Abstract
A multilocus sequence typing (MLST) scheme has been developed for the unambiguous characterization of encapsulated and noncapsulated Haemophilus influenzae isolates. The sequences of internal fragments of seven housekeeping genes were determined for 131 isolates, comprising a diverse set of 104 serotype a, b, c, d, e, and f isolates and 27 noncapsulated isolates. Many of the encapsulated isolates had previously been characterized by multilocus enzyme electrophoresis (MLEE), and the validity of the MLST scheme was established by the very similar clustering of isolates obtained by these methods. Isolates of serotypes c, d, e, and f formed monophyletic groups on a dendrogram constructed from the differences in the allelic profiles of the isolates, whereas there were highly divergent lineages of both serotype a and b isolates. Noncapsulated isolates were distinct from encapsulated isolates and, with one exception, were within two highly divergent clusters. The relationships between the major lineages of encapsulated H. influenzae inferred from MLEE data could not be discerned on a dendrogram constructed from differences in the allelic profiles, but were apparent on a tree reconstructed from the concatenated nucleotide sequences. Recombination has not therefore completely eliminated phylogenetic signal, and in support of this, for encapsulated isolates, there was significant congruence between many of the trees reconstructed from the sequences of the seven individual loci. Congruence was less apparent for noncapsulated isolates, suggesting that the impact of recombination is greater among noncapsulated than encapsulated isolates. The H. influenzae MLST scheme is available at www.mlst.net, it allows any isolate to be compared with those in the MLST database, and (for encapsulated isolates) it assigns isolates to their phylogenetic lineage, via the Internet.
In many parts of the world encapsulated (typeable) strains of Haemophilus influenzae are an important cause of serious childhood invasive diseases, including meningitis and septicemia (31). The great majority of invasive disease is caused by encapsulated isolates of H. influenzae serotype b (Hib), and in those countries where Hib conjugate vaccines have been introduced, there have been very dramatic reductions in Hib disease and reduced carriage of Hib, among both vaccinated and nonvaccinated children (31). In the United States, Hib disease has been reduced by >95% (2, 42) and now only occurs in a small number of vaccinated children who do not produce a protective immune response (3, 16) or as a consequence of ongoing Hib transmission in communities where vaccine uptake has been low (15, 20). In the United Kingdom and other countries where there are high levels of vaccine uptake, there have been similarly dramatic reductions in Hib disease (31, 37). Although Hib conjugate vaccines have been introduced into the childhood vaccination programs of many developed countries, the effect on global levels of Hib disease is relatively modest, since vaccination has not been introduced into many of those countries that have the highest burden of Hib disease (31).
Invasive H. influenzae disease in some countries using the conjugate vaccines is now more commonly caused by non-Hib isolates than Hib isolates (17). Prior to the introduction of the Hib vaccines, isolates of serotype a, c, d, e, and f were far less commonly recovered from patients with invasive disease than Hib isolates. Of these non-Hib serotypes, serotype f has most commonly been associated with invasive disease, and in some vaccinated populations it has been reported that the incidence of serotype f disease may have increased in recent years (40). Noncapsulated (nontypeable) H. influenzae is commonly carried in the pharynx, is one of the major causes of acute otitis media, and can cause diseases of the upper and lower respiratory tract, including sinusitis and pneumonia. These isolates may also cause invasive disease, and serious non-Hib disease in children is more commonly due to noncapsulated isolates than to encapsulated isolates of the non-Hib serotypes (1, 2, 10, 17). In some countries where the Hib conjugate vaccines are used (e.g., the United Kingdom) there has been an increase in adult invasive disease due to non-Hib isolates, most of which is due to noncapsulated isolates (34).
Precise methods for characterizing isolates of bacterial pathogens are required to understand disease transmission, to track the spread of virulent or antibiotic-resistant strains, and to monitor the impact of vaccines on bacterial populations and can be used to probe the nature of virulence and the population and evolutionary biology of bacterial species. Several methods have been used to characterize strains of H. influenzae, but by far the greatest insights have been obtained using multilocus enzyme electrophoresis (MLEE) (24-28). These studies have shown that encapsulated H. influenzae populations are highly clonal, with limited evidence of any major impact of recombination within housekeeping loci, and that isolates of a single serotype typically have restricted genetic diversity (28). Thus, isolates of serotypes c, d, e, and f form monophyletic clusters and isolates of both serotypes a and b fall into only two highly divergent phylogenetic groups (group I and II isolates). Noncapsulated isolates are more diverse than encapsulated isolates, and their population structure may be more influenced by recombination (25, 32).
Although MLEE is a powerful and valid technique, it is not ideal, since comparisons of the results obtained in different laboratories are problematic, and the relationship between nucleotide sequence variation and isoenzyme variation typically is unknown. Multilocus sequence typing (MLST) provides an unambiguous and precise method for characterizing isolates of bacterial pathogens by sequencing internal fragments of seven housekeeping genes (7, 23). The different sequences at a locus are assigned different allele numbers, and the allele numbers at each of the seven loci define the allelic profile, which precisely and accurately characterizes each isolate. MLST schemes have been described for a number of important bacterial pathogens, including Neisseria meningitidis (23), Streptococcus pneumoniae (6), Staphylococcus aureus (8), Campylobacter jejuni (5), Streptococcus pyogenes (9), Escherichia coli (33), and Enterococcus faecium (19). MLST has a great advantage over other typing methods; isolates characterized in different laboratories can be readily compared, and the allelic profiles of isolates and associated epidemiological information can be held in a single database that can be investigated over the Internet (7, 23). We describe an MLST scheme for the precise characterization of encapsulated and noncapsulated isolates of H. influenzae that should be of value for the continuing surveillance of disease caused by this important pathogen. We also show that recombination has not completely eliminated deep phylogenetic signal in this species and demonstrate the ability of the MLST data to reconstruct some of the phylogenetic relationships between H. influenzae lineages.
MATERIALS AND METHODS
Bacterial isolates.
A total of 131 H. influenzae isolates were included in this study, collected from patients and carriers from 13 countries worldwide in the period from 1947 to 2000. Of those, 104 were encapsulated and 27 were noncapsulated. Sixty-eight isolates of serotypes a, b, c, d, e, and f were from the collections of Richard Moxon (University of Oxford, Oxford, United Kingdom) and J. S. Kroll and were included in this study as they had been used in the MLEE studies of Musser et al. (28). They included representatives that spanned the genetic diversity within each serotype and examples of each of the major lineages of these serotypes that previously were assigned by MLEE (28) and were used to relate the clusters of isolates obtained with MLST and MLEE. The designations for these isolates begin with the prefix “RM.” To investigate the utility of the MLST scheme in outbreak investigations, a further 26 isolates were recovered between December 1999 and February 2000 during a study of ongoing Hib transmission among rural communities in Pennsylvania where vaccine uptake has been low (15). Eight of the latter isolates were from patients with cases of invasive disease; the others were from asymptomatic carriers in two of the communities in which there were cases of Hib disease. One of the eight isolates from patients with disease was subsequently found to be noncapsulated. A group of 11 Hib biotype IV isolates were collected in 1990 from the Baltimore, Md., area and were from the blood or cerebrospinal fluid (CSF) of patients with meningitis. Three pairs of these isolates were from blood and CSF samples taken from the same patient during a single disease episode. An isolate of the H. influenzae biogroup aegyptius clone (29) from a patient with Brazilian purpuric fever in Brazil during 1986 was also characterized. The biotype IV isolates and the isolate of the latter clone were from the collection of J. S. Kroll.
A group of 25 noncapsulated isolates were from a large collection recovered in 1994 and 1995 from the middle ear fluid of Finnish children with acute otitis media during vaccine-related studies undertaken by the Finnish Otitis Media Group and have been characterized by ribotyping in the laboratory of R. Goldstein. The 25 noncapsulated isolates were selected from across a dendrogram constructed from the ribotyping data obtained for these Finnish isolates and other noncapsulated and encapsulated isolates and were chosen for study by MLST as a diverse sample of the noncapsulated H. influenzae population. Four pairs of isolates, each from the left and right ear of the same child, were included (isolates nt-1158 and nt-1159, isolates nt-1207 and nt-1209, isolates nt-1231 and nt-1232, and isolates nt-1180 and nt-1181); a further isolate from middle ear fluid of the last of these children was obtained one month later (nt-1292).
The serotypes of isolates were provided by the originating laboratory but were redetermined for those isolates that clustered anomalously, as previously described (11).
Bacterial growth and preparation of chromosomal DNA.
Isolates were grown on tryptic soy chocolate agar at 37°C in an atmosphere of 95% air-5% CO2. A single colony was selected and spread across a second plate of the same agar, and after incubation overnight, the bacterial growth from one quarter of the plate was collected with a sterile cotton wool swab and resuspended in a 1.5-ml microcentrifuge tube containing 1 ml of tryptic soy broth. After centrifugation at 5,000 × g for 10 min the supernatant was removed, and chromosomal DNA was prepared from the bacterial pellet using a Qiagen DNeasy Tissue Kit (Qiagen Inc., Valencia, Calif.).
MLST.
The following primers were used for the amplification of the housekeeping gene fragments, based on the genome sequence of H. influenzae Rd (14): adk-up (5′-GGTGCACCGGGTGCAGGTAA-3′) and adk-dn (5′-CCTAAGATTTTATCTAACTC-3′);atpG-up (5′-ATGGCAGGTGCAAAAGAGAT-3′) and atpG-dn (5′-TTGTACAACAGGCTTTTGCG-3′);frdB-up (5′-CTTATCGTTGGTCTTGCCGT-3′) and frdB-dn, 5′-TTGGCACTTTCCACTTTTCC-3′);fucK-up (5′-ACCACTTTCGGCGTGGATGG-3′) and fucK-dn (5′-AAGATTTCCCAGGTGCCAGA-3′);mdh-up (5′-TCATTGTATGATATTGCCCC-3′) and mdh-dn (5′-ACTTCTGTACCTGCATTTTG-3′);pgi-up (5′-GGTGAAAAAATCAATCGTAC-3′) and pgi-dn (5′-ATTGAAAGACCAATAGCTGA-3′); and recA-up (5′-ATGGCAACTCAAGAAGAAAA-3′) and recA-dn (5′-TTACCAAACATCACGCCTAT-3′.
PCRs (50-μl reaction volumes) were carried out in a 96-well microtiter plate format using a PTC-200 DNA engine (MJ Research Inc., Waltham, Mass.) with initial denaturation at 95°C for 4 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s. The samples were then maintained at 72°C for a further 10 min, cooled to 4°C, and stored at −20°C. The amplified DNA fragments were precipitated using 20% polyethylene glycol 8000-2.5 M NaCl, washed twice in 70% ethanol, dried, and resuspended in sterile water. The DNA fragments were sequenced on each strand with the primers used in the initial PCR amplification, ABI PRISM BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.), and an ABI3700 DNA sequencer. The forward and reverse sequences were trimmed to the correct length and edited.
Data analysis.
For each of the seven loci, every different sequence obtained from the H. influenzae isolates was assigned as a distinct allele using the Macintosh program, Sequence Output (available from www.mlst.net). Each isolate is defined by a string of seven integers (the allelic profile), which corresponds to the allele numbers at the seven loci, in the order adk, atpG, frdB, fucK, mdh, pgi, recA. Each unique allelic profile is considered to be a clone and is assigned as a sequence type (ST), which also provides a convenient descriptor for the clone. Clusters of related STs that appear to be descended from a common ancestor are defined as clonal complexes or lineages. The MLST database containing the allelic profiles and information about the H. influenzae strains, together with interrogation and analysis software, can be found on the H. influenzae pages of the MLST website (http://haemophilus.mlst.net/). The relatedness of isolates was displayed by cluster analysis, using the matrix of pairwise differences in the allelic profiles, and the unweighted pair-group method with arithmetic averages (UPGMA) (Statistica; StatSoft Inc., Tulsa, Okla.).
For each of the 68 identified STs, the sequences at the seven loci were concatenated using the facility available at the H. influenzae MLST web pages (address given above). The fragments of the housekeeping genes used in the H. influenzae MLST scheme all start at the first nucleotide of a codon and end at the third position; the +1 reading frame was therefore maintained throughout the entire concatenated sequence. A minimum evolution tree was constructed from the concatenated sequences (3,057 bp), using all nucleotide sites, and the Kimura two-parameter method for estimating pairwise genetic distances. An initial tree was obtained using the neighbor-joining method, and the minimum evolution method was used to search for the tree which minimizes the sums of the branch length estimates by branch-swapping using closest-neighbor interchange (39). The level of statistical support for the nodes on the tree was evaluated by examining their percentage of recovery in 1,000 resampled trees using the bootstrap test (36). The minimum evolution tree, the sequence diversity at each locus, and the ratios of synonymous and nonsynonymous sites (dS/dN) were obtained using MEGA, version 2.1 (22).
Sawyer's test (35) was used to evaluate whether the distribution of synonymous polymorphisms among the sequences were randomly distributed, thus providing an indication of the extent of recombination, and was implemented using a Macintosh program available from www.mlst.net.
Congruence between loci.
To obtain a set of distantly related isolates, a UPGMA dendrogram constructed using the differences in the allelic profiles of all H. influenzae isolates was truncated at a genetic distance of 0.5, and one ST descending from each of the resulting 24 lineages was selected at random. The allelic profile of each selected ST differed from those of all others at a genetic distance of at least 0.55 (corresponding to different alleles at four or more of the seven loci). Twelve of these 24 lineages included encapsulated isolates, and 12 contained noncapsulated isolates, and the analysis of congruence was performed separately for the sequences of the seven loci from these two groups of 12 STs. The statistical tests for congruence between all pairs of loci were carried out as described previously (12). Briefly, the method computes the maximum likelihood tree using the sequence data from locus A and then calculates the log likelihood value (−ln L, a measure of fit) of this tree as a fit to the sequence data from locus B. A distribution of −ln L values is obtained for each of 200 random tree topologies as a fit to the sequence data from locus B. If the −ln L value for the tree from locus A, as a fit to the sequence data from locus B, is outside the 99th percentile value of the −ln L values for the 200 random tree topologies, the two loci are considered to be significantly congruent; otherwise, they are incongruent. Maximum likelihood trees were reconstructed using PAUP* version 4.0 (Sinauer Associates, Sunderland, Mass.). The HKY85 model of DNA substitution was used, and the optimal ratio of transitions to transversions (Ts/Tv) and of the α parameter, which describes the extent of rate variation among nucleotide sites (assuming a discrete gamma distribution with eight categories), were estimated from the empirical data during tree reconstruction. Branch lengths and the values of Ts/Tv and α were reoptimized to maximize the likelihood of each tree topology on the reference sequence data.
RESULTS
Selection of housekeeping loci for MLST.
Genes involved in general metabolic processes (housekeeping genes) were selected from the H. influenzae Rd genome sequence (14). Regions of the genome were selected that included several contiguous housekeeping genes and which appeared to be devoid of genes under diversifying selection (e.g., genes encoding cell envelope components) or genes associated with virulence. A central housekeeping gene within such a region was selected as a candidate MLST locus. For each candidate locus, oligonucleotide primers were designed that could amplify an approximately 500- to 600-bp internal fragment. Primers were based on regions of the gene products that were well conserved among bacterial species. The likelihood that the primers would amplify the desired gene fragment from any encapsulated or noncapsulated H. influenzae isolate was evaluated using a diverse subset of the 12 isolates, which included an isolate of each of the known serotypes, and a selection of noncapsulated H. influenzae isolates that (on the basis of ribotyping data) were considered to cover the diversity of these isolates. H. influenzae is a relatively diverse species, and some primers failed to amplify the desired fragment from all isolates. Additional primers were designed for the gene, and if these also failed to amplify the fragment from all of the isolates, an alternative housekeeping gene was selected until a final set of seven loci and primers were obtained. The seven selected housekeeping genes were distributed around the H. influenzae chromosome (Table 1) and were separated by at least 120-kb, except for recA and fucK, which were only 22 kb apart.
TABLE 1.
Details of loci used in MLST scheme
| Locus | Gene product | Length of sequenced fragment (bp) | No. of alleles | No. of variable sites | % Sequence diversity [range (mean)] | dS/dN ratio | TIGR identifiera | Chromosomal location (kb) |
|---|---|---|---|---|---|---|---|---|
| adk | Adenylate kinase | 477 | 23 | 50 | 0.2-5.8 (2.5) | 23.2 | HI0349 | 376 |
| atpG | ATP synthase F1 subunit gamma | 447 | 26 | 35 | 0.2-4.2 (1.9) | 9.1 | HI0480 | 502 |
| frdB | Fumarate reductase iron-sulfur protein | 489 | 26 | 50 | 0.2-5.1 (2.6) | 119 | HI0834 | 883 |
| fucK | Fuculokinase | 345 | 22 | 32 | 0.3-5.5 (2.5) | 5.3 | HI0613 | 644 |
| mdh | Malate dehydrogenase | 405 | 36 | 50 | 0.2-6.7 (3.3) | 29.5 | HI1210 | 1,277 |
| pgi | Glucose-6-phosphate isomerase | 468 | 32 | 73 | 0.2-6.8 (4.3) | 15.7 | HI1576 | 1,645 |
| recA | RecA protein | 426 | 23 | 36 | 0.2-5.9 (2.6) | >100 | HI0600 | 622 |
Identifier number of the open reading frame in the genome sequence of H. influenzae strain Rd (14). TIGR, The Institute for Genomic Research.
The seven sets of primers were used to amplify and sequence the seven gene fragments from the complete set of 131 H. influenzae isolates. The numbers of alleles per locus, the numbers of variable sites, and the sequence diversity are shown in Table 1. All seven loci appeared to be under stabilizing selection pressure, as most of the substitutions were at synonymous sites, as indicated by the positive ratios of synonymous to nonsynonymous substitutions (dS/dN).
The allelic profiles and properties of the 131 H. influenzae isolates are provided in Table 2, where they are listed in the order of the position of their STs on the minimum evolution tree obtained from the concatenated sequences (see below). On average there were 27 alleles per locus, and the MLST scheme has the potential to resolve about 10 billion distinct STs. The 131 isolates were resolved into 68 different STs, and the similarities between the isolates were examined by cluster analysis, using the matrix of pairwise differences between their allelic profiles (Fig. 1).
TABLE 2.
Properties of H. influenzae isolates used in this study
| Serotype-isolate namea | Geographic origin | Date of isolation | Allelic profile
|
ST | MLEE lineageb | ETb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| adk | atpG | frdB | fucK | mdh | pgi | recA | ||||||
| BPF-F3028 | Brazil | 1986 | 24 | 26 | 24 | 21 | 36 | 33 | 21 | 65 | ||
| b-RM7017 | Ghana | 1980s | 10 | 14 | 5 | 7 | 26 | 22 | 8 | 31 | A1e | 52 |
| b-RM7419 | Kenya | 1980s | 10 | 14 | 5 | 7 | 4 | 7 | 8 | 53 | A2a | 60 |
| b-RM7118 | Australia | 1985 | 9 | 14 | 12 | 3 | 4 | 3 | 8 | 22 | A1a | 1.7 |
| b-RM7717 | Australia | 1989 | 9 | 14 | 12 | 3 | 4 | 3 | 8 | 22 | ||
| b-Eagan | United States | 1950s | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | A1a | 6 |
| b-RM6107 | England | 1985 | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | A1a | 31 |
| b-RM7020 | Ghana | 1983 | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | A1d | 50 |
| b-RM7430 | Kenya | 1980s | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | A1a | 8 |
| b-M6587 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | ||
| b-M6594 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | ||
| b-M6603 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | ||
| b-M6604 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | ||
| b-M6605 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | ||
| b-M6606 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | ||
| b-M6607 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 3 | 4 | 3 | 8 | 44 | ||
| b-RM8012 | United States | 1984 | 10 | 14 | 4 | 5 | 4 | 31 | 8 | 55 | A2e | 122 |
| b-RM6094 | England | 1983 | 23 | 20 | 21 | 5 | 4 | 7 | 8 | 64 | A2a | 71 |
| b(iv)-7853 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7854 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7863 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7868 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7871 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7884 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7885 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7887 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7894 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7909 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b(iv)-7910 | United States | Before 1991 | 10 | 14 | 22 | 5 | 4 | 7 | 22 | 54 | ||
| b-M6596 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6597 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6598 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6599 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6600 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6612 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6613 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6614 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6615 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6616 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6617 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6618 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6619 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6624 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-M6894 | Pennsylvania | 1999-2000 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-RM7651 | Norway | 1976 | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | ||
| b-RM7109 | Sweden | 1985 | 10 | 14 | 4 | 5 | 4 | 7 | 20 | 24 | A2a | 21.9 |
| b-RM7414 | Kenya | 1980s | 6 | 20 | 23 | 1 | 33 | 29 | 23 | 50 | B1b | 140 |
| b-M6588 | Pennsylvania | 1999-2000 | 6 | 20 | 20 | 1 | 33 | 29 | 7 | 45 | ||
| b-M6589 | Pennsylvania | 1999-2000 | 6 | 20 | 20 | 1 | 33 | 29 | 7 | 45 | ||
| b-M6755 | Pennsylvania | 1999-2000 | 6 | 20 | 20 | 1 | 33 | 29 | 7 | 45 | ||
| d-RM7271 | Malaysia | 1972 | 5 | 15 | 10 | 15 | 8 | 5 | 11 | 48 | B1g | 162 |
| d-RM7429 | Kenya | 1986 | 5 | 15 | 10 | 8 | 8 | 5 | 11 | 49 | B1g | 163 |
| d-RM6137 | England | 1963 | 5 | 15 | 10 | 9 | 8 | 5 | 11 | 10 | B1f | 157 |
| d-RM7033 | Papua New Guinea | 1980s | 5 | 15 | 10 | 9 | 8 | 5 | 11 | 10 | B1f | 161 |
| d-RM1168 | United States | 1983 | 5 | 15 | 7 | 9 | 7 | 5 | 11 | 47 | B1f | 158 |
| d-RM6150 | England | 1985 | 5 | 15 | 7 | 9 | 7 | 5 | 11 | 47 | B1f | 159 |
| d-RM8039 | United States | 1984 | 5 | 15 | 7 | 9 | 7 | 5 | 11 | 47 | B1f | 160 |
| d-Rd | United States | Before 1952 | 5 | 15 | 7 | 9 | 7 | 5 | 11 | 47 | ||
| nt-486 | Finland | 1994-1995 | 17 | 4 | 17 | 1 | 7 | 12 | 11 | 41 | ||
| a-RM1042 | United States | 1981 | 12 | 5 | 5 | 2 | 3 | 11 | 7 | 5 | B2a | 164 |
| a-RM7190 | Malaysia | 1973 | 13 | 16 | 5 | 2 | 3 | 11 | 7 | 23 | B2c | 169 |
| a-RM7191 | Malaysia | 1973 | 13 | 16 | 5 | 2 | 3 | 11 | 7 | 23 | B2b | 167 |
| a-RM7031 | Papua New Guinea | 1980s | 13 | 16 | 5 | 18 | 3 | 11 | 7 | 56 | B2b | 165 |
| a-RM7032 | Papua New Guinea | 1980s | 13 | 16 | 5 | 18 | 3 | 11 | 7 | 56 | B2b | 166 |
| a-RM7198 | Malaysia | 1974 | 13 | 16 | 5 | 18 | 3 | 11 | 7 | 56 | B2b | 168 |
| a-RM7205 | The Gambia | 1980s | 4 | 17 | 4 | 1 | 2 | 9 | 6 | 4 | B4 | 173 |
| a-RM7416 | Kenya | 1986 | 4 | 17 | 4 | 1 | 2 | 9 | 6 | 4 | B4 | 174/PICK> |
| nt-1008 | Finland | 1994-1995 | 1 | 22 | 19 | 19 | 32 | 17 | 2 | 43 | ||
| c-RM6134 | England | 1975 | 7 | 11 | 6 | 7 | 6 | 10 | 10 | 8 | D4 | 185 |
| c-RM7422 | Kenya | 1986 | 7 | 11 | 6 | 7 | 6 | 10 | 10 | 8 | D5 | 186 |
| c-RM6132 | England | 1964 | 7 | 11 | 6 | 6 | 18 | 16 | 9 | 19 | D2 | 183 |
| c-RM8032 | United States | 1983 | 14 | 20 | 6 | 8 | 6 | 16 | 21 | 58 | D3 | 184 |
| c-RM7267 | Malaysia | 1973 | 7 | 2 | 6 | 6 | 6 | 16 | 9 | 51 | D1b | 180 |
| c-RM7270 | Malaysia | 1975 | 7 | 11 | 6 | 6 | 6 | 16 | 9 | 7 | D1a | 178 |
| c-RM7424 | Kenya | 1986 | 7 | 11 | 6 | 6 | 6 | 16 | 9 | 7 | E1 | 187 |
| c-RM1167 | United States | Before 1983 | 7 | 11 | 6 | 8 | 6 | 16 | 9 | 9 | D1a | 177 |
| c-RM1271 | Probably United States | 1968 | 7 | 11 | 6 | 8 | 6 | 16 | 9 | 9 | D1b | 179 |
| nt-M6593 | Pennsylvania | 1999-2000 | 1 | 25 | 1 | 14 | 15 | 1 | 5 | 46 | ||
| nt-981 | Finland | 1994-1995 | 2 | 24 | 18 | 18 | 27 | 1 | 5 | 42 | ||
| nt-477 | Finland | 1994-1995 | 1 | 1 | 1 | 14 | 15 | 1 | 5 | 1 | ||
| nt-432 | Finland | 1994-1995 | 1 | 3 | 1 | 1 | 31 | 1 | 5 | 40 | ||
| nt-1124 | Finland | 1994-1995 | 1 | 1 | 1 | 13 | 13 | 25 | 16 | 12 | ||
| nt-1247 | Finland | 1994-1995 | 1 | 10 | 1 | 1 | 1 | 6 | 5 | 33 | ||
| nt-375 | Finland | 1994-1995 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 3 | ||
| nt-723 | Finland | 1994-1995 | 5 | 1 | 1 | 1 | 1 | 2 | 5 | 14 | ||
| nt-1180 | Finland | 1994-1995 | 14 | 7 | 1 | 15 | 16 | 4 | 1 | 2 | ||
| nt-1181 | Finland | 1994-1995 | 14 | 7 | 1 | 15 | 16 | 4 | 1 | 2 | ||
| nt-1292 | Finland | 1994-1995 | 14 | 7 | 1 | 15 | 16 | 4 | 1 | 2 | ||
| nt-1200 | Finland | 1994-1995 | 16 | 21 | 14 | 16 | 29 | 14 | 3 | 36 | ||
| nt-1268 | Finland | 1994-1995 | 16 | 21 | 14 | 16 | 29 | 14 | 3 | 36 | ||
| nt-285 | Finland | 1994-1995 | 15 | 23 | 16 | 16 | 30 | 1 | 3 | 39 | ||
| nt-1231 | Finland | 1994-1995 | 11 | 2 | 15 | 8 | 28 | 26 | 3 | 34 | ||
| nt-1232 | Finland | 1994-1995 | 11 | 2 | 15 | 8 | 28 | 26 | 3 | 34 | ||
| nt-176 | Finland | 1994-1995 | 10 | 2 | 15 | 17 | 26 | 26 | 3 | 38 | ||
| nt-1158 | Finland | 1994-1995 | 1 | 8 | 1 | 14 | 9 | 14 | 13 | 11 | ||
| nt-1159 | Finland | 1994-1995 | 1 | 8 | 1 | 14 | 9 | 14 | 13 | 11 | ||
| nt-1207 | Finland | 1994-1995 | 3 | 9 | 8 | 4 | 14 | 8 | 4 | 13 | ||
| nt-1209 | Finland | 1994-1995 | 3 | 9 | 8 | 4 | 14 | 8 | 4 | 13 | ||
| nt-1233 | Finland | 1994-1995 | 3 | 9 | 8 | 4 | 14 | 8 | 4 | 13 | ||
| nt-162 | Finland | 1994-1995 | 14 | 7 | 13 | 4 | 1 | 21 | 1 | 37 | ||
| nt-667 | Finland | 1994-1995 | 14 | 7 | 13 | 7 | 17 | 13 | 17 | 57 | ||
| a-RM6064 | England | 1966 | 20 | 12 | 1 | 7 | 20 | 23 | 19 | 21 | H1b | 223 |
| a-RM6068 | England | 1968 | 20 | 12 | 25 | 7 | 20 | 23 | 19 | 30 | H1b | 226 |
| a-RM6083 | England | 1977 | 20 | 12 | 21 | 20 | 34 | 30 | 19 | 60 | H1a | 220 |
| a-RM6070 | England | 1962 | 20 | 12 | 1 | 8 | 20 | 23 | 19 | 59 | H1b | 224 |
| a-RM6073 | England | 1966 | 20 | 12 | 2 | 1 | 21 | 23 | 19 | 25 | H1a | 221 |
| a-RM7115 | Dominican Republic | 1980s | 21 | 12 | 9 | 7 | 19 | 24 | 19 | 62 | I1b | 230 |
| a-RM6062 | England | 1965 | 21 | 13 | 9 | 7 | 19 | 24 | 19 | 20 | I1a | 227 |
| a-RM6069 | England | 1963 | 21 | 13 | 9 | 7 | 19 | 24 | 19 | 20 | I1a | 228 |
| a-RM6080 | England | 1967 | 21 | 13 | 9 | 7 | 19 | 24 | 19 | 20 | I1a | 229 |
| b-RM1324 | United States | 1947 | 21 | 12 | 9 | 4 | 5 | 20 | 18 | 61 | J3 | 239 |
| e-RM6158 | England | 1962 | 7 | 6 | 3 | 7 | 10 | 27 | 10 | 52 | F2a | 197 |
| e-RM6169 | England | 1964 | 18 | 6 | 3 | 8 | 23 | 28 | 19 | 27 | F1 | 189 |
| e-RM6181 | England | 1965 | 18 | 6 | 3 | 10 | 11 | 28 | 14 | 17 | F2b | 202 |
| e-RM6167 | England | 1963 | 18 | 6 | 3 | 7 | 25 | 28 | 12 | 67 | F2c | 204 |
| e-RM7066 | Papua New Guinea | 1980s | 18 | 6 | 3 | 7 | 24 | 28 | 19 | 28 | F2b | 203 |
| e-RM6185 | England | 1965 | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | F1 | 190 |
| e-RM6229 | England | 1977 | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | F2c | 215 |
| e-RM7280 | Malaysia | 1973 | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | F2c | 206 |
| e-RM6210 | England | 1972 | 18 | 6 | 26 | 7 | 10 | 28 | 12 | 68 | F2a | 196 |
| e-RM7423 | Kenya | 1986 | 19 | 6 | 3 | 10 | 10 | 28 | 14 | 32 | G1 | 218 |
| e-RM1018 | United States | Before 1983 | 18 | 6 | 3 | 11 | 10 | 28 | 12 | 66 | F2a | 191 |
| e-RM8031 | United States | 1983 | 18 | 6 | 3 | 22 | 10 | 28 | 12 | 69 | F2a | 192 |
| f-RM7290 | Malaysia | 1970s | 22 | 19 | 11 | 9 | 22 | 19 | 15 | 29 | K1c | 251 |
| f-RM7283 | Malaysia | 1970s | 22 | 18 | 11 | 11 | 22 | 19 | 15 | 26 | K1c | 249 |
| f-RM7298 | Malaysia | 1970s | 22 | 18 | 11 | 11 | 22 | 19 | 15 | 26 | K1c | 250 |
| f-RM7417 | Kenya | 1986 | 22 | 19 | 11 | 11 | 35 | 32 | 15 | 63 | L1 | 267 |
| f-RM6237 | England | 1963 | 22 | 6 | 11 | 12 | 1 | 18 | 15 | 16 | K2a | 265 |
| f-RM6244 | England | 1964 | 22 | 6 | 11 | 12 | 1 | 18 | 15 | 16 | K2a | 263 |
| f-RM6255 | England | 1967 | 22 | 6 | 11 | 12 | 1 | 18 | 15 | 16 | K2a | 257 |
| f-RM6252 | England | 1966 | 22 | 6 | 11 | 11 | 12 | 18 | 15 | 15 | K1b | 241 |
The prefixes a-, b-, c-, d-, e-, f- and nt- indicate the serotype of the isolate, or that it is nontypeable (noncapsulated [nt]). Isolates beginning with the prefix “b(iv)” are serotype b, biotype iv. STs are listed in their order on the minimum evolution tree (Fig. 3). Isolates of group I are separated by a blank line from those of group II.
The MLEE lineage and ET are shown, where known, for isolates previously characterized by MLEE (28).
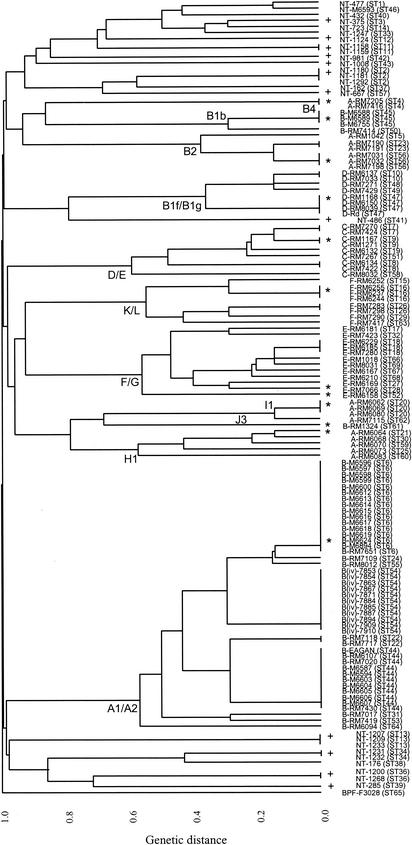
FIG. 1.
Clustering of encapsulated and noncapsulated H. influenzae isolates constructed using UPGMA from the matrix of pairwise differences in the allelic profiles of all 131 isolates. The prefix to the isolate name indicates the serotype of the isolate or whether it is noncapsulated (NT-). The relationship between STs and clonal complexes defined by MLST and the MLEE lineages defined by Musser et al. (28) are indicated alongside the branches of the dendrogram. The STs marked with an asterisk (encapsulated isolates) or a plus sign (noncapsulated isolates) were used in the statistical analysis of congruence between loci.
Characterization of encapsulated H. influenzae isolates by MLST.
All 54 serotype a, c, d, e, and f isolates and 14 of the Hib isolates have previously been characterized by MLEE (28). The STs and clonal complexes assigned by MLST could therefore be related to the electrophoretic types (ETs) and lineages defined by MLEE; the latter are shown in Table 2. The overall similarity in the clustering of isolates was excellent. Using both methods, isolates of serotypes c, d, e, and f each formed single monophyletic clusters of related genotypes on the dendrogram, which were assigned as clonal complexes or lineages (isolates that are likely to be descended from the same ancestral genotype). Similar clustering of isolates of these serotypes has been observed using MLEE (26, 28) and pulsed-field gel electrophoresis (PFGE) (30).
Serotype c isolates were all linked at a genetic distance of 0.58, and this cluster corresponded to MLEE lineages D1, D2, D3, D4, D5, and E1 as defined by Musser et al. (28). Serotype d isolates were very similar in genotype as shown by MLST and were linked at a genetic distance of 0.35. These isolates were also closely related by MLEE and were within lineages B1f and B1g (28). Serotype e isolates were linked at a genetic distance of 0.55 by MLST and were within MLEE lineages F1, F2, and G1 (28). Serotype f isolates were tightly clustered (genetic distance of 0.53 by MLST) and were all within MLEE lineages K1, K2, and L1 (28).
The serotype a isolates were at two main locations on the UPGMA dendrogram (Fig. 1), and these corresponded to isolates of MLEE lineages B2/B4 and H1/I1 (28). With one exception (allele 1 of fucK was present in an ST of both lineage B4 and H1), serotype a isolates from these two locations on the tree had different alleles at all seven loci. A similar division of serotype a isolates into two highly divergent clusters has been observed by MLEE and by other typing methods (26, 28, 30), and these clusters are within the different phylogenetic divisions within H. influenzae (group I and group II) that have been proposed from MLEE studies (26). Isolates of MLEE lineages B2/B4 were clearly resolved further into two divergent clusters by MLST, which corresponded to lineages B2a, B2b, and B2c (STs 5, 23, and 56), and lineage B4 (ST4); isolates in these different clusters shared no alleles in common. Isolates in MLEE lineage H1/I1 were also further resolved into two clusters, which corresponded to MLEE lineages H1a and H1b (STs 21, 25, 30, 59, and 60) and lineages I1a and I1b (STs 20 and 62); isolates in these two clusters shared up to three alleles in common.
The majority of the Hib isolates were assigned to two divergent clusters by MLST (Fig. 1). Several of these isolates have been studied by MLEE, which allowed one cluster (STs 6, 22, 24, 31, 44, 53, 54, 55, and 64) to be assigned as MLEE lineages A1/A2, which include about 90% of Hib isolates from patients with invasive disease (28). This cluster included the 11 isolates of Hib biotype IV, which were all from the blood or CSF of patients with meningitis in Maryland. Three pairs of isolates were from blood and CSF taken from the same patient, and all 11 were identical by MLST (ST54). The other Hib cluster (STs 45 and 50) corresponded to MLEE lineage B1, which includes most of the other Hib isolates recovered from invasive disease (28). A small number of Hib isolates (group II isolates) are highly divergent in genotype from those of the major Hib lineages A1/A2 and B1 and are assigned to MLEE lineage J (28). One Hib isolate assigned to MLEE lineage J was available for characterization by MLST. Consistent with this assignment, isolate b-RM1324 (ST61) clustered with the serotype a isolates assigned to group II MLEE lineage I1.
Hib isolates from ongoing transmission in Pennsylvania communities.
MLST was used to characterize eight Hib isolates from patients with invasive disease occurring during 1999 and 2000 in rural Pennsylvania communities (15). The eight isolates from disease had four different allelic profiles; analysis of the same isolates by PFGE resolved five DNA fragment patterns (15). Two isolates, from two different communities, were ST6, and another three, from three other communities, were ST45. These STs differ at all seven loci and based on the MLST data can be assigned to the MLEE lineages A1/A2 (ST6) and B1 (ST45). Two further isolates (ST44) clustered with ST6 but differed from it at two of the seven loci. The last of the eight disease Hib isolates (M6953) was ST46, which was not closely related to any of the Hib isolates that were characterized by MLST and clustered among a group of noncapsulated isolates. The serotype of M6953 was reexamined, and the isolate was shown by both slide agglutination and PCR serotyping (11) to be noncapsulated and was renamed nt-M6953. Eighteen carriage Hib isolates were found in two of the Pennsylvania communities in which a case of invasive disease occurred. Five carried Hib isolates from one of these communities were all ST44, as was the disease isolate from the same community (b-M6587). However, all 13 carried isolates from the second community were identical (ST6), but the disease isolate from the same community was of ST44. The identities and similarities of the carriage and disease isolates from these communities correspond well to those found using PFGE (15).
Characterization of noncapsulated H. influenzae isolates by MLST.
The noncapsulated isolates recovered from the middle ear fluid of Finnish children with acute otitis media were expected to be diverse, as they were selected from across a dendrogram produced from ribotyping data on a much larger collection of noncapsulated isolates from Finnish children and other sources. However, four pairs of isolates were included that were recovered from the left and right ear of the same child during the same episode of acute otitis media. As expected, each of the four pairs of isolates from the same child were identical in allelic profile (STs 2, 11, 13, and 34); the third isolate of ST2 was from the same child as the other two but was recovered from middle ear fluid 1 month later. The 20 epidemiologically unlinked isolates were different in genotype, except that one isolate was of ST13 (nt-1233) although it was from a different child than the other two isolates of this ST, and nt-1200 and nt-1268 were both of ST36 but were from different children.
One noncapsulated isolate (nt-486; ST41) clustered with the serotype d isolates, but the others formed two highly divergent clusters (Fig. 1). Isolate nt-486 shared alleles at two of the seven loci with isolates of serotype d; the noncapsulated status of this strain was confirmed using PCR (11). The allelic profiles of all other noncapsulated isolates typically differed from those of encapsulated isolates at all seven loci.
Phylogenetic signal and recombination in encapsulated and noncapsulated H. influenzae.
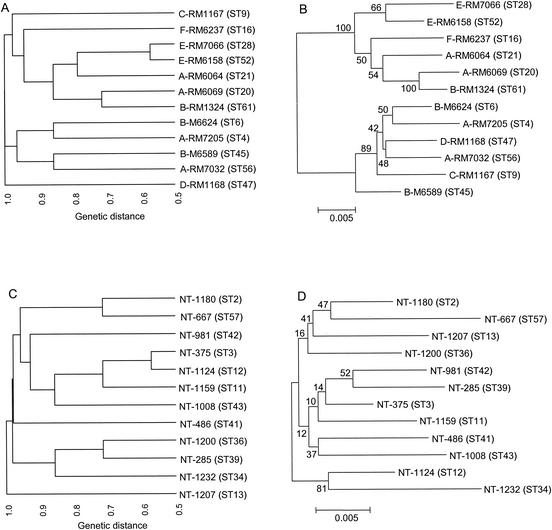
Differences in allelic profiles are used in MLST and MLEE to identify clusters of related isolates. MLST provides the sequences at seven housekeeping loci, which potentially can be used to probe the deeper relationships among isolates. However, this is only a valid approach if recombination, over the long term, has not eliminated phylogenetic signal (13). Significant congruence between gene trees provides an indication of the presence of phylogenetic signal and has been observed in H. influenzae (12), which may allow meaningful trees to be constructed from the concatenated sequences at the seven MLST loci. However, it has been proposed that noncapsulated isolates are less clonal and have higher rates of recombination than encapsulated isolates (25, 32). We therefore compared the congruence between loci separately for encapsulated and noncapsulated isolates. Isolates at the tips of the tree (Fig. 1) are almost certainly descended from a common ancestor and share alleles at several loci. Some significant congruence could therefore be introduced if multiple isolates of the same clonal complex, or lineage, were included in the analysis, and for both encapsulated and noncapsulated H. influenzae, 12 divergent STs were selected that differed from each other at a genetic distance of ≥0.55 (Fig. 2).
FIG. 2.
Phylogenetic relationships among diverse STs. A UPGMA tree was constructed from the differences in the allelic profiles of the 12 diverse STs of encapsulated (A) and noncapsulated (C) H. influenzae isolates (these STs are identified in Fig. 1). All isolates differed from each other at a genetic distance of ≥0.55 (N.B.: the tree is truncated at a genetic distance of 0.5). The concatenated sequences were used to construct a minimum evolution tree from the same sets of diverse encapsulated (B) and noncapsulated (D) STs. The percentage of recovery of the nodes in 1,000 replicate trees using the bootstrap test are shown on the minimum evolution trees.
The STs used in the analysis of congruence are marked on the tree shown in Fig. 1, and the apparent relationships between these isolates are shown on the UPGMA tree derived from the differences in their allelic profiles (Fig. 2). Of the 42 pairwise tree comparisons, there were 18 (43%) comparisons for which the gene trees obtained from the 12 encapsulated isolates were significantly congruent (Table 3). For the noncapsulated isolates, only 3 of the 42 comparisons (7%) showed a statistically significant level of congruence (Table 3).
TABLE 3.
Log likelihood values for the maximum likelihood tree for each reference locus and for this tree as a fit to the data from the other locia
| Isolate group and reference locus | −In L | αb | Ts/Tv | 99th percentile of −ln L for random trees | −ln L value for other loci
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| adk | atpG | frdB | fucK | mdh | pgi | recA | |||||
| Encapsulated | |||||||||||
| adk | 944.2 | 0.006 | 2.83 | 1,002.0 | 1,021.8 | 1,020.6 | 1,024.3 | 1,053.1 | 1,021.7 | 987.9 | |
| atpG | 762.8 | 0.069 | 2.63 | 828.8 | 837.4 | 828.2 | 827.6 | 835.2 | 779.5 | 826.4 | |
| frdB | 913.6 | 0.243 | 2.82 | 979.6 | 1,011.5 | 985.6 | 992.4 | 978.4 | 983.8 | 985.6 | |
| fucK | 589.3 | 783.7 | 4.34 | 624.3 | 624.4 | 589.3 | 628.1 | 624.4 | 589.3 | 589.3 | |
| mdh | 945.3 | 0.006 | 2.91 | 965.0 | 963.6 | 963.8 | 965.1 | 965.6 | 960.3 | 965.1 | |
| pgi | 1,152.7 | 0.074 | 1.93 | 1,200.7 | 1,248.9 | 1,202.8 | 1,209.2 | 1,247.1 | 1,219.4 | 1,242.9 | |
| recA | 790.5 | 0.003 | 2.19 | 856.7 | 801.2 | 823.2 | 849.6 | 823.2 | 840.6 | 823.2 | |
| Noncapsulated | |||||||||||
| adk | 810.7 | 0.006 | 9.21 | 824.3 | 832.7 | 849.2 | 840.0 | 848.3 | 849.3 | 814.9 | |
| atpG | 791.9 | 0.069 | 3.44 | 835.4 | 849.9 | 868.3 | 874.9 | 867.5 | 869.8 | 850.1 | |
| frdB | 914.6 | 0.004 | 2.58 | 947.8 | 971.5 | 984.6 | 971.5 | 988.8 | 1,000.0 | 982.0 | |
| fucK | 599.2 | 0.111 | 4.38 | 619.7 | 638.8 | 655.7 | 653.0 | 654.5 | 652.1 | 646.6 | |
| mdh | 893.3 | 0.051 | 4.3 | 947.7 | 984.3 | 967.0 | 990.5 | 990.5 | 980.9 | 975.1 | |
| pgi | 1,058.8 | 0.052 | 4.56 | 1,112.0 | 1,191.5 | 1,176 | 1,195.2 | 1,195.2 | 1,187.2 | 1,184.5 | |
| recA | 745.1 | 0.002 | 1.21 | 785.8 | 755.6 | 820.6 | 822.8 | 783.2 | 820.9 | 842.3 | |
Those values that are closer to the -ln L value of the reference locus than the 99th percentile of values for the 200 random tree topologies are shown in boldface type and denote significant congruence between the loci.
α, a parameter describing the rate variation among sites. (see Materials and Methods).
Recombination has clearly occurred in H. influenzae housekeeping genes. Analysis of the distribution of polymorphic synonymous sites within the alleles at each locus using Sawyer's test (35) indicated significant mosaic gene structure (an indication that different regions of the gene fragments have different phylogenetic histories) for pgi, recA, mdh, and frdB (P values of 0.02 to 0.0008). The substantially higher levels of congruence between loci in encapsulated isolates, compared to that in noncapsulated, indicates that recombination may have been less frequent among encapsulated isolates, and Sawyer's test was rerun using only those alleles found among encapsulated isolates; significant mosaic structure was still evident at all of these loci (P values of 0.03 to 0.002). This is perhaps not surprising, as even in the encapsulated isolates, over half the comparisons between gene trees were noncongruent.
Phylogenetic relationships within H. influenzae.
Although the clustering of isolates by MLEE and MLST was very similar, some of the relationships between the major lineages discerned in the MLEE studies were not apparent using MLST. For example, the sharing of alleles between isolates of the major A1/A2 and B1 lineages of Hib, which is clearly apparent in the MLEE data (28), was not seen using MLST. Instead, Hib isolates assigned to these two major MLEE lineages appeared to be only distantly related using MLST, as they differed from each other at all seven loci. Similarly, isolates of the other different clonal complexes defined by MLST differed at all loci, preventing any comparison of the deeper relationships between lineages with those proposed from the MLEE data, including the deep separation of H. influenzae into group I and II isolates (26).
The significant congruence in nearly half of all pairwise tree comparisons indicates that recombination, although evident, has not eliminated all phylogenetic signal in the gene trees constructed for encapsulated H. influenzae. The construction of a tree based on the concatenated nucleotide sequences from the seven MLST loci is therefore justified and could provide some indication of the phylogenetic relationships between the major clusters of encapsulated isolates, which cannot be obtained using the pairwise differences between their allelic profiles.
Initially minimum evolution trees were constructed using the concatenated sequences from the set of 12 diverse STs from the encapsulated and the noncapsulated populations (Fig. 2B and D). The encapsulated isolates were resolved into two major groups. This major division was strongly supported and was demonstrated in 100% of bootstrap replicates, but the relationships between the diverse isolates within these two groups were less strongly supported. Only the association of ST20 and ST61 and the separation of a group of five STs from ST45 were strongly supported (bootstrap support of 100 and 89%, respectively), although the majority of bootstrap values were >50%. For the noncapsulated isolates, there was very little structure in the minimum evolution tree, and only two of the interior branches were recovered in more than 50% of the bootstrap replicates. The analysis of congruence and the minimum evolution tree obtained from the concatenated sequences indicate that there is some phylogenetic signal in trees obtained using the sequences from the encapsulated isolates, but little is apparent for the noncapsulated isolates.
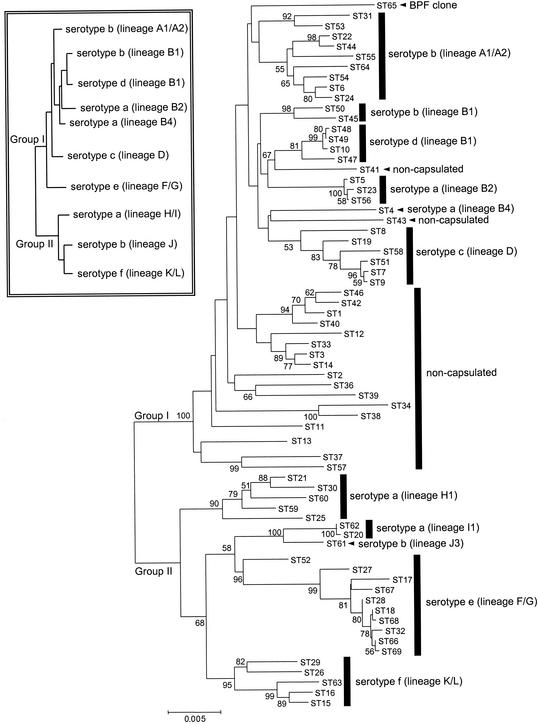
Figure 3 shows a minimum evolution tree based on the concatenated sequences of all 68 STs represented among the encapsulated and noncapsulated isolates. Not surprisingly, the clustering of encapsulated isolates was similar to that obtained using the pairwise differences in allelic profiles. Isolates of serotypes c, d, e, and f formed monophyletic groupings, and the branches leading to these groups of STs were strongly supported in the minimum evolution tree (81 to 96% of bootstrap replicates), except for the inclusion of ST8 with the other STs that are serotype c, which was supported by only 53% of replicates.
FIG. 3.
Phylogenetic relationships among STs of encapsulated and noncapsulated H. influenzae. A minimum evolution tree was reconstructed using the concatenated sequences from the seven MLST loci (3,057 bp) for each of the 68 STs. The percent recoveries of the nodes in 1,000 bootstrap replicates are shown where these are ≥50%. The serotypes and MLEE lineages of the STs and the major phylogenetic division of isolates into group I and group II are shown. The tree is rooted at the midpoint of the longest distance between the STs. The box (inset) shows a simplified tree illustrating the relationships between major lineages inferred from the pairwise differences in the electrophoretic profiles of isolates obtained using MLEE. Tree shown in inset adapted from reference 28 with permission of the publisher.
Serotype a isolates were separated into four clusters (Fig. 3). Within group I, the serotype a isolates of MLEE lineage B2 were clearly resolved from those of lineage B4. Similarly, within group II, the serotype a isolates of MLEE lineages H1 and I1 were resolved. Some resolution of these four serotype a lineages can also be observed in the tree obtained from the differences in the allelic profiles (Fig. 1).
The two major clusters of Hib isolates that were identified in Fig. 1 were also recovered on the minimum evolution tree and corresponded to the major MLEE lineages A1/A2 and B2 lineages (Fig. 3). However, support for inclusion of two STs 31 and 53 within the A1/A2 cluster was not strong. Only one Hib isolate clustered outside the A1/A2 or B2 lineages. This isolate (b-RM1324; ST61) has been assigned to MLEE lineage J3 and was strongly associated with the serotype a isolates of MLEE lineage I1 (100% of bootstrap replicates) within phylogenetic group II (Fig. 3).
The deeper relationships between the major clusters that could not be discerned on the tree constructed from the allelic profiles were more apparent on the minimum evolution tree and were consistent with those proposed from MLEE studies (Fig. 3). A deep division among H. influenzae isolates was observed in the minimum evolution tree and had strong statistical support (Fig. 2 and 3); the branch leading to the group I isolates was recovered in 100% of bootstrap replicates. However, the division differed slightly from the major phylogenetic division proposed from the MLEE data, as isolates of serotype e were clearly within group II on the minimum evolution tree, whereas by MLEE they were placed within group I (26, 28). The STs within group I and group II on the minimum evolution tree had a similar level of sequence diversity (average of 2.0% for group I and 1.9% for group II, using the concatenated sequences), but the average diversity between the groups was 3.7%. However, although the average diversity between group I and II sequences was substantial, for five of the seven MLST loci, there were identical alleles in STs of both group I and II. For example, among the 58 different alleles present in the STs of group II, 10 were also found in STs of group I.
The relatedness of the Hib STs that correspond to the two major MLEE lineages, A1/A2 and B1 (26), was not apparent in the tree obtained using the differences in the allelic profile but was apparent using the concatenated sequences (Fig. 3). Similarly, isolates of MLEE lineages A1/A2 (serotype b), B1 (serotypes b and d), B2 and B4 (both serotype a), and D (serotype c) appeared to be resolved from other isolates (Fig. 3). However, most of the relationships between the different MLEE lineages were poorly supported by the bootstrap test. Indeed, only the association between MLEE lineage I1 and J3 and the deep separation of the group I isolates from group II isolates, were strongly supported. Weaker support was found for some other associations, including the separation of isolates of MLEE lineages F/G, K/L, I1, and J from those of lineage H1 (68% of replicates), and the branch leading to lineages F/G, I1, and J (58% of replicates).
The minimum evolution tree placed all of the noncapsulated isolates within group I (Fig. 3). Some clusters of noncapsulated isolates were apparent, but the much lower levels of congruence between gene trees within this population suggests that there is little phylogenetic signal and that the deeper relationships among encapsulated isolates on the tree are likely to be unreliable.
An isolate of the Brazilian purpuric fever clone (H. influenzae biogroup aegyptius) was characterized by MLST and appeared from cluster analysis to be unrelated to any of the other isolates (Fig. 1). The alleles at six of the loci were unique to this isolate, but the recA allele was found in one serotype c isolate (RM8032). MLEE has suggested that this clone is most closely allied to isolates of serotype c (29) but this was not apparent on the minimum evolution tree obtained from the concatenated sequences, where this clone was firmly placed within the group I lineages, but was not significantly associated with any of the clusters of encapsulated isolates (Fig. 3).
DISCUSSION
The H. influenzae MLST scheme has been developed using a diverse collection of isolates to increase the likelihood that the seven housekeeping gene fragments can be amplified from any encapsulated or noncapsulated isolate. H. influenzae is a relatively diverse species and several candidate loci were eliminated since the chosen gene fragments could not readily be amplified from the initial subset of strains. The internal fragments of the seven loci that were selected for the final MLST scheme could be amplified from all encapsulated and noncapsulated H. influenzae isolates that we examined, including the Brazilian purpuric fever clone, and allow a single pair of primers to be used for both the initial amplification step from chromosomal DNA and for the DNA sequencing reactions.
The advantages of MLST over other typing schemes have been discussed elsewhere (7, 23). Of particular importance are the precision of MLST and the unambiguous nature of the genetic variation that is indexed, which arises from the use of nucleotide sequence data, and the ease with which strains analyzed in different laboratories can be compared by the interrogation of a central database via the Internet. The latter feature makes the procedure ideal for national or international surveillance programs that involve multiple laboratories, such as those for monitoring the spread of drug-resistant clones or of strains causing invasive disease or for monitoring changes in a bacterial population following vaccine implementation. For example, MLST could be used to address issues arising from the implementation of the Hib conjugate vaccine, for example, ongoing transmission of Hib disease in some communities, the nature of Hib isolates recovered from fully vaccinated children, or the characterization of invasive non-Hib isolates from vaccinated populations. MLST can also be used to uncover the diversity of noncapsulated H. influenzae that any successful vaccine would have to encompass and any differences between isolates associated with invasive disease compared to carriage (41).
The initial MLST database is sufficient to demonstrate that the MLST scheme provides a valid and discriminatory method for the precise and unambiguous characterization of H. influenzae isolates. The validity of the scheme was established by the excellent correlation between the clustering of the encapsulated isolates of serotypes a to f obtained using MLST and MLEE. In addition, MLST indicated very similar genetic relationships among the isolates of Hib from disease and carriage in Pennsylvania communities to those previously inferred using MLEE (15). Also, isolates that were known, or expected, to be indistinguishable in genotype, such as the pairs of noncapsulated isolates from the left and right ear of children with acute otitis media, were identical in ST when characterized by MLST.
This is the first use of MLST to characterize isolates of H. influenzae and demonstrates how the STs and clusters of related genotypes defined by MLST could be related to those defined previously by MLEE (28). The comparative level of resolution achieved by MLST and MLEE is not yet established, and further studies are required to clarify those situations where the precision and ease of interlaboratory comparison make MLST superior to other methods and whether it provides the required level of discrimination between isolates for the particular surveillance activities or epidemiological studies that need to be undertaken. The study of isolates from ongoing Hib transmission showed that MLST provided the same epidemiological information as MLEE, but larger evaluation studies are required. One problem that limits the utility of many typing procedures is that many isolates of Hib are very similar in genotype. Thus, the extensive MLEE studies of Musser et al. (24-28) show that the great majority of Hib isolates from invasive disease are members of only three clusters of closely related genotypes (lineages A1a, A2a, and B1b), and that 70% of all Hib isolates recovered over more than twenty years from worldwide sources belong to only four ETs (clones) within these three lineages (28). Many isolates assigned to these four major clones need to be characterized by MLST to establish whether this technique provides any increased resolution compared to MLEE, but it is very likely that other molecular typing procedures which index more rapidly evolving variation will be needed in cases in which epidemiological investigations require further resolution within these major Hib clones.
As found previously, most noncapsulated isolates appear to be distinct in genotype from encapsulated isolates and appear to be a separate and diverse population, rather than being recently derived from encapsulated strains by loss or inactivation of the capsular biosynthetic genes (25, 32).
Most epidemiological studies are concerned with the clustering of similar isolates rather than the relationships between the clusters. Whether the implied relationships between clusters identified using a typing procedure have any phylogenetic meaning depends on the impact of recombination. Even relatively high rates of recombination will not obscure the clustering of very similar genotypes, as these have emerged by the recent diversification of the founding genotype, but over the longer term, recombination will obscure the phylogenetic relationships between the major clusters (13). Thus, if rates of recombination are high, MLEE and MLST (and any other valid typing method) should identify the same clusters of closely related genotypes, but they will give different (and meaningless) apparent relationships between the clusters. If rates of recombination are lower, the relationships between the major lineages may be meaningful, and these can be assessed by using the concatenated sequences from the seven MLST loci.
MLST and MLEE resolved the same clusters of related genotypes among the encapsulated H. influenzae isolates (e.g., the clustering of isolates of serotypes c, d, e, and f, or the Hib isolates of MLEE lineages A1/A2), but the deeper relationships between these clusters could not be resolved on a UPGMA dendrogram based on the differences in allelic profiles, since most isolates of the different clusters differed at all seven loci (Fig. 1). On the UPGMA dendrogram the lack of resolution between major lineages occurs because the accumulation of even a single nucleotide difference at each of the seven loci results in two strains that are different at all loci. In contrast, a change in an allele assigned by MLEE requires a nonsynonymous substitution that results in a change in the electrophoretic mobility of the gene product, and multiple genetic events at a locus typically will be required before this occurs. Alleles therefore change more slowly by MLEE than by MLST, and as more loci are examined in MLEE, it will take far longer for two isolates to diverge so that they differ at most or all loci by MLEE, compared to MLST. MLEE therefore allows deeper relationships between lineages to be probed, although whether these are meaningful depends on the impact of recombination.
The rate of recombination in H. influenzae is not clear. The species is naturally transformable and the genome contains about 1,500 copies of an uptake sequence, which allows recognition, and selective uptake, of Haemophilus DNA (38). Several naturally transformable bacterial species have been shown to have relatively high rates of recombination, which may not prevent the emergence of clones or the ability to recognize clusters of related isolates (clonal complexes) but which has a major impact on clonal diversification and, over the long term, leads to a loss of phylogenetic signal and a lack of congruence between gene trees (12, 13). Recombination was clearly apparent in H. influenzae housekeeping genes, as significant mosaic structure was found in four of the MLST loci, and there appeared to have been repeated transfer of alleles between group I and II lineages, since 10 of the 58 alleles present in group II STs were also present in group I STs, although these STs differed on average at 3.7% of nucleotide sites. Furthermore, analysis of very closely related isolates indicated that allelic changes at a locus are more commonly occurring by recombination than point mutation (data not shown; see reference 12); a more extensive database is required to produce a reliable estimate of the ratio of recombination to mutation during clonal diversification (12, 13).
Despite this evidence for recombination at housekeeping loci, the process appears to have had somewhat less impact in encapsulated H. influenzae than in the other naturally transformable species that have been studied, and there was statistically significant congruence between gene trees for nearly half (43%) of the pairwise tree comparisons, although there was much less congruence between gene trees for the noncapsulated isolates. Using the same approach, there were no loci that showed significant congruence in S. pneumoniae and only 3 of 42 significant tree comparisons in N. meningitidis (the same as in the noncapsulated H. influenzae), both of which are naturally transformable species (12). The strong association between capsular serotype and genotype observed in studies using both MLST and MLEE also indicates that capsular genes have very rarely become established in new lineages as a result of horizontal transfer and recombination at the capsular biosynthetic locus (24, 26-28). The presence of the type a and b capsular genes in the distantly related lineages of group I and group II indicates that these genes were distributed horizontally at some time in the past (21, 28), but this process appears to be rarer than in S. pneumoniae and N. meningitidis, where isolates with very different genotypes frequently have the same capsular serotype (serogroup), and variation in capsular serotype (serogroup) is found even within isolates of the same clone (4, 6).
The extent of congruence between trees from different loci observed for the encapsulated isolates suggests that recombination in H. influenzae has not completely eliminated the phylogenetic signal (12), and this is supported by the similar relationships between the major lineages of encapsulated isolates obtained using the concatenated nucleotide sequences and the tree derived from MLEE (Fig. 3). However, the statistical support for most of the deeper relationships was low, presumably due to the distorting effect of recombination (Fig. 2 and 3), and it is unclear to what extent this tree approximates a phylogeny of the species. The minimum evolution tree constructed from the concatenated sequences appears to be useful for characterizing H. influenzae by MLST; isolates with identical or similar isolates within a population can be identified using a dendrogram based on the differences in the allelic profiles, but where unambiguous assignment of an isolate to a major lineage is required, or the deeper relationships between lineages need to be probed, this can be more readily achieved by the reconstruction of the tentative phylogeny using the concatenated sequences at the seven MLST loci. It should be stressed that the latter approach to analyzing MLST data is not justified in species such as the meningococcus or pneumococcus, where there is little or no significant congruence between gene trees and there is a high rate of recombination (12, 18), and is also probably not valid for noncapsulated H. influenzae isolates where very little congruence between loci was observed.
A facility for concatenating the sequences at the seven loci, maintaining the reading frame, and for comparing the concatenated sequence of a query isolate with those of a reference set of isolates of the major lineages is available at the H. influenzae pages of the MLST website (http://haemophilus.mlst.net/). In this way, by interrogating the MLST website, the ST of an isolate can be assigned, any identical or similar isolates in the MLST database can be identified, and a dendrogram which displays the relatedness of the isolate to similar isolates in the database can be obtained using the differences in allelic profiles; its assignment to a major lineage, and to phylogenetic group I or II, can then be obtained by using the concatenated sequences to determine the position of the isolate on a minimum evolution tree.
Recombination appears to have had more impact on the divergence of noncapsulated isolates. Congruence between loci was poor, and the deeper relationships between STs on the minimum evolution tree were poorly supported (Fig. 2 and 3). This is compatible with previous studies, which indicate that the population structure of these isolates is much less clonal than that of encapsulated isolates (4a, 25, 32). The increased impact of recombination in noncapsulated, compared to encapsulated, isolates could be due to their increased ability to be transformed and/or increased opportunities for noncapsulated isolates to meet other H. influenzae within the nasopharynx.
Finally, the deep division into group I and group II strains has been noted using both MLEE and MLST data and raises the possibility that there are two cryptic species within H. influenzae. The average sequence diversity between the STs of group I and II is about twice that within each group, suggesting a significant phylogenetic separation. However, as discussed above, there is evidence of considerable recent movement of alleles between these two groups and of more ancient events that distributed the serotype a and b capsular genes between them. In the absence of any evidence for more substantial genetic isolation between the isolates of phylogenetic group I and II there seems no valid reason to assign them as distinct species.
Acknowledgments
This work was supported by grants from the Wellcome Trust (to B.G.S.) and the NIH NIDCD (to R.G.).
We thank the members of the Finnish Otitis Media Study Group at the National Finnish Public Health Institute for the provision of the noncapsulated H. influenzae isolates; Richard Moxon for providing encapsulated isolates; Derek Hood, Paul Langford, and Ming-Shi Li for providing DNA of H. influenzae isolates; and Linda Allen, Jean Bennetch, Richard Berman, Padget Burkey, Geoffrey Cantor, Nancy Caruso, Vicki Gordon, Stephanie Laubach, Perrianne Lurie, Susan Miller, Janey Schear, and Suzanne Yeager for the use of isolates from epidemiological studies of Hib transmission among Pennsylvania communities.
E.M and E.J.F. contributed equally to this work.
REFERENCES
- 1.Anderson, E. C., N. T. Begg, S. C. Crawshaw, R. M. Hargreaves, A. J. Howard, and M. P. Slack. 1995. Epidemiology of invasive Haemophilus influenzae infections in England and Wales in the pre-vaccination era (1990-2). Epidemiol. Infect. 115:89-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2002. Progress toward elimination of Haemophilus influenzae type b invasive disease among infants and children—United States, 1998-2000. Morb. Mortal. Wkly. Rep. 51:234-237. [PubMed] [Google Scholar]
- 3.Breukels, M. A., L. Spanjaard. L.A. Sanders, and G. T. Rijkers. 2001. Immunological characterization of conjugated Haemophilus influenzae type b vaccine failure in infants. Clin. Infect. Dis. 32:1700-1705. [DOI] [PubMed] [Google Scholar]
- 4.Caugant, D. A., L. F. Mocca, C. E. Frasch, L. O. Froholm, W. D. Zollinger, and R. K. Selander. 1987. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J. Bacteriol. 169:2781-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Cody, A. J., D. Field, E. F. Feil, S. Stringer, M. E. Deadman, A. G. Tsolaki, B. Gratz, V. Bouchet, R. Goldstein, D. W. Hood, and E. R. Moxon. Infect. Genet. Evol., in press. [DOI] [PMC free article] [PubMed]
- 5.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing: tracking the global spread of virulent and antibiotic-resistant strains of bacteria. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for the characterization of methicillin-resistant (MRSA) and methicillin-susceptible (MSSA) clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falla, T. J., S. R., Dobson, D. W. Crook, W. A. Kraak, W. W. Nichols, E. C. Anderson, J. Z. Jordens, M. P. Slack, D. Mayon-White, and E. R. Moxon. 1993. Population-based study of non-typable Haemophilus influenzae invasive disease in children and neonates. Lancet 341:851-854. [DOI] [PubMed] [Google Scholar]
- 11.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feil, E. J., E. C. Holmes, D. E. Bessen, M.-S. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population biology of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 14.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 15.Fry, A. M., P. Lurie, M. Gidley, S. Schmink, J. Lingappa, M. Fischer, and N. E. Rosenstein. 2001. Haemophilus influenzae type b disease among Amish children in Pennsylvania: reasons for persistent disease. Pediatrics 108:e60. [DOI] [PubMed]
- 16.Heath, P. T., R. Booy, H. Griffiths, E. Clutterbuck, H. J. Azzopardi, M. P. Slack, J. Fogarty, A. C. Moloney, and E. R. Moxon. 2000. Clinical and immunological risk factors associated with Haemophilus influenzae type b conjugate vaccine failure in childhood. Clin. Infect. Dis. 31:973-980. [DOI] [PubMed] [Google Scholar]
- 17.Heath, P. T., R. Booy, H. J. Azzopardi, M. P. Slack, J. Fogarty, A. C. Moloney, M. E. Ramsay, and E. E. Moxon. 2001. Non-type b Haemophilus influenzae disease: clinical and epidemiologic characteristics in the Haemophilus influenzae type b vaccine era. Pediatr. Infect. Dis. J. 20:300-305. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, E. C., R. Urwin, and M. C. J. Maiden. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 19.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. Van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jafari, H. S., W. G. Adams, K. A. Robinson, B. D. Plikaytis, J. D. Wenger, et al. 1999. Efficacy of Haemophilus influenzae type b conjugate vaccines and persistence of disease in disadvantaged populations. Am. J. Public Health 89:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroll, J. S., and E. R. Moxon. 1990. Capsulation in distantly related strains of Haemophilus influenzae type b: genetic drift and gene transfer at the capsulation locus. J. Bacteriol. 172:1374-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 23.Maiden, M. C. J., J. A. Bygraves, E. J. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musser, J. M., D. M. Granoff, P. E. Pattison, and R. K. Selander. 1985. A population genetic framework for the study of invasive diseases caused by serotype b strains of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 82:5078-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotypable strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musser, J. M., J. S. Kroll, E. R. Moxon, and R. K. Selander, R. K. 1988. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 85:7758-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musser, J. M., J. S. Kroll, E. R. Moxon, and R. K. Selander. 1988. Clonal population structure of encapsulated Haemophilus influenzae. Infect. Immun. 56:1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, H. Dabernat, W. Frederiksen, J. Hamel, G. Hammond, E. A. Hoiby, K. E. Jonsdottir, M. Kabeer, I. Kallings, W. N. Khan, M. Kilian, K. Knowles, H. J. Koornhof, B. Law, K. I. Li, J. Montgomery, P. E. Pattison, J.-C. Piffaretti, A. K. Takala, M. L. Thong, R. A. Wall, J. I. Ward, and R. K. Selander. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 29.Musser, J. M., and R. K. Selander. 1990. Brazilian purpuric fever: evolutionary genetic relationships of the case clone of Haemophilus influenzae biogroup aegyptius to encapsulated strains of Haemophilus influenzae. J. Infect. Dis. 161:130-133. [DOI] [PubMed] [Google Scholar]
- 30.Omikunle, A., S. Takahashi, C. L. Ogilvie, Y. Wang, C. A. Rodriguez, J. W. St Geme, and E. E. Adderson. 2002. Limited genetic diversity of recent invasive isolates of non-serotype b encapsulated Haemophilus influenzae. J. Clin. Microbiol. 40:1264-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peltola, H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 13:302-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porras, O., D. A. Caugant, B. Gray, T. Lagergard, B. R. Levin, and C. Svanborg-Eden. 1986. Difference in structure between type b and nontypable Haemophilus influenzae populations. Infect. Immun. 53:79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 34.Sarangi, J., K. Cartwright, J. M. Stuart, S. Brookes, R. Morris, and M. P. Slack. 2000. Invasive Haemophilus influenzae disease in adults. Epidemiol. Infect. 124:441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 36.Sitnikova, T., A. Rzhetsky, and M. Nei. 1995. Interior-branch and bootstrap tests of phylogenetic trees. Mol. Biol. Evol. 12:319-333. [DOI] [PubMed] [Google Scholar]
- 37.Slack, M. P., H. J. Azzopardi, R. M. Hargreaves, and M. E. Ramsay. 1998. Enhanced surveillance of invasive Haemophilus influenzae disease in England, 1990 to 1996: impact of conjugate vaccines. Pediatr. Infect. Dis. J. 17:S204-207. [DOI] [PubMed] [Google Scholar]
- 38.Smith, H. O., J. F. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538-540. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, K., and M. Nei. 2000. Efficiencies of fast algorithms of phylogenetic inference under the criteria of maximum parsimony, minimum evolution, and maximum likelihood when a large number of sequences are used. Mol. Biol. Evol. 17:1251-1258. [DOI] [PubMed] [Google Scholar]
- 40.Urwin, G., J. A. Krohn, K. Deaver-Robinson, J. D. Wenger, M. M. Farley, et al. 1996. Invasive disease due to Haemophilus influenzae serotype f: clinical and epidemiological characteristics in the H. influenzae serotype b vaccine era. Clin. Infect. Dis. 22:1069-1076. [DOI] [PubMed] [Google Scholar]
- 41.Vitovski, S., K. T. Dunkin, A. J. Howard, and J. R. Sayers. 2002. Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels. JAMA 287:1699-1705. [DOI] [PubMed] [Google Scholar]
- 42.Wenger, J. D. 1998. Epidemiology of Haemophilus influenzae type b disease and impact of Haemophilus influenzae type b conjugate vaccines in the United States and Canada. Pediatr. Infect. Dis. J. 17:S132-S136. [DOI] [PubMed]