Abstract
Bordetella avium is an avian respiratory disease pathogen responsible for substantial economic losses to the turkey industry. The inability to distinguish isolates has hampered outbreak investigations and prevents a complete understanding of transmission mechanisms. Isolates of Bordetella hinzii, often referred to as B. avium-like or as Alcaligenes faecalis type II prior to 1995, have also been acquired from the respiratory tracts of diseased poultry but are not believed to be pathogenic for birds. Therefore, differentiating between B. avium and B. hinzii is of importance for veterinary diagnostic laboratories. It was recently reported that both PvuII ribotyping and HinfI/DdeI restriction endonuclease analysis (REA) show promise for distinguishing isolates of B. avium and B. hinzii. Here we compare the ability of these techniques to discern inter- and intraspecies differences. While both approaches distinguished numerous types within a species, only REA was sufficiently discriminatory for routine use as an epidemiologic tool. Both techniques clearly distinguish between B. avium and B. hinzii, although the results of ribotyping are more easily interpreted. Ribotyping and REA identified numerous, previously unrecognized B. hinzii strains from a collection of bordetella isolates, including one acquired from a rabbit. This is the first report of B. hinzii isolation from a nonhuman mammalian species. At least some of the newly recognized B. hinzii isolates have been previously reported to cause disease in poults, suggesting that the pathogenicity of this agent for poultry should be more rigorously examined.
Bordetella avium is the etiologic agent of coryza or rhinotracheitis in poultry, a highly contagious disease resulting in substantial economic losses to the turkey industry (25). Epidemiologic investigation of outbreaks is not generally undertaken, since no typing system is available for differentiating among isolates. A recent report demonstrated that restriction endonuclease analysis (REA), based on restriction fragment polymorphisms resulting from digestion with HinfI and, separately, with DdeI, may be a useful discriminatory tool (23). Data from the same study suggested that ribotyping with PvuII, previously used to distinguish Bordetella bronchiseptica isolates (19, 20), might also be employed for categorizing strains of B. avium, although only a few isolates were evaluated.
Bordetella hinzii, referred to as B. avium-like or as Alcaligenes faecalis type II prior to 1995 (27), has also been acquired from the respiratory tracts of diseased poultry but has not been demonstrated to be pathogenic for birds (2, 11, 12). Consequently, differentiating between B. avium and B. hinzii is of importance for veterinary diagnostic laboratories. Several human isolates have also been reported recently, and most were documented to cause disease, including one fatality (6, 7, 8, 14, 27). Therefore, accurate identification of B. hinzii is also of importance for human diagnostic laboratories. A few biochemical tests have been shown to delineate B. avium and B. hinzii, but they do not correctly identify all strains (27) and results of some may vary depending on inoculum size and the procedure used (1, 3, 9). Although these species have been shown to possess unique cellular fatty acid profiles (6, 7, 13, 27), the results are affected by culture conditions (14, 27). Discriminatory methods based on stable genetic elements, rather than variable phenotypic characteristics, are likely to provide a more reliable approach. Both REA analysis, using either HinfI or DdeI (23), and 16S rRNA sequence analysis (14) have been shown to differentiate between B. avium and B. hinzii. Preliminary results, based on a comparison of the patterns obtained from seven B. avium isolates and eight B. hinzii isolates, indicate that PvuII ribotyping may likewise be useful for identification of these species (23). A comparison of ribotyping and REA for typing isolates of B. hinzii is also of interest, given the increasing importance of this organism as an opportunistic human pathogen.
In the present study, 57 B. avium isolates, a B. avium vaccine strain, and 11 B. hinzii isolates were evaluated by PvuII ribotyping to more fully assess the discriminatory capacity of this method for distinguishing these two species. Additionally, 5 isolates classified as B. avium-like, 18 isolates identified as A. faecalis type II prior to the establishment of B. hinzii as a species, and 5 isolates previously identified as atypical B. bronchiseptica were analyzed by REA and ribotyping to ascertain their proper classification. The discriminatory power of REA and ribotyping for intraspecies classification was quantitated to determine the suitability of these methods as epidemiologic tools.
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
The isolates studied are listed in Table 1 according to their original designations. Isolates with the prefix DBL were kindly provided by Linda Schroeder-Tucker, National Veterinary Services Laboratories, Diagnostic Bacteriology Laboratory, Ames, Iowa. B. avium 197N and B. avium-like isolate GOBL110 (provided by Louise Temple, Drew University, Madison, N.J.) were originally acquired by the laboratory of Y. M. Saif, The Ohio State University, Wooster. The B. avium vaccine strain Art-VaxJ (4) (Schering-Plough Animal Health, Union, N.J.) was purchased from a commercial vendor. Other isolates generously provided by colleagues include B. hinzii DMMZ 1277 and DMMZ 1280 (7) (Reinhard Zbinden, Institute for Medical Microbiology, Zurich, Switzerland), B. hinzii 96025473 (8) (Ignacio Gadea, Department of Medical Microbiology, Fundación Jiménez Díaz, Madrid, Spain), B. bronchiseptica 5132 (Tibor Magyar, Veterinary Medical Research Institute, Hungarian Academy of Sciences, Budapest), and B. hinzii TR96-1212 (Louise Temple, Drew University). All remaining isolates were obtained from a collection at the National Animal Disease Center. Bacteria were grown on sheep's blood agar at 37°C for 18 to 36 h.
TABLE 1.
B. avium isolates evaluated in this study
| Species | Strain | Yr of isolationb | Geographic origin | Host | Ribotypea | Profile
|
|
|---|---|---|---|---|---|---|---|
| HinfIa | DdeIa | ||||||
| B. avium | ATCC 35086T | 1977 | Germany | Turkey | BA5 | BA007 | BA002 |
| B. avium | 197N | 1983 | Ohio | Turkey | BA1 | BA002 | BA005 |
| B. avium | 4084 | 1979 or prior | North Carolina | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4085 | 1979 or prior | North Carolina | Turkey | BA2 | BA006 | BA010 |
| B. avium | 4087 | 1979 or prior | North Carolina | Turkey | BA4 | BA002 | BA001 |
| B. avium | 4089 | 1979 or prior | Germany | Turkey | BA5 | BA007 | BA002 |
| B. avium | 4091 | 1979 or prior | Germany | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4092 | 1979 or prior | Germany | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4093 | 1979 or prior | Germany | Turkey | BA6 | BA007 | BA003 |
| B. avium | 4094 | 1979 or prior | Germany | Turkey | BA6 | BA007 | BA003 |
| B. avium | 4095 | 1979 or prior | North Carolina | Turkey | BA1 | BA003 | BA005 |
| B. avium | 4139 | 1979 or prior | Minnesota | Turkey | BA1 | BA001 | BA011 |
| B. avium | 4142 | 1979 or prior | Minnesota | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4143 | 1979 or prior | Iowa | Turkey | BA1 | BA003 | BA005 |
| B. avium | 4148 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA004 |
| B. avium | 4149 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4150 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4151 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4152 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA004 |
| B. avium | 4153 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4154 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4155 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4156 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4157 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4158 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4163 | 1979 or prior | Ohio | Turkey | BA1 | BA002 | BA001 |
| B. avium | 4164 | 1979 or prior | Ohio | Turkey | BA1 | BA002 | BA001 |
| B. avium | 4165 | 1979 or prior | Ohio | Turkey | BA1 | BA002 | BA001 |
| B. avium | 4166 | 1979 or prior | Ohio | Turkey | BA1 | BA002 | BA001 |
| B. avium | 4167 | 1979 or prior | Ohio | Turkey | BA1 | BA002 | BA001 |
| B. avium | 4168 | 1979 or prior | Ohio | Turkey | BA1 | BA002 | BA001 |
| B. avium | 4169 | 1979 or prior | Ohio | Turkey | BA1 | BA001 | BA001 |
| B. avium | 4480 | 1981 or prior | Iowa | Turkey | BA1 | BA008 | BA012 |
| B. avium | 4481 | 1981 or prior | Iowa | Turkey | BA1 | BA002 | BA001 |
| B. avium | 4482 | 1981 or prior | Iowa | Turkey | BA1 | BA001 | BA008 |
| B. avium | 4483 | 1981 or prior | Iowa | Turkey | BA1 | BA001 | BA009 |
| B. avium | 4484 | 1981 or prior | Iowa | Turkey | BA1 | BA002 | BA001 |
| B. avium | 4485 | 1981 or prior | Iowa | Turkey | BA1 | BA002 | BA005 |
| B. avium | 4486 | 1981 or prior | Iowa | Turkey | BA1 | BA003 | BA005 |
| B. avium | 4506 | 1981 or prior | South Africa | Turkey | BA3 | BA004 | BA007 |
| B. avium | 4507 | 1981 or prior | South Africa | Turkey | BA3 | BA001 | BA006 |
| B. avium | 4508 | 1981 or prior | South Africa | Turkey | BA3 | BA004 | BA006 |
| B. avium | T4 | 1998 | New Jersey | Turkey | BA1 | BA011 | NTc |
| B. avium | D4 | 1998 | New Jersey | Duck | BA1 | BA007 | NT |
| B. avium | D10 | 1998 | New Jersey | Duck | BA3 | BA009 | NT |
| B. avium | D23 | 1998 | New Jersey | Duck | BA3 | BA009 | NT |
| B. avium | D24 | 1998 | New Jersey | Duck | BA3 | BA009 | NT |
| B. avium | D25 | 1998 | New Jersey | Duck | BA3 | BA009 | NT |
| B. avium | D26 | 1998 | New Jersey | Duck | BA3 | BA009 | NT |
| B. avium | D27 | 1998 | New Jersey | Duck | BA3 | BA009 | NT |
| B. avium | G24 | 1998 | New Jersey | Goose | BA7 | BA012 | BA014 |
| B. avium | DBL-239 | 1997 | Minnesota | Turkey | BA1 | BA014 | BA004 |
| B. avium | DBL-254-1 | 1997 | Minnesota | Turkey | BA1 | BA010 | BA014 |
| B. avium | DBL-260 | 1997 | Minnesota | Turkey | BA8 | BA010 | BA004 |
| B. avium | DBL-971 | 1997 | Minnesota | Turkey | BA1 | BA010 | BA004 |
| B. avium | DBL-O1067 | 2001 | Iowa | Turkey | BA1 | BA010 | BA011 |
| B. avium | DBL-191 | 1989 | New York | Turkey | BH2 | BH008 | BH002 |
| B. avium | Art-Vax | Prior to 1980 | North Carolina | Turkey | BA1 | BA005 | BA001 |
| B. avium-like | DBL-019 | 2000 | Pennsylvania | Turkey | UKd | UKd | UKe |
| B. avium-like | DBL-245 | 1997 | Minnesota | Turkey | BA1 | BA013 | BA015 |
| B. avium-like | DBL-254-3 | 1997 | Minnesota | Turkey | BA1 | BA004 | BA013 |
| B. avium-like | DBL-243-2 | 1997 | Minnesota | Turkey | BH2 | BH004 | NT |
| B. avium-like | GOBL110 | 1985 or prior | United States | Turkey | BH2 | BH009 | BH001 |
| B. hinzii | L60 (ATCC 51730) | 1994 | Washington | Human | BH1 | BH003 | BH001 |
| B. hinzii | ATCC 51783T | UK | Australia | Chicken | BH1 | BH003 | BH001/PICK> |
| HinfIa | DdeIa | ||||||
| B. hinzii | ATCC 51784 | UK | Belgium | Chicken | BH2 | BH004 | NT |
| B. hinzii | 1277 | 1992 | Switzerland | Human | BH1 | BH006 | BH004 |
| B. hinzii | 1280 | 1993 | Switzerland | Human | BH1 | BH006 | BH004 |
| B. hinzii | 96025473 | 1999 | Spain | Human | BH1 | NDf | ND |
| B. hinzii | 4134 | 1979 or prior | Minnesota | Turkey | BH1 | BH003 | BH001 |
| B. hinzii | 4159 | 1979 or prior | Ohio | Turkey | BH1 | BH003 | BH001 |
| B. hinzii | 4161 | 1979 or prior | Ohio | Turkey | BH1 | BH003 | BH001 |
| B. hinzii | 4509 | 1981 or prior | South Africa | Chicken | BH2 | BH004 | BH001 |
| B. hinzii | TR96-1212 | 1996 | United States | Turkey | BH1 | BH005 | BH001 |
| A. faecalis type II | 4137 | 1979 or prior | Minnesota | Turkey | UKd | UKd | UKd |
| A. faecalis type II | 4138 | 1979 or prior | Minnesota | Turkey | UKd | UKd | UKd |
| A. faecalis type II | 4140 | 1979 or prior | Minnesota | Turkey | BH1 | BH005 | BH003 |
| A. faecalis type II | 4141 | 1979 or prior | Minnesota | Turkey | BH1 | BH005 | BH003 |
| A. faecalis type II | 4147 | 1979 or prior | Ohio | Turkey | BH1 | BH004 | BH001 |
| A. faecalis type II | 4160 | 1979 or prior | Ohio | Turkey | BH1 | BH002 | BH002 |
| A. faecalis type II | 4162 | 1979 or prior | Ohio | Turkey | BH1 | BH002 | BH002 |
| A. faecalis type II | 4445 | 1981 or prior | Minnesota | Turkey | BH1 | BH005 | BH001 |
| A. faecalis type II | 4449 | 1981 or prior | Minnesota | Turkey | BH3 | BH007 | BH001 |
| A. faecalis type II | 4595 | 1982 or prior | Iowa | Turkey | BH1 | BH003 | BH001 |
| A. faecalis type II | 4596 | 1982 or prior | Iowa | Turkey | BB15 | BB010 | ND |
| A. faecalis type II | 4597 | 1982 or prior | Iowa | Turkey | BH2 | BH004 | BH001 |
| A. faecalis type II | 4598 | 1982 or prior | Iowa | Turkey | BH2 | BH004 | BH006 |
| A. faecalis type II | 4599 | 1982 or prior | Iowa | Turkey | BH2 | BH004 | BH005 |
| A. faecalis type II | 4081 | 1979 or prior | North Carolina | Turkey | UKd | UKd | UKe |
| A. faecalis type II | 4086 | 1979 or prior | North Carolina | Turkey | UKd | UKd | UKd |
| A. faecalis type II | 4135 | 1979 or prior | Minnesota | Turkey | BH1 | BH001 | BH001 |
| A. faecalis type II | 4136 | 1979 or prior | Minnesota | Turkey | BH3 | BH007 | BH001 |
| B. bronchiseptica | 4444 | 1981 or prior | Minnesota | Turkey | UKd | UKd | UKe |
| B. bronchiseptica | 4447 | 1981 or prior | Minnesota | Turkey | BH3 | BH004 | BH001 |
| B. bronchiseptica | 4450 | 1981 or prior | Minnesota | Turkey | BH3 | BH007 | BH001 |
| B. bronchiseptica | 4451 | 1981 or prior | Minnesota | Turkey | BH2 | BH004 | BH001 |
| B. bronchiseptica | 5132 | 1990 | Hungary | Rabbit | BH2 | BH004 | BH001 |
B. avium ribotypes and REA patterns have the prefix BA, B. hinzii ribotypes and REA patterns have the prefix BH, and B. bronchiseptica ribotypes and REA patterns have the prefix BB.
Where a year is followed by “or prior,” isolates were received at the National Animal Disease Center during the year indicated; the actual year of isolation is unknown.
NT, not typeable.
Unknown; pattern does not match or cluster within B. avium, B. hinzii, or B. bronchiseptica profiles.
Unknown; pattern does not match, but does cluster within, previously identified B. avium profiles.
ND, not determined.
Ribotyping.
Genomic DNA was isolated using a commercially available kit (Gentra Systems, Minneapolis, Minn.). Ribotyping analysis was based on hybridization of PvuII digestion fragments with a portion of the Escherichia coli rRNA operon rrnB followed by chemiluminescent detection, as described previously (19, 20). The ribotypes of six B. avium isolates included in this study have been previously reported (17).
REA.
DNA isolation and REA were carried out as previously described (23). Briefly, bacterial cells were harvested in 0.85 M NaCl, pelleted by centrifugation and stored at −70°C. DNA was isolated using a commercially available kit (DNAzol; Gibco-BRL, Gaithersburg, Md.) according to recommendations of the manufacturer. Fragments resulting from digestion with HinfI or DdeI were separated by electrophoresis in 0.7% agarose gels using TBE buffer (0.089 M Tris, 0.089 M boric acid, 2 mM EDTA, pH 8.0). Gels were stained with ethidium bromide and photographed. Profiles of some isolates included in this study have been previously determined (23).
Data analysis.
Photographs or chemilumigraphs were scanned for computer analysis using a ScanJet IIcx with DeskScan software (Hewlett-Packard, Boise, Idaho). GelCompar software (Applied Maths, Kortrijk, Belgium) was used for comparison of fingerprint profiles. Ribotypes are designated by Arabic numerals, with B. avium types given the prefix BA, B. hinzii ribotypes given the prefix BH, and B. bronchiseptica types given the prefix BB. REA profiles are designated using the previously proposed DIE code (23), based on three-place Arabic numerals preceded by BA, BH, or BB, as appropriate. Strains that exhibited single-band differences were assigned to different ribotypes or REA profiles. Similarity between all possible pairs of ribotypes or REA profiles using the coefficient of Dice (26) was calculated by the cluster analysis module of the software. Dendrograms were derived from a matrix of similarity values by the unweighted pair group method using arithmetic averages, based on a tolerance of 0.5% for ribotyping and 1.0% for REA. The discriminatory power of ribotyping and/or REA analysis was defined by calculating discrimination indices as described (10).
RESULTS
Ribotyping.
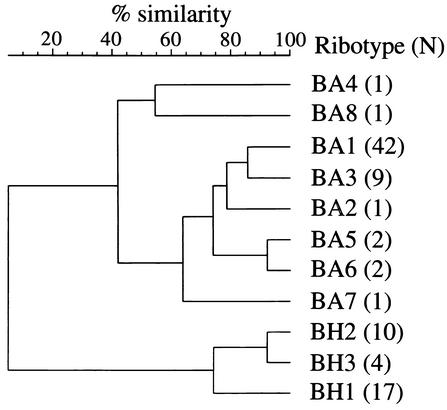
The ribotype of individual isolates is indicated in Table 1. A total of nine unique patterns was observed among the B. avium isolates examined based on combinations of 15 fragments ranging in size from approximately 2 to 16 kb. Four patterns were identical to previously reported B. avium ribotypes (17, 23) (Fig. 1A, lanes 1, 2, 3, and 7), while four others constitute novel types (Fig. 1A, lanes 4, 5, 6, and 8). The ninth pattern, associated only with isolate DBL-191, does not display the cluster of bands greater than 11 kb characteristic of B. avium isolates and shares only 5.9% similarity with the remaining eight B. avium profiles. This pattern was later found to be identical to one associated with some isolates of B. hinzii (Fig. 1B, lane 2). Accordingly, isolate DBL-191 was reclassified as B. hinzii. Similarity among the 8 true B. avium profiles, designated BA1 to BA8, ranges from 41.9 to 92.3% (Fig. 2).
FIG. 1.
Ribotype patterns observed following digestion of B. avium (A) or B. hinzii (B) genomic DNA with PvuII. Lane numbers correspond to the assigned ribotype profile.
FIG. 2.
Dendrogram and cluster analysis of the B. avium and B. hinzii PvuII ribotype patterns identified from isolates in Table 1 (tolerance, 0.5%). B. avium patterns are assigned the prefix BA; B. hinzii patterns are assigned the prefix BH. The number of isolates identified for each ribotype is indicated in parentheses to its right.
Two ribotype patterns, designated BH1 and BH2, were identified from the 11 B. hinzii isolates analyzed based on eight fragments of roughly 1.8 to 6 kb (Table 1; Fig. 1B, lanes 1 and 2). These profiles share only 5% similarity with B. avium profiles and are immediately recognizable as unique by visual comparison (Fig. 1 and 2). BH1 and BH2 profiles have a calculated similarity of 74.2% (Fig. 2).
Since ribotyping readily distinguishes between B. avium and B. hinzii, this technique was used to evaluate 5 isolates characterized as B. avium-like and 18 previously identified as A. faecalis type II (22). Recent research suggests that isolates in these categories are more properly classified as B. hinzii (27). Thirteen of the 23 isolates displayed either BH1 or BH2 profiles. Two isolates have a profile with 92.3% similarity to BH2, which was designated BH3 (Fig. 1B, lane 3; Fig. 2). Of the remaining eight isolates examined, two possess BA1 ribotypes, one has a B. bronchiseptica ribotype 15 profile (19, 20), and five displayed patterns that bear no resemblance to previously defined B. avium, B. hinzii, or B. bronchiseptica patterns. However, the profile of one (strain 4081) is nearly identical to the pattern of fragments observed for A. faecalis strain 4615, obtained from the Centers for Disease Control and Prevention (K. B. Register, unpublished data).
These results, combined with previous analysis of over 250 B. bronchiseptica isolates (19, 20, 21), demonstrate that ribotype profiles of B. avium, B. hinzii, and B. bronchiseptica are readily distinguishable from one another. Therefore, this technique was used to evaluate a group of five isolates originally identified by other laboratories as B. bronchiseptica but which were found in our laboratory to have one or more atypical phenotypic characteristics (Table 1). These isolates were acquired prior to the description of B. hinzii as a species. Four isolates displayed B. hinzii ribotype profiles. The profile of isolate 4444 did not resemble those of either B. avium, B. hinzii, or B. bronchiseptica and was also unique compared to the A. faecalis profile observed above.
Distribution of ribotypes.
B. avium ribotypes BA1 and BA3 are the most prevalent among the isolates examined here (71 and 15%, respectively) and are also among the most closely related, with 85.7% similarity (Fig. 2). No geographic or chronologic association was evident for these isolates. They were acquired from several different regions of the United States or from Germany during a period of at least 22 years, extending through 2001. The remaining B. avium ribotypes are represented by only one or two. The single goose isolate evaluated is the only type BA7 identified and shares the lowest degree of relatedness to other ribotypes in its cluster. Types BA4 and BA8 comprise a separate cluster but are not obviously related to one another based on geographic origin or date of isolation.
Approximately 54.8% of all B. hinzii isolates displayed a BH1 profile, while 32.2% were identified as BH2 and 12.9% were identified as BH3. Animal hosts from which B. hinzii isolates were identified include turkey, chicken, human and rabbit. The distribution of ribotype profiles among host species is shown in Table 2. The two major B. hinzii clusters include isolates from different geographic locations representing four continents. Although most isolates were obtained at least 20 years ago, more recent isolates are represented in both clusters. The BH3 profile was found only in isolates acquired in 1981 or earlier. However, this observation may simply reflect the limited number of more recent isolates available for analysis and the lower frequency with which BH3 strains appear to occur. All mammalian isolates have a BH1 profile, but this profile is also frequently found in avian isolates.
TABLE 2.
B. hinzii ribotype frequency and distribution within host species
| Ribotype | No. of strains (%) from:
|
|||
|---|---|---|---|---|
| Turkey | Chicken | Human | Rabbit | |
| BH1 | 12 (52.2) | 1 (33.3) | 4 (100) | 1 (100) |
| BH2 | 7 (30.4) | 2 (66.7) | ||
| BH3 | 4 (17.4) | |||
| Total | 23 | 3 | 4 | 1 |
REA.
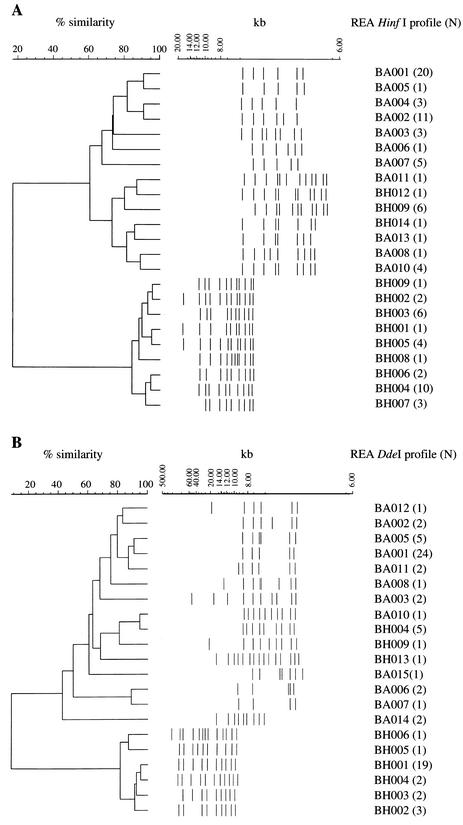
HinfI and DdeI REA was carried out on all isolates evaluated by ribotyping. Profiles are indicated in Table 1. As previously reported using a smaller group of isolates, REA with either HinfI or DdeI clearly discriminated between B. avium and B. hinzii (Fig. 3). Depending on the enzyme used, REA fingerprints share 8.5 to 17.1% interspecies similarity. Eight B. avium isolates and two B. hinzii isolates could not be typed using DdeI, since the fragments generated were too large to be resolved under the conditions used.
FIG. 3.
Dendrogram and cluster analysis of the B. avium and B. hinzii HinfI (A) or DdeI (B) REA patterns identified from isolates in Table 1 (tolerance, 1%). Individual REA patterns are represented graphically to the right of each dendrogram. B. avium patterns are assigned the prefix BA; B. hinzii patterns are assigned the prefix BH. The number of isolates identified for each REA pattern is indicated in parentheses to its right.
All isolates which displayed B. avium ribotypes, including those not originally identified as B. avium using biochemical methods, displayed B. avium-specific REA patterns. Among these isolates, 14 unique HinfI profiles (Fig. 3A) and 15 unique DdeI profiles (Fig. 3B) could be distinguished. HinfI profiles BA009 to BA014 and DdeI profiles BA013 to BA015 have not been previously reported and are associated only with isolates acquired since 1997. Depending on the enzyme used, B. avium profiles share a minimum of 60.5 to 42.6% similarity. The majority of isolates (52.5%) was represented either by HinfI profile BA001 or BA002. Other HinfI profiles each comprised 1.7 to 10.2% of the remaining isolates. The most common B. avium DdeI profile was BA001 (47.0%), with the remaining DdeI profiles representing 2.0 to 9.8% of isolates. Twenty-three of the 31 isolates in the two most common HinfI groups also displayed the most common DdeI pattern. Identification of less prevalent DdeI profiles from the remaining eight isolates demonstrates that additional discriminatory benefit is achieved when analysis is carried out with both enzymes. DdeI REA profiles also further distinguished B. avium isolates with HinfI BA004, BA007, and BA010 profiles (Table 1).
B. hinzii-specific REA patterns were observed for all isolates previously shown to possess B. hinzii ribotypes. Nine HinfI and six DdeI profiles were identified from these isolates (Fig. 3). HinfI profiles BH008 and BH009 and DdeI profiles BH005 and BH006 have not been previously reported. B. hinzii REA patterns share a similarity of at least 81.5% (for DdeI) to 84.2% (for HinfI). The most common B. hinzii HinfI profiles are BH004 and BH003, comprising 33.3 and 20.0% of all isolates, respectively. The remaining seven profiles each account for 3.3 to 13.3% of isolates. The DdeI pattern associated with the majority of B. hinzii isolates is BH001 (67.9%). Other DdeI patterns account for 3.6 to 10.7% of isolates. As noted for B. avium, nearly all B. hinzii isolates in the most common HinfI groups were also members of the most common DdeI group. However, a few isolates could be further distinguished when results with both enzymes were considered.
The six isolates originally identified as B. avium-like, A. faecalis type II, or B. bronchiseptica but found not to possess bordetella-specific ribotypes were also evaluated by REA. HinfI and DdeI patterns of isolates 4137, 4138, and 4086 do not match or cluster within the group of patterns associated with either B. avium or B. hinzii (Register, unpublished data). Patterns of isolates DBL-019, 4081, and 4444, all unique from one another, fall within the B. avium cluster when results with DdeI are considered (Register, unpublished data). However, their HinfI profiles do not match or cluster within either B. avium or B. hinzii patterns, nor do they cluster with the 48 HinfI patterns previously described for B. bronchiseptica (24). Thus, the sum of evidence indicates none of these six isolates are B. avium, B. hinzii, or B. bronchiseptica, and their identities remain unknown.
Discriminatory power of ribotyping and REA.
Discrimination indices were calculated for PvuII ribotyping, REA using HinfI, and REA using DdeI, both separately and in combination, for B. avium isolates and B. hinzii isolates (Table 3). A method is generally considered sufficiently discriminatory for typing purposes if an index of 0.900 or greater is obtained (10). No single analysis evaluated here appears sufficiently discriminatory for routine use as a typing tool. However, combined application of HinfI and DdeI REA does yield a level of confidence suitable for routine use in epidemiologic investigations. The discriminatory power is slightly increased when ribotyping results are also considered. Two B. avium REA types, HinfI BA002-DdeIBA001 and HinfI BA010-DdeIBA004, could be further distinguished with ribotyping. Isolates with the B. hinzii fingerprint profile HinfI BH004-DdeIBH001were comprised of three different ribotypes.
TABLE 3.
Discrimination indices associated with single and combined typing methods
| Species | Discrimination index obtained by indicated method(s)
|
||||
|---|---|---|---|---|---|
| RTa | REA
|
RT + REA HinfI/DdeI | |||
| HinfI | DdeI | HinfI/DdeI | |||
| B. avium | 0.474 | 0.835 | 0.764 | 0.894 | 0.901 |
| B. hinzii | 0.598 | 0.837 | 0.534 | 0.902 | 0.926 |
RT, ribotyping.
DISCUSSION
The results reported here establish the validity of combined HinfI/DdeI REA as an epidemiologic tool for both B. avium and B. hinzii, as proposed in an earlier preliminary study (23). Although the discrimination index associated with HinfI REA alone falls just short of the recommended limit, it may be advantageous to adopt this method as a preliminary tool in outbreak investigations since some isolates cannot be typed by DdeI REA and the additional discriminatory power provided is relatively low. While the highest level of discrimination is achieved using a combination of ribotyping and REA, in most cases the modest increase over REA alone is unlikely to justify the added time and expense.
In some instances, isolates that share a high degree of similarity by ribotyping also cluster tightly based on REA. However, in many cases there is no concurrence between ribotype and REA clustering patterns of individual strains. This observation is not surprising, considering that REA is a whole-genome method while ribotyping specifically targets rRNA genes. The superior discriminatory power of REA justifies its use in monitoring the spread of strains during outbreaks. Nonetheless, the phenotypic implications of base pair changes resulting in distinct patterns are not clear and may be relatively minor. Ribotyping may be a more accurate indicator of meaningful phylogenetic relationships, given the high degree of conservation found among rRNA genes. Overall, both REA and ribotyping indicate greater genetic diversity among B. avium isolates than among B. hinzii isolates, suggesting B. hinzii may have evolved more recently than B. avium. Although B. hinzii has only been formally recognized as a unique species since 1995, our data confirm its existence since at least the late 1970s. It is currently unclear whether B. hinzii truly has a more recent origin than B. avium or whether difficulties related to accurate identification precluded awareness of its existence.
REA results for B. avium suggest new variants may have recently emerged. HinfI profiles BA009 to BA014, which cluster together in one of the two major B. avium HinfI groups, are associated exclusively with isolates acquired since 1997. None of the 43 isolates obtained in 1983 or earlier fall within this cluster. The only additional strain in this group was isolated during the interim, in 1989. The majority of the most recent isolates are either nontypeable using DdeI, which was not the case for any pre-1997 isolate, or have profiles BA013 to BA015, also not found in older isolates. The DdeI dendrogram shows profiles BA013 to BA015 to be the least similar to other profiles in their respective clusters. Analysis of a larger group of recent isolates is needed to better assess the degree of genetic divergence actually occurring. There is strong evidence suggestive of vaccine-driven evolution in the human pathogen Bordetella pertussis (15, 16, 28). It has been proposed that clonal expansion of strains began occurring within 10 to 20 years after the initiation of mass vaccination (28). Since widespread vaccination against B. avium in poultry has only been implemented in the previous 15 to 20 years, it is tempting to speculate about its potential contribution to the emergence of new variants. Interestingly, REA results indicate the vaccine strain included in this study is among those isolates sharing the least similarity with those obtained recently.
B. hinzii colonizes the respiratory tract of poultry but is generally believed to be nonpathogenic. Thus, it is of importance for veterinary diagnostic laboratories to distinguish this bacterium from B. avium, which causes significant losses in the poultry industry. Previous reports have indicated that most B. avium isolates agglutinate guinea pig erythrocytes, while A. faecalis type II and B. avium-like isolates, most of which now appear more properly classified as B. hinzii, are hemagglutination negative (12, 25). However, some strains display weak reactions, and the results are not always consistent. We have likewise observed inconsistent results for hemagglutination of sheep and turkey erythrocytes by B. avium and B. hinzii and found that the outcome is affected by the method used (Register, unpublished data). Most B. hinzii isolates are weakly positive when a 96-well plate procedure is employed (18) but negative when tested by a slide agglutination method (22). Currently, there is no readily available technique for reliably distinguishing between B. avium and B. hinzii. The results reported here demonstrate that both ribotyping and REA are equally useful for this purpose. Ribotyping may be preferable since a quick visual examination of the resulting patterns is the only postassay analysis needed. In contrast, the complexity of REA patterns requires computer-assisted analysis for interpretation. 16S rRNA gene sequencing has recently been proposed as a method for definitive identification of B. hinzii (14). Only a single B. avium isolate was included in that study, but the numerous base pair differences found between B. avium and B. hinzii suggest this technique would also detect other B. avium isolates, although this has yet to be demonstrated. While 16S rRNA sequencing is a high-resolution technique, it requires amplification of genomic DNA and takes considerably longer to complete than ribotyping. No PvuII sites are present in the B. avium and B. hinzii 16S rRNA sequences reported, indicating that the sequence variability responsible for distinguishing these species by ribotyping must be located in other regions of the rrnB operon included in the probe.
Since the establishment of B. hinzii as a separate taxon, relatively few isolates have been reported, and all were derived either from humans or poultry (5-8, 14, 23, 27). Additional, but unrecognized, isolations are likely to have been made by some laboratories, as we discovered numerous additional isolates in our own collection following ribotyping and REA. These were previously identified as B. avium-like or misidentified as B. avium, B. bronchiseptica, or A. faecalis type II. Included among these is a rabbit isolate representing the first B. hinzii strain found in a nonhuman mammal. The difficulty of distinguishing B. hinzii from closely related bacteria may have prevented the discovery of other rabbit isolates, as well as isolates from additional hosts, in the past. Whether B. hinzii is pathogenic in rabbits is unclear, since no further information is available regarding the host animal.
The findings presented here suggest that the pathogenicity of B. hinzii for poultry should be more rigorously examined. Studies from other laboratories, using isolates described at the time as B. avium-like, failed to show virulence in poults (2, 12). More recent attempts to induce disease in 1-day-old chickens and turkey poults with B. hinzii have also failed (27). However, at least three isolates (4147, 4159, and 4161), formerly classified as A. faecalis type II based on biochemical testing, have been previously reported to cause 100% morbidity in poults with a severity indistinguishable from that of B. avium (22). Furthermore, the B. hinzii isolate DBL-191, originally misidentified as B. avium, is believed to have been the cause of respiratory disease in the flock from which it was obtained. Nonetheless, it is apparent that several other isolates identified in this report as B. hinzii are incapable of causing disease in poultry (22). Potential virulence factors of this bacterium have not yet been described. Additional studies may reveal particular bacterial products, produced by only a subset of isolates, that are required for virulence in poultry and/or other hosts.
Acknowledgments
The technical skills of Pamala Beery are greatly appreciated.
REFERENCES
- 1.Berkhoff, H. A., and G. D. Riddle. 1984. Differentiation of Alcaligenes-like bacteria of avian origin and comparison with Alcaligenes spp. reference strains. J. Clin. Microbiol. 19:477-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackall, P. J., and C. M. Doheny. 1987. Isolation and characterisation of Bordetella avium and related species and an evaluation of their role in respiratory disease in poultry. Aust. Vet. J. 64:235-239. [DOI] [PubMed] [Google Scholar]
- 3.Blackall, P. J., and J. G. Farrah. 1986. An evaluation of two methods of substrate alkalinization for the identification of Bordetella avium and other similar organisms. Vet. Microbiol. 11:301-306. [DOI] [PubMed] [Google Scholar]
- 4.Burke, D. S., and M. M. Jensen. 1980. Immunization against turkey coryza by colonization with mutants of Alcaligenes faecalis. Avian Dis. 24:726-733. [PubMed] [Google Scholar]
- 5.Coenye, T., J. Goris, T. Spilker, P. Vandamme, and J. J. LiPuma. 2002. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J. Clin. Microbiol. 40:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cookson, B. T., P. Vandamme, L. C. Carlson, A. M. Larson, J. V. Sheffield, K. Kersters, and D. H. Spach. 1994. Bacteremia caused by a novel Bordetella species, “B. hinzii.” J. Clin. Microbiol. 32:2569-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funke, G., T. Hess, A. von Graevenitz, and P. Vandamme. 1996. Characteristics of Bordetella hinzii strains isolated from a cystic fibrosis patient over a 3-year period. J. Clin. Microbiol. 34:966-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadea, I., M. Cuenca-Estrella, N. Benito, A. Blanco, M. L. Fernandez-Guerrero, P. L. Valero-Guillen, and F. Soriano. 2000. Bordetella hinzii, a “new” opportunistic pathogen to think about. J. Infect. 40:298-299. [DOI] [PubMed] [Google Scholar]
- 9.Hinz, K.-H. Glünder, G. 1986. Identification of Bordetella avium sp. nov. by the API 20NE system. Avian Pathol. 15:611-614. [DOI] [PubMed] [Google Scholar]
- 10.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackwood, M. W., S. M. McCarter, and T. P. Brown. 1995. Bordetella avium: an opportunistic pathogen in Leghorn chickens. Avian Dis. 39:360-367. [PubMed] [Google Scholar]
- 12.Jackwood, M. W., Y. M. Saif, P. D. Moorhead, and R. N. Dearth. 1985. Further characterization of the agent causing coryza in turkeys. Avian Dis. 29:690-705. [PubMed] [Google Scholar]
- 13.Jackwood, M. W., M. Sasser, and Y. M. Saif. 1986. Contribution to the taxonomy of the turkey coryza agent: cellular fatty acid analysis of the bacterium. Avian Dis. 30:172-178. [PubMed] [Google Scholar]
- 14.Kattar, M. M., J. F. Chavez, A. P. Limaye, S. L. Rassoulian-Barrett, S. L. Yarfitz, L. C. Carlson, Y. Houze, S. Swanzy, B. L. Wood, and B. T. Cookson. 2000. Application of 16S rRNA gene sequencing to identify Bordetella hinzii as the causative agent of fatal septicemia. J. Clin. Microbiol. 38:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooi, F. R., Q. He, H. van Oirschot, and J. Mertsola. 1999. Variation in the Bordetella pertussis virulence factors pertussis toxin and pertactin in vaccine strains and clinical isolates in Finland. Infect. Immun. 67:3133-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mooi, F. R., H. van Oirschot, K. Heuvelman, H. G. van der Heide, W. Gaastra, and R. J. Willems. 1998. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immun. 66:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raffel, T. R., K. B. Register, S. A. Marks, and L. Temple. 2002. Prevalence of Bordetella avium infection in selected wild and domesticated birds in the Eastern USA. J. Wildl. Dis. 38:40-46. [DOI] [PubMed] [Google Scholar]
- 18.Register, K. B. 2001. Novel genetic and phenotypic heterogeneity in Bordetella bronchiseptica pertactin. Infect. Immun. 69:1917-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Register, K. B., A. Boisvert, and M. R. Ackermann. 1997. Use of ribotyping to distinguish Bordetella bronchiseptica isolates. Int. J. Syst. Bacteriol. 47:678-683. [DOI] [PubMed] [Google Scholar]
- 20.Register, K. B., and T. Magyar. 1999. Optimized ribotyping protocol applied to Hungarian Bordetella bronchiseptica isolates: identification of two novel ribotypes. Vet. Microbiol. 69:277-285. [DOI] [PubMed] [Google Scholar]
- 21.Register, K. B., R. E. Sacco, and G. Foster. 2000. Ribotyping and restriction endonuclease analysis reveal a novel clone of Bordetella bronchiseptica in seals. J. Vet. Diagn. Investig. 12:535-540. [DOI] [PubMed] [Google Scholar]
- 22.Rimler, R. B., and D. G. Simmons. 1983. Differentiation among bacteria isolated from turkeys with coryza (rhinotracheitis). Avian Dis. 27:491-500. [PubMed] [Google Scholar]
- 23.Sacco, R. E., K. B. Register, and G. E. Nordholm. 2000. Restriction enzyme analysis and ribotyping distinguish Bordetella avium and Bordetella hinzii isolates. Epidemiol. Infect. 124:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacco, R. E., K. B. Register, and G. E. Nordholm. 2000. Restriction endonuclease analysis discriminates Bordetella bronchiseptica isolates. J. Clin. Microbiol. 38:4387-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skeeles, J. K., and L. H. Arp. 1997. Bordetellosis (turkey coryza), p. 275-287, In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougald, and Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames.
- 26.Sneath, P. H. A., and R. R. Sokal. 1973. The principle and practice of numerical classification. W. H. Freeman, San Francisco, Calif.
- 27.Vandamme, P., J. Hommez, M. Vancanneyt, M. Monsieurs, B. Hoste, B. Cookson, C. H. Wirsing von Konig, K. Kersters, and P. J. Blackall. 1995. Bordetella hinzii sp. nov., isolated from poultry and humans. Int. J. Syst. Bacteriol. 45:37-45. [DOI] [PubMed] [Google Scholar]
- 28.Van Loo, I. H., and F. R. Mooi. 2002. Changes in the Dutch Bordetella pertussis population in the first 20 years after the introduction of whole-cell vaccines. Microbiology 148:2011-2018. [DOI] [PubMed] [Google Scholar]