferred or poor carbon and/or nitrogen sources (data not shown). Hyphal fusion was normal, as assayed by formation of forced heterokaryons (using strains with different auxotrophic markers) and by microscopic analysis of formation and fusion of conidial anastomosis tubes (55). Growth in VM-sucrose-submerged cultures was normal, as was sensitivity to 0.75 M NaCl, 0.75 M KCl, 1.5 M sorbitol, 5% ethanol, 1% sorbose, or high agar concentration (4%). In fact, the only defect observed for Δgpr-4 mutants when cultured on VM-sucrose medium was a slower apical extension rate (∼70% of wild type) and less dry mass accumulation at the elevated growth temperature of 42°C.
FIG. 3.
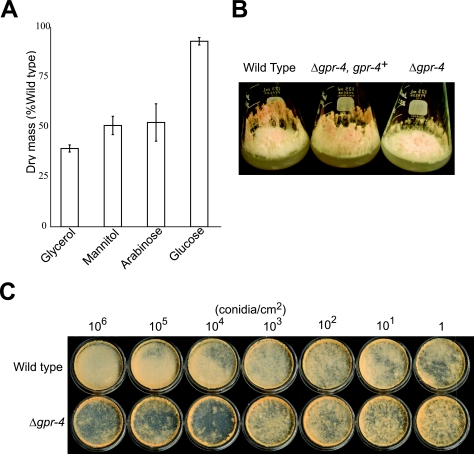
Phenotypes of Δgpr-4 mutants. (A) Dry mass using 100 mM glucose, glycerol, mannitol, or arabinose as a sole carbon source in solid medium. One-microliter aliquots containing 1 × 109 conidia were inoculated in the center of cellophane-overlaid plates. Tissues were collected after incubation for 3 days at 30°C in the dark, and dry mass was measured as described in Materials and Methods. Strains were 74A (wild type) and 7-32 (Δgpr-4 mutant). Data are from three independent experiments, with three replicates/experiment. Errors are calculated as standard errors. (B) Δgpr-4 mutants have fewer hyphae than the wild type when cultured on glycerol medium. VM-glycerol flasks were inoculated in the center using 1 × 109 conidia (in 1 μl) and then incubated for 3 days in the dark at 30°C, followed by four more days under light at 25°C. Strains are the same as for Fig. 2D. (C) Cell density-dependent mass formation on glycerol solid medium. The indicated amounts of conidia were spread evenly onto the surface of 35-mm-diameter VM-glycerol plates, followed by incubation in the dark at 30°C. Plates were photographed after 8 days. Strains are 74A (wild type) and 7-32 (Δgpr-4 mutant).
Since GPR-4 is homologous to putative carbon sensory receptors found in yeasts, we performed further detailed phenotypic analysis in the presence of different carbon sources. When grown on poor carbon sources (100 mM glycerol, mannitol, or arabinose), the dry mass of Δgpr-4 strains was significantly less than that of the wild type (Fig. 3A). The relative defect was most severe on glycerol, where the dry mass was approximately one-third that of the wild type (Fig. 3A). The reduced mass accumulation of Δgpr-4 mutants cultured on glycerol solid medium cannot be explained by defects in conidial germination or hyphal fusion. In contrast, we consistently observed that wild-type strains produced more aerial hyphae than the Δgpr-4 mutant at the inoculation point on VM-glycerol plates (data not shown). The Δgpr-4 mutant also had fewer aerial hyphae than the wild type during growth in agar flasks or in standing liquid cultures with 100 mM glycerol as a carbon source (Fig. 3B; data not shown). The reduction in the quantity of aerial hyphae likely explains the reduced biomass accumulation observed in Δgpr-4 mutants relative to the wild type. On the other hand, Δgpr-4 mutants form the same amount of conidia as wild type (data not shown). Thus, Δgpr-4 mutants produce more conidia per aerial hypha, a result which is also supported by microscopic observation.
To investigate the possible cause of the aerial hypha production defect in Δgpr-4 mutants, we next examined the effect of spreading VM-glycerol plates with different amounts of conidia from Δgpr-4 and wild-type strains. At a lower conidial density (1 to 1,000 conidia/cm2 agar medium), wild-type and Δgpr-4 strains produce similar amounts of basal and aerial hyphae when cultured on VM-glycerol plates (Fig. 3C). However, at higher amounts of conidia (106, 105, or 104/cm2 agar medium), the wild-type strain formed more aerial hyphae than the Δgpr-4 mutant (Fig. 3C). The defects in hypha formation of Δgpr-4 strains led to decreased biomass (Fig. 3C). Taken together, our results suggest that biomass accumulation in Δgpr-4 mutants when cultured on VM-glycerol medium is dependent on the initial density of conidia used for inoculation, with defects only observed at levels at or above 104/cm2. Thus, GPR-4 appears to negatively regulate a previously uncharacterized pathway involving formation of aerial hyphae at high inoculation cell densities in N. crassa.
The Gα gene gna-1 is epistatic to gpr-4 with regards to mass accumulation on glycerol medium.
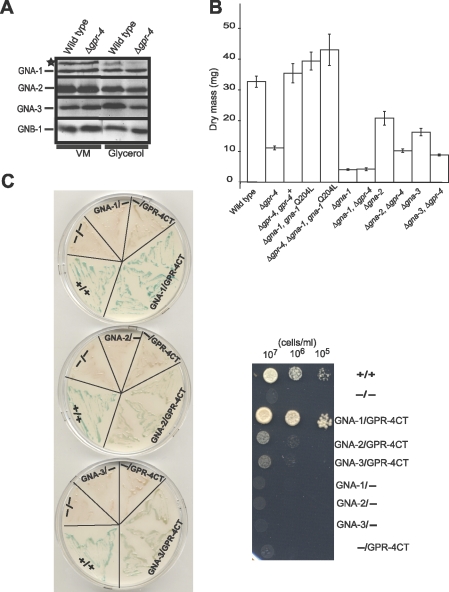
As mentioned above, N. crassa possesses three Gα protein genes, gna-1, gna-2, and gna-3. Of these, the protein encoded by gna-3 is most similar to the yeast Gα proteins represented by S. cerevisiae Gpa2p. Our laboratory has previously demonstrated roles for GNA-3 in regulation of adenylyl cyclase protein levels and conidiation (31, 33). However, other studies have shown that GNA-1, not GNA-3, regulates the activity of adenylyl cyclase in N. crassa (29, 31). GNA-2 appears to play a compensatory role in relation to GNA-1 and GNA-3 (3, 31). To elucidate which Gα protein(s) operates downstream of GPR-4, we conducted epistasis analyses of gpr-4 and the three Gα genes. To control for possible effects of the Δgpr-4 mutation on G protein stability, we first analyzed levels of the three Gα proteins and the Gβ protein in the Δgpr-4 background. Western blot analyses were performed using tissues from VM-sucrose and VM-glycerol plates (Fig. 4A). Levels of GNA-1, GNA-2, GNA-3, and GNB-1 were similar in wild-type and Δgpr-4 strains cultured with the two carbon sources. Thus, G protein levels are similar with sucrose or glycerol as a carbon source and are also not affected by loss of gpr-4 in N. crassa.
FIG. 4.
GPR-4 is coupled to GNA-1. (A) Levels of heterotrimeric G protein subunits. A cell fraction enriched for plasma membranes was isolated from VM-glycerol and VM-sucrose plate cultures, and samples containing 30 μg protein were analyzed by Western blot using G protein antisera (see Materials and Methods for details). Strains are 74A (wild type) and 7-32 (Δgpr-4 mutant). The asterisk denotes a nonspecific band recognized by the GNA-1 antiserum. (B) Epistasis analyses between gpr-4 and the three Gα genes. Experimental conditions and dry mass determination are the same as for Fig. 3A. Strains are 74A (wild type), 7-32 (Δgpr-4), 1B8 (Δgna-1), a1r54 (Δgna-1 Δgpr-4), a29-1 (Δgna-2), a2r2 (Δgna-2 Δgpr-4), 2a1 (Δgna-3), a3r1 (Δgna-1 Δgpr-4), Rmi2 (Δgpr-4 gpr-4+), Δ1gna-1Q204L (Δgna-1 gna-1Q204L), and a1rQ204L-25 (Δgpr-4 Δgna-1 gna-1Q204L). Data are the averages of results from two representative independent experiments, and errors are calculated as standard errors. (C) Yeast two-hybrid assay. Left panel: β-galactosidase assay. Yeast transformants carrying the indicated ORFs or domains in the pGBKT7 (left of slash) or pGAD424 (right of slash) vectors were cultured on SD minus leucine and tryptophan plates for ∼2 days. β-Galactosidase activity was determined using filter assays (see Materials and Methods for details), and photos were taken 2 h after application of the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside substrate solution. Right panel: growth assay. Serial dilutions were made of SD minus leucine and tryptophan liquid cultures of yeast strains with the indicated ORFs or protein domains. Aliquots corresponding to each concentration of cells (107, 106, or 105 cells/ml) were spotted onto SD plates lacking adenine, histidine, leucine, and tryptophan, supplemented with 5 mM 3-aminotriazole. The plate was photographed after incubation at 30°C for 3 days. GNA-1, GNA-1 ORF; GNA-2, GNA-2 ORF; GNA-3, GNA-3 ORF; CT, carboxy terminus of GPR-4; +/+, positive control containing pGBKT7-53 and pGADT7-Rec (Clontech); −, pGBKT7 (left) or pGAD424 (right) empty vector.
We next examined the phenotype of the three Gα mutants on solid medium with glycerol as a carbon source. All of the Gα single mutants possessed approximately the same dry mass as the wild type when cultured with glucose as a carbon source (data not shown). However, the Δgna-1, Δgna-2, and Δgna-3 mutants accumulated less mass than the wild type when grown on VM-glycerol (Fig. 4B). Levels in Δgna-1 strains showed the greatest reduction, with only 13% of the mass of the wild type, while accumulation in Δgna-2 and Δgna-3 mutants was 51 to 65% of that of the wild type, respectively (Fig. 4B). The observation of reduced mass in Δgna-2 strains cultivated on VM-glycerol is the first report of a phenotype for mutants lacking only the gna-2 gene.
To analyze the relationship between gpr-4 and the three Gα genes, we constructed Δgna-1 Δgpr-4, Δgna-2 Δgpr-4, and Δgna-3 Δgpr-4 double mutants. Examination of mass accumulation on glycerol solid medium showed that Δgpr-4 Δgna-2 and Δgpr-4 Δgna-3 mutants have a more severe defect than the Δgna-2 or Δgna-3 single mutant (Fig. 4B). This result suggests that GNA-2 and GNA-3 do not function downstream of GPR-4 to regulate mass accumulation on glycerol medium. In contrast, Δgna-1 Δgpr-4 double mutants possess the same reduced dry mass as Δgna-1 strains. This finding indicates that gna-1 is epistatic to gpr-4 (Fig. 4B).
We further probed the epistatic relationship between gna-1 and gpr-4 through analysis of a Δgpr-4 strain containing either of two previously characterized, GTPase-deficient, constitutively activated gna-1 alleles, gna-1Q204L and gna-1R178C (79). If GPR-4 is a GPCR coupled to GNA-1, then mutational activation of gna-1 should suppress defects caused by the Δgpr-4 mutation. To obtain N. crassa strains containing a gna-1-activated allele in the Δgpr-4 background, the gna-1Q204L and gna-1R178C constructs were targeted to the his-3 locus of a Δgna-1 Δgpr-4 his-3 strain and the desired transformants were selected and purified (see Materials and Methods). The resulting homokaryotic Δgpr-4 Δgna-1 gna-1Q204L or Δgpr-4 Δgna-1 gna-1R178C strains are identical to Δgna-1 gna-1Q204L or Δgna-1 gna-1R178C strains with respect to mass accumulation and colony morphology (Fig. 4B; data not shown), demonstrating that introduction of a constitutively activated gna-1 allele can suppress the defects of the Δgpr-4 mutation. This result further supports the hypothesis that GNA-1 acts downstream of GPR-4.
Our previous results demonstrated that GNA-1 is required for GTP-stimulated adenylyl cyclase (encoded by cr-1) activity in N. crassa (79). Based on the epistatic relationship between GPR-4 and GNA-1, we also compared the dry mass of cr-1 mutants to that of Δgpr-4 and wild-type strains. It has previously been reported that cr-1 mutants grow extremely poorly in the presence of several poor carbon sources, including glycerol, mannitol, and arabinose (66). Therefore, we compared the dry mass of cr-1 and wild-type strains cultured with these carbon sources on solid medium. With glucose as the carbon source, the cr-1 mutant has a dry mass that is 44.0 ± 2.8% of that of wild-type strains (data not shown). When grown in the presence of glycerol, mannitol, or arabinose, mass accumulation in the cr-1 strain was only 18.8 ± 2.2%, 25.4 ± 8.0%, or 21.6 ± 3.1% of that of the wild type cultured on the same medium, respectively (Fig. 3A; also data not shown). Correcting for the decreased mass of cr-1 mutants relative to the wild type on glucose (44%), the relative reduction in mass accumulation on glycerol, mannitol, or arabinose is similar for cr-1 and Δgpr-4 mutants. Furthermore, on solid VM-glycerol medium, the dry weight of the cr-1 mutant is similar to that observed for Δgna-1 and Δgpr-4 Δgna-1 strains (Fig. 4B; also data not shown). Thus, the mass accumulation pattern of Δgpr-4, Δgna-1, and cr-1 mutants is consistent with a cAMP-dependent pathway regulating growth on solid medium with glycerol as the carbon source.
GPR-4 physically interacts with GNA-1 in the yeast two-hybrid assay.
Since the results of epistasis experiments suggested that GPR-4 acts upstream of GNA-1 during mass accumulation on glycerol medium, we utilized the yeast two-hybrid assay to examine a possible physical interaction between GPR-4 and GNA-1. Assays with GNA-2 and GNA-3 were included as controls. For these tests, the entire ORF for each Gα gene was cloned in frame behind the GAL4 activation domain in pGAD424, while a carboxy-terminal fragment of GPR-4 (amino acids 527 to 654) was inserted in frame behind the GAL4 DNA binding domain in pGBKT7. The corresponding carboxy-terminal region of S. cerevisiae Gpr1p has been demonstrated to interact with Gpa2p in yeast two-hybrid assays (78, 82). We performed the two hybrid assays with the N. crassa proteins using two methods. The activity of the β-galactosidase reporter was measured using filter assays on plates (Fig. 4C, left panel). Expression of the ADE-2 and HIS-3 reporters was monitored by assessing growth on medium lacking adenine and histidine (Fig. 4C, right panel).
In β-galactosidase assays of cells containing GNA-1 and the carboxy terminus of GPR-4, a dark blue color developed within 30 min (similar to the positive control), representative of a strong interaction. However, the color was much paler and took 1 to 2 h to develop in assays of cells containing the GPR-4 carboxy terminus and GNA-2 or GNA-3, indicating a very weak interaction. The growth assays on medium lacking histidine and adenine showed that strains with the GNA-1 and carboxy-terminal GPR-4 vectors grew very well, similar to the positive control (Fig. 4C, right panel). In contrast, cells containing the GPR-4 carboxy terminus and GNA-2 or GNA-3 exhibited growth only at higher cell densities and grew only slightly better than negative controls (Fig. 4). Thus, the two-hybrid assay provided evidence for a direct interaction between GPR-4 and all three Gα proteins, with the strongest binding to GNA-1. These results further strengthen the notion that GPR-4 is coupled to GNA-1. To our knowledge, there have been no previous reports of a GPCR that physically interacts with a Gα protein related to GNA-1 in filamentous fungi.
GPR-4 is required for a carbon source-dependent transient increase in cAMP levels.
A common downstream effector pathway of fungal heterotrimeric G proteins involves cAMP signaling. The enzyme adenylyl cyclase catalyzes the conversion of ATP to cAMP. As mentioned above, our laboratory has previously shown that GNA-1 is required for normal GTP-stimulated adenylyl cyclase activity. The results from epistatic analysis and two-hybrid assays support an interaction between GPR-4 and GNA-1. Taken together, these findings raise the possibility that GPR-4 may regulate adenylyl cyclase activity. Therefore, we next analyzed the relationship between GPR-4 and levels of cAMP and adenylyl cyclase protein in N. crassa.
We first tested whether addition of 1 mM exogenous cAMP to VM-glycerol medium would affect the dry mass phenotype of Δgpr-4 strains. As shown in Fig. 3A and 4B, the dry mass of Δgpr-4 mutants was only about one-third of that of the wild type in the absence of cAMP. The addition of cAMP increased the mass of both wild-type and Δgpr-4 strains, but it had the greatest effect on Δgpr-4. In the presence of 1 mM cAMP, the dry mass of Δgpr-4 was two-thirds of that of the wild type, the same level accumulated by the wild type without exogenous cAMP (data not shown). Thus, cAMP addition achieved a partial rescue of the dry mass defects of Δgpr-4 mutants on VM-glycerol medium.
We next measured steady-state cAMP levels in wild-type, Δgpr-4, and Δgpr-4 gpr-4+-complemented strains when cultured on VM-sucrose or VM-glycerol plates. Interestingly, cAMP amounts did not vary greatly whether sucrose or glycerol was the carbon source (Table 3). The level of cAMP was also similar in all three strains, indicating that the Δgpr-4 mutation does not significantly affect steady-state intracellular cAMP levels in N. crassa (Table 3).
TABLE 3.
Steady-state intracellular cAMP levels
| Genotype | Strain | Mean ± SE intracellular cAMP level [pmol/mg protein (% of wild type)] on VM plates with indicated carbon sourcea:
|
|
|---|---|---|---|
| Sucrose | Glycerol | ||
| Wild type | 74A | 4.66 ± 0.20 (100) | 5.65 ± 0.50 (100) |
| Δgpr-4 | 7-32 | 4.77 ± 0.18 (102.4) | 4.62 ± 0.81 (81.8) |
| Δgpr-4 gpr-4+ | Rmi2 | 3.59 ± 0.66 (77.0) | 6.01 ± 0.76 (106.4) |
Data are from two independent experiments.
It has been shown that S. cerevisiae Gpr1p is required for a brief increase in cAMP levels after the addition of glucose to glucose-starved cultures (37, 81). In addition, our laboratory has proposed that GNA-1 may be required for a transient cAMP increase during growth and development, as Δgna-1 mutants have low adenylyl cyclase activity but normal steady-state cAMP levels in shaken submerged cultures (29). In light of these findings, we next analyzed cAMP levels after the transition from a poor carbon source (glycerol) to glucose in wild-type and Δgpr-4 strains.
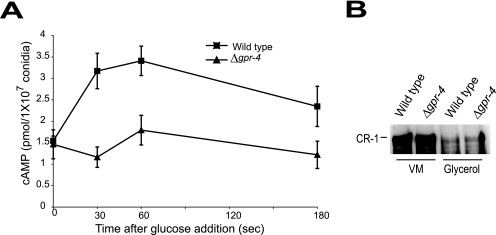
Consistent with the results presented in Table 3, levels of cAMP were similar in glycerol-grown submerged cultures of wild-type and Δgpr-4 strains at time zero (Fig. 5A). The addition of glucose led to more than a twofold increase in cAMP levels within 30 to 60 s in wild-type cells. In contrast, no significant difference in cAMP amount was observed for the Δgpr-4 mutant over this same time period (Fig. 5A). These results indicate that GPR-4 is required for the glucose-dependent transient increase in cAMP levels, supporting a role for GPR-4 in glucose sensing.
FIG. 5.
Measurement of cAMP levels after glucose addition and comparison of adenylyl cyclase (CR-1) protein levels. (A) cAMP levels after glucose addition. Conidia were inoculated in VM-glycerol liquid medium and cultured with shaking at 30°C for 16 h. Glucose was added to a final concentration of 100 mM (time = 0 s), and samples were collected at the indicated times (see Materials and Methods for details). Data are the averages of results from four independent experiments. Strains are 74A (wild type; squares) and 7-32 (Δgpr-4 mutant; triangles). Errors are indicated as standard errors. (B) Levels of CR-1 protein. Whole-cell extracts were prepared from shaken submerged VM-sucrose and VM-glycerol cultures, and aliquots containing 90 μg of protein were subjected to Western blot analysis using a CR-1 antibody. Strains are the same as for Fig. 2D.
We also examined adenylyl cyclase (CR-1) protein levels in VM-sucrose- and VM-glycerol-submerged cultures of wild-type and Δgpr-4 strains using a CR-1 antiserum (28) (Fig. 5B). CR-1 levels are similar in the wild type and Δgpr-4 mutants cultured on the same medium. This observation is consistent with previous results from our laboratory, indicating that GNA-1 does not influence levels of CR-1 protein (28). Interestingly, the carbon source did affect CR-1 levels, as CR-1 amounts in both strains are greater with VM-sucrose than with VM-glycerol (Fig. 5B).
DISCUSSION
Glucose, sucrose, maltose, fructose, and mannose are among the best carbon sources for supporting the growth of N. crassa, while arabinose, mannitol, glycerol, and sorbose only allow slow growth and low biomass accumulation (15). The finding that Δgpr-4 mutants have reduced mass accumulation compared to the wild type when cultured with poor carbon sources and lack the transient increase in cAMP levels normally observed during the shift from glycerol to glucose-rich medium suggests that GPR-4 may act as a carbon sensor in N. crassa. GPR-4 is most similar to several hypothetical seven-transmembrane helix proteins from pathogenic filamentous fungi. This indicates that the ScGpr1p GPCR superfamily (22, 42) is widely present in pathogenic filamentous fungi and that a carbon-sensing GPCR-Gα protein-adenylyl cyclase-cAMP-PKA pathway is likely to be functionally conserved in these organisms. In the filamentous fungus Aspergillus nidulans, the heterotrimeric G-protein GanB(α)-SfaD(β)-GpgA(γ) has been implicated in a carbon-sensing cAMP/PKA pathway that regulates conidial germination (12, 20, 41), but as yet, no GPCR has been reported for this pathway. The closest match to N. crassa GPR-4 in A. nidulans is GprC (23) (AN3765; E = 6e−3). However, GprC is actually more similar to N. crassa GPR-1 (E = 5e−05), GPR-2 (E = 3e−03), and GPR-3 (E = 4e−05), predicted GPCRs with similarity to cAMP receptor-like proteins found in D. discoideum (8, 54).
The results from epistasis analysis and yeast two-hybrid assays support the hypothesis that GNA-1 interacts with and operates downstream of GPR-4 to regulate the growth and development of N. crassa in the presence of poor carbon sources (Fig. 6). The coupling between GPR-4 and GNA-1 illustrates the mechanistic diversity of the G protein signaling pathways that are involved in carbon sensing in fungi. As mentioned above, GPR-4 is homologous to Gpr1p in S. cerevisiae and Git3 in S. pombe. However, the N. crassa homologue of the yeast Gα proteins coupled to these GPCRs is GNA-3, not GNA-1 (33). This variation could stem from several reasons, including regulatory differences due to the presence of three instead of two Gα subunits in ascomycete filamentous fungi versus yeasts. In addition, as mentioned above, both GNA-1 and GNA-3 regulate adenylyl cyclase in N. crassa, and it has also been demonstrated that the homologue of either GNA-1 or GNA-3 is required for pathogenesis in filamentous fungal species (reviewed in references 7, 32, and 43). Since it has been shown in many cases that modulation of cAMP levels by the Gα protein is an important aspect of pathogenesis, the relative importance of GNA-1 versus GNA-3 to cAMP levels (and pathogenesis) may result from the impact of adenylyl cyclase activity versus protein levels in various species.
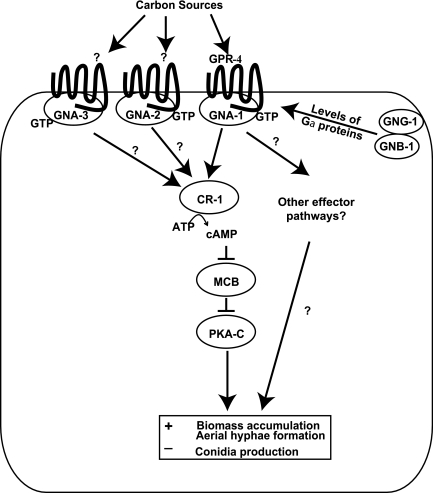
FIG. 6.
Model for control of carbon source-dependent growth and development by GPR-4 and heterotrimeric G proteins in N. crassa. During growth on poor carbon sources such as glycerol, ligand binding to GPR-4 activates a GNA-1 signaling pathway to modulate growth (mass accumulation), conidiation, and aerial hypha formation. A likely effector is adenylyl cyclase, but other downstream targets may also be regulated by the GPR-4/GNA-1 pathway. GNA-2 and GNA-3 also regulate mass accumulation on glycerol medium, through a GPR-4-independent mechanism. The ligands that bind to GPR-4 are currently unknown but may include sugars or other cellular metabolites.
The GPCRs that are coupled to GNA-2 and GNA-3 during growth on glycerol are currently unknown. Although our results support a scenario in which GPR-4 acts through GNA-1 to regulate mass accumulation on glycerol, we cannot rule out that, under certain circumstances, GPR-4 may also interact with GNA-2 and GNA-3. There are reports of GPCRs that interact with multiple Gα proteins, particularly in mammals (for examples, see references 11, 14, 19, 39, 40, 44, 51, 58, 62, and 76). As mentioned above, GPR-4 is expressed under a variety of growth conditions and, thus, potentially may respond to different environmental signals through coupling to more than one G protein. Such a scenario has been proposed to explain the patterns of G protein coupling to prokineticin receptors in humans (14).
Although the response to glucose is lost in Δgpr-4 mutants, we do not know whether glucose is the in vivo ligand for GPR-4. The correlation between cAMP responses and elicitation of a phenotype is not absolute for the related group of receptors in fungi. In S. cerevisiae, data support sucrose and glucose as agonist ligands for Gpr1p, while mannose is an antagonist (42). However, maltose stimulates hyphal growth but does not elicit a cAMP transient after addition (56). In S. pombe, Git3 is required for increased cAMP levels after the transfer from glucose starvation to glucose-rich conditions (10, 73). C. albicans CaGpr1 has been variously reported to respond to glucose (48) or amino acids, including alanine and methionine (46, 47), for activation of cAMP synthesis. However, cAMP levels are not affected by proline addition, even though this amino acid causes morphological phenotypes (4). In the filamentous fungus A. nidulans, a transient increase in cAMP amount has been reported after glucose addition to starved cultures, although, as mentioned above, the GPCR required for this carbon-sensing cAMP signaling pathway has not been identified (41). In C. neoformans, the GPCR Gpr4 is required to sustain a short-lived increase in cAMP levels in response to methionine but not glucose (77). We did not observe any alteration in growth or colony morphology in N. crassa after the addition of methionine to media with high or low carbon content (data not shown), suggesting that GPR-4 responds to carbon source(s) and not this amino acid.
Wild-type strains produce much more aerial hyphae than Δgpr-4 mutants when plated at a higher cell density (≥104 conidia/cm2) on glycerol medium. This result is consistent with a scenario in which GPR-4 negatively regulates a cell density-dependent system involving aerial hypha formation in N. crassa. Cell density-dependent control of aerial hypha production by GPR-4 could be regulated by small molecules. In C. albicans, tyrosol, farnesol, and farnesoic acid have been recently shown to act as quorum-sensing molecules that regulate the switch between yeast and filamentous forms (13, 26, 52). The filamentous fungus A. nidulans does not produce farnesol, but exogenous farnesol causes apoptosis in this species (61). Based on these observations, it has been proposed that farnesol production by C. albicans may lead to a competitive advantage relative to other fungal species in the environment (26, 61). It will be interesting to determine if quorum-sensing molecules control aerial hypha formation in a pathway involving GPR-4 in N. crassa. It is possible that GPR-4 directly binds both the carbon and cell density signal ligands and even that the same chemical is shared between the two responses.
cAMP is an important regulator of vegetative growth and development in N. crassa and other filamentous fungi. Our experiments showed that the cr-1 and Δgpr-4 mutants grow poorly in comparison to the wild type in plate cultures with glycerol, mannitol, or arabinose as the carbon source and that exogenous cAMP can partially reverse the dry mass defect of Δgpr-4 mutants on glycerol solid medium. Although the Δgpr-4 mutation did not lead to a significant difference in steady-state intracellular cAMP levels on VM-glycerol plates, Δgpr-4 mutants do lack the cAMP transient increase observed in wild-type cells when transferred from glycerol to glucose-rich medium. Taken together, these findings are consistent with a role for GPR-4 in the regulation of cAMP levels in N. crassa.
Three glucose sensing/signaling pathways have been identified in S. cerevisiae, including the Snf1p protein kinase/Mig1p repressor pathway, the Snf3p and Rgt2p glucose sensors/Rgt1 repressor system, and the Gpr1p/Gpa2p/cAMP pathway (30, 57). Our work supports alignment of GPR-4 with the third pathway, presumably functioning in the presence of poor carbon sources in N. crassa. Previous studies have provided evidence for a second carbon sensory pathway, which involves rco-3 (45). RCO-3 is required for high- and low-affinity glucose transport in N. crassa (45). Glucose deprivation triggers conidiation in wild-type N. crassa cultures; however, rco-3 null mutants conidiate inappropriately in submerged culture in the presence of abundant glucose, suggesting that they have lost sensory capabilities (45). Moreover, the sequence and functional similarity observed between RCO-3 and S. cerevisiae Snf3p and Rtg2p supports the hypothesis that RCO-3 functions as a carbon sensor in N. crassa (45). Future studies will probe possible cross talk between the GPR-4 and RCO-3 carbon sensory pathways in N. crassa.
Δgpr-4 mutants exhibit their most severe mass defects during growth on glycerol, which may have implications for the understanding and control of plant pathogenic fungi. Many studies indicate that glycerol participates in numerous pathways and serves diverse cellular roles (9). Glycerol has recently been shown to be a major nutrient obtained by the fungal pathogen from the host plant (72). On the other hand, Δgpr-4 mutants are also defective during growth on arabinose; this sugar is a main component of the monocot cell wall and thus likely to be encountered by fungal plant pathogens in nature (63, 68). As GPR-4 has many close homologues in pathogenic filamentous fungi, the study of GPR-4 may shed light on a possible function for these proteins during pathogenesis, growth, and development. Future work will focus on identification of the stimulatory ligands and downstream signaling pathway(s) controlled by GPR-4 during carbon sensing in N. crassa.
Acknowledgments
We thank Svetlana Krystofova, Hyojeong Kim, Gyungsoon Park, Suzanne Phillips, Carol Jones, and Sara Martinez for comments on the manuscript and/or many helpful discussions. We acknowledge Hyojeong Kim and Sara Martinez for two-hybrid vector construction and advice on procedures. We thank Jingxiao Ye for assistance with certain experimental procedures.
This work was supported by grant no. GM 48626 from the National Institutes of Health to K.A.B.
REFERENCES
- 1.Ansari, K., S. Martin, M. Farkasovsky, I.-M. Ehbrecht, and H. Kuntzel. 1999. Phospholipase C binds to the receptor-like GPR1 protein and controls pseudohyphal differentiation in Saccharomyces cerevisiae. J. Biol. Chem. 274:30052-30058. [DOI] [PubMed] [Google Scholar]
- 2.Aramayo, R., and R. L. Metzenberg. 1996. Gene replacements at the his-3 locus of Neurospora crassa. Fungal Genet. Newsl. 43:9-13. [Google Scholar]
- 3.Baasiri, R. A., X. Lu, P. S. Rowley, G. E. Turner, and K. A. Borkovich. 1997. Overlapping functions for two G protein α subunits in Neurospora crassa. Genetics 147:137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batlle, M., A. Lu, D. A. Green, Y. Xue, and J. P. Hirsch. 2003. Krh1p and Krh2p act downstream of the Gpa2p Gα subunit to negatively regulate haploid invasive growth. J. Cell Sci. 116:701-710. [DOI] [PubMed] [Google Scholar]
- 5.Bieszke, J. A., E. L. Braun, L. E. Bean, S. Kang, D. O. Natvig, and K. A. Borkovich. 1999. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. USA 96:8034-8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieszke, J. A., E. N. Spudich, K. L. Scott, K. A. Borkovich, and J. L. Spudich. 1999. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry 38:14138-14145. [DOI] [PubMed] [Google Scholar]
- 7.Bolker, M. 1998. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 25:143-156. [DOI] [PubMed] [Google Scholar]
- 8.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brisson, D., M.-C. Vohl, J. St-Pierre, T. J. Hudson, and D. Gaudet. 2001. Glycerol: a neglected variable in metabolic processes? BioEssays 23:534-542. [DOI] [PubMed] [Google Scholar]
- 10.Byrne, S. M., and C. S. Hoffman. 1993. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 105:1095-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabre, O., B. R. Conklin, S. Brandon, H. R. Bourne, and L. E. Limbird. 1994. Coupling of the alpha 2A-adrenergic receptor to multiple G-proteins. A simple approach for estimating receptor-G-protein coupling efficiency in a transient expression system. J. Biol. Chem. 269:5730-5734. [PubMed] [Google Scholar]
- 12.Chang, M. H., K. S. Chae, D. M. Han, and K. Y. Jahng. 2004. The GanB Gα-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans. Genetics 167:1305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H., M. Fujita, Q. Feng, J. Clardy, and G. R. Fink. 2004. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. USA 101:5048-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, J., C. Kuei, S. Sutton, S. Wilson, J. Yu, F. Kamme, C. Mazur, T. Lovenberg, and C. Liu. 2005. Identification and pharmacological characterization of prokineticin 2β as a selective ligand for prokineticin receptor 1. Mol. Pharmacol. 67:2070-2076. [DOI] [PubMed] [Google Scholar]
- 15.Davis, R. H. 2000. Neurospora: contributions of a model organism. Oxford University Press, New York, N.Y.
- 16.Davis, R. H., and F. J. D. Serres. 1970. Genetic and microbiological research techniques in Neurospora crassa. Methods Enzymol. 71A:79-143. [Google Scholar]
- 17.Dohlman, H. G., J. Thorner, M. G. Caron, and R. J. Lefkowitz. 1991. Model systems for the study of seven-transmembrane-segment receptors. Annu. Rev. Biochem. 60:653-688. [DOI] [PubMed] [Google Scholar]
- 18.Ebbole, D., and M. S. Sachs. 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37:17-18.
- 19.Fernandez-Fernandez, J. M., F. C. Abogadie, G. Milligan, P. Delmas, and D. A. Brown. 2001. Multiple pertussis toxin-sensitive G-proteins can couple receptors to GIRK channels in rat sympathetic neurons when expressed heterologously, but only native Gi-proteins do so in situ. Eur. J. Neurosci. 14:283-292. [DOI] [PubMed] [Google Scholar]
- 20.Fillinger, S., M. K. Chaveroche, K. Shimizu, N. Keller, and C. d'Enfert. 2002. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44:1001-1016. [DOI] [PubMed] [Google Scholar]
- 21.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L.-J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. C. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 22.Graul, R. C., and W. Sadee. 2001. Evolutionary relationships among G protein-coupled receptors using a clustered database approach. AAPS Pharm. Sci. 3:E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, K.-H., J.-A. Seo, and J.-H. Yu. 2004. A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol. Microbiol. 51:1333-1345. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman, C. S. 2005. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 33:257-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornby, J. M., E. C. Jensen, A. D. Lisec, J. J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K. W. Nickerson. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivey, F. D., P. N. Hodge, G. E. Turner, and K. A. Borkovich. 1996. The Gαi homologue gna-1 controls multiple differentiation pathwyas in Neurospora crassa. Mol. Biol. Cell 7:1283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivey, F. D., A. M. Kays, and K. A. Borkovich. 2002. Shared and independent roles for a Gαi protein and adenylyl cyclase in regulating development and stress responses in Neurospora crassa. Eukaryot. Cell 1:634-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivey, F. D., Q. Yang, and K. A. Borkovich. 1999. Positive regulation of adenylyl cyclase activity by a Gαi homologue in Neurospora crassa. Fungal Genet. Biol. 26:48-61. [DOI] [PubMed] [Google Scholar]
- 30.Kaniak, A., Z. Xue, D. Macool, J. H. Kim, and M. Johnston. 2004. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 3:221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kays, A. M., and K. A. Borkovich. 2004. Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all heterotrimeric Gα proteins. Genetics 166:1229-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kays, A. M., and K. A. Borkovich. 2004. Signal transduction pathways mediated by heterotrimeric G proteins. In R. Brambl and G. A. Marzluf (ed.), The mycota, vol. III. Biochemistry and molecular biology, 2nd ed. Springer-Verlag, Berlin, Germany.
- 33.Kays, A. M., P. S. Rowley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the Gα protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20:7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, H., and K. A. Borkovich. 2004. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol. Microbiol. 52:1781-1798. [DOI] [PubMed] [Google Scholar]
- 35.Kim, H., and K. A. Borkovich. 2006. Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot. Cell 5:544-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kore-eda, S., T. Murayama, and I. Uno. 1991. Suppression of the cr-1 mutation in Neurospora crassa. Jpn. J. Genet. 66:77-83. [DOI] [PubMed] [Google Scholar]
- 37.Kraakman, L., K. Lemaire, P. Ma, A. W. R. H. Teunissen, M. C. V. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32:1002-1012. [DOI] [PubMed] [Google Scholar]
- 38.Krystofova, S., and K. A. Borkovich. 2005. The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gβγ dimer required for normal female fertility, asexual development, and Gα protein levels in Neurospora crassa. Eukaryot. Cell 4:365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kukkonen, J. P. 2004. Explicit formulation of different receptor-G-protein interactions and effector regulation. Bioinformatics 20:2411-2420. [DOI] [PubMed] [Google Scholar]
- 40.Kukkonen, J. P. 2004. Regulation of receptor-coupling to (multiple) G proteins. A challenge for basic research and drug discovery. Receptors Channels 10:167-183. [DOI] [PubMed] [Google Scholar]
- 41.Lafon, A., J. A. Seo, K. H. Han, J. H. Yu, and C. d'Enfert. 2005. The heterotrimeric G-protein GanB(α)-SfaD(β)-GpgA(γ) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics 171:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemaire, K., S. Van de Velde, P. Van Dijck, and J. M. Thevelein. 2004. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16:293-299. [DOI] [PubMed] [Google Scholar]
- 43.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, F., I. Usui, L. G. Evans, D. A. Austin, P. L. Mellon, J. M. Olefsky, and N. J. G. Webster. 2002. Involvement of both Gq/11 and Gs proteins in gonadotropin-releasing hormone receptor-mediated signaling in Lbeta T2 cells. J. Biol. Chem. 277:32099-32108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madi, L., S. A. McBride, L. A. Bailey, and D. J. Ebbole. 1997. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics 146:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maidan, M. M., L. De Rop, J. Serneels, S. Exler, S. Rupp, H. Tournu, J. M. Thevelein, and P. Van Dijck. 2005. The G protein-coupled receptor Gpr1 and the Gα protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16:1971-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maidan, M. M., J. M. Thevelein, and P. Van Dijck. 2005. Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem. Soc. Trans. 33:291-293. [DOI] [PubMed] [Google Scholar]
- 48.Miwa, T., Y. Takagi, M. Shinozaki, C.-W. Yun, W. A. Schell, J. R. Perfect, H. Kumagai, and H. Tamaki. 2004. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3:919-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neves, S. R., P. T. Ram, and R. Iyengar. 2002. G protein pathways. Science 296:1636-1639. [DOI] [PubMed] [Google Scholar]
- 50.Nocero, M., T. Isshiki, M. Yamamoto, and C. S. Hoffman. 1994. Glucose repression of fbp1 transcription in Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein {alpha} subunit encoded by gpa2 (git8). Genetics 138:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noda, M., H. Higashida, S. Aoki, and K. Wada. 2004. Multiple signal transduction pathways mediated by 5-HT receptors. Mol. Neurobiol. 29:31-39. [DOI] [PubMed] [Google Scholar]
- 52.Oh, K.-B., H. Miyazawa, T. Naito, and H. Matsuoka. 2001. Purification and characterization of an autoregulatory substance capable of regulating the morphological transition in Candida albicans. Proc. Natl. Acad. Sci. USA 98:4664-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pierce, K. L., R. T. Premont, and R. J. Lefkowitz. 2002. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3:639-650. [DOI] [PubMed] [Google Scholar]
- 54.Raisley, B., M. Zhang, D. Hereld, and J. A. Hadwiger. 2004. A cAMP receptor-like G protein-coupled receptor with roles in growth regulation and development. Dev. Biol. 265:433-445. [DOI] [PubMed] [Google Scholar]
- 55.Roca, M. G., J. Arlt, C. E. Jeffree, and N. D. Read. 2005. Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot. Cell 4:911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rolland, F., J. H. de Winde, K. Lemaire, E. Boles, J. M. Thevelein, and J. Winderickx. 2000. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38:348-358. [DOI] [PubMed] [Google Scholar]
- 57.Rolland, F., J. Winderickx, and J. M. Thevelein. 2002. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2:183-201. [DOI] [PubMed] [Google Scholar]
- 58.Rosenkilde, M. M., K. A. McLean, P. J. Holst, and T. W. Schwartz. 2004. The CXC chemokine receptor encoded by herpesvirus saimiri, ECRF3, shows ligand-regulated signaling through Gi, Gq, and G12/13 proteins but constitutive signaling only through Gi and G12/13 proteins. J. Biol. Chem. 279:32524-32533. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Sciascia, Q. L., P. A. Sullivan, and P. C. Farley. 2004. Deletion of the Candida albicans G-protein-coupled receptor, encoded by orf19.1944 and its allele orf19.9499, produces mutants defective in filamentous growth. Can. J. Microbiol. 50:1081-1085. [DOI] [PubMed] [Google Scholar]
- 61.Semighini, C. P., J. M. Hornby, R. Dumitru, K. W. Nickerson, and S. D. Harris. 2006. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol. Microbiol. 59:753-764. [DOI] [PubMed] [Google Scholar]
- 62.Sidhu, A., and H. B. Niznik. 2000. Coupling of dopamine receptor subtypes to multiple and diverse G proteins. Int. J. Dev. Neurosci. 18:669-677. [DOI] [PubMed] [Google Scholar]
- 63.Solomon, P. S., K.-C. Tan, and R. P. Oliver. 2003. The nutrient supply of pathogenic fungi; a fertile field for study. Mol. Plant Pathol. 4:203-210. [DOI] [PubMed] [Google Scholar]
- 64.Staben, C., B. Jensen, M. Singer, J. Pollock, M. Schechtman, J. Kinsey, and E. Selker. 1989. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newsl. 36:79-81. [Google Scholar]
- 65.Tamaki, H., T. Miwa, M. Shinozaki, M. Saito, C.-W. Yun, K. Yamamoto, and H. Kumagai. 2000. GPR1 regulates filamentous growth through FLO11 in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 267:164-168. [DOI] [PubMed] [Google Scholar]
- 66.Terenzi, H. F., J. A. Jorge, J. E. Roselino, and R. H. Migliorini. 1979. Adenylyl cyclase deficient cr-1 (Crisp) mutant of Neurospora crassa: cyclic AMP-dependent nutritional deficiencies. Arch. Microbiol. 123:251-258. [DOI] [PubMed] [Google Scholar]
- 67.Thevelein, J. M., L. Cauwenberg, S. Colombo, J. H. De Winde, M. Donation, F. Dumortier, L. Kraakman, K. Lemaire, P. Ma, and D. Nauwelaers. 2000. Nutrient-induced signal transduction through the protein kinase A pathway and its role in the control of metabolism, stress resistance, and growth in yeast. Enzyme Microb. Technol. 26:819-825. [DOI] [PubMed] [Google Scholar]
- 68.Tonukari, N. J., J. S. Scott-Craig, and J. D. Walton. 2000. The Cochliobolus carbonum SNF1 gene is required for cell wall-degrading enzyme expression and virulence on maize. Plant Cell 12:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turner, G. E., and K. A. Borkovich. 1993. Identification of a G protein alpha subunit from Neurospora crassa that is a member of the Gi family. J. Biol. Chem. 268:14805-14811. [PubMed] [Google Scholar]
- 70.Versele, M., K. Lemaire, and J. M. Thevelein. 2001. Sex and sugar in yeast: two distinct GPCR systems. EMBO Rep. 2:574-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogel, H. J. 1964. Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat. 98:435-446. [Google Scholar]
- 72.Wei, Y., W. Shen, M. Dauk, F. Wang, G. Selvaraj, and J. Zou. 2004. Targeted gene disruption of glycerol-3-phosphate dehydrogenase in Colletotrichum gloeosporioides reveals evidence that glycerol is a significant transferred nutrient from host plant to fungal pathogen. J. Biol. Chem. 279:429-435. [DOI] [PubMed] [Google Scholar]
- 73.Welton, R. M., and C. S. Hoffman. 2000. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ and the git3 putative glucose receptor. Genetics 156:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wess, J. 1997. G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition. FASEB J. 11:346-354. [PubMed] [Google Scholar]
- 75.Westergaard, M., and H. K. Mitchell. 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34:573-577.
- 76.Wong, S. K. 2003. G protein selectivity is regulated by multiple intracellular regions of GPCRs. Neurosignals 12:1-12. [DOI] [PubMed] [Google Scholar]
- 77.Xue, C., Y. S. Bahn, G. M. Cox, and J. Heitman. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xue, Y., M. Batlle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17:1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang, Q., and K. A. Borkovich. 1999. Mutational activation of a Gαi causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics 151:107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang, Q., S. I. Poole, and K. A. Borkovich. 2002. A G-protein β subunit required for sexual and vegetative development and maintenance of normal Gα protein levels in Neurospora crassa. Eukaryot. Cell 1:378-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yun, C.-W., H. Tamaki, R. Nakayama, K. Yamamoto, and H. Kumagai. 1998. Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 252:29-33. [DOI] [PubMed] [Google Scholar]
- 82.Yun, C.-W., H. Tamaki, R. Nakayama, K. Yamamoto, and H. Kumagai. 1997. G-protein coupled receptor from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 240:287-292. [DOI] [PubMed] [Google Scholar]