Abstract
The hepatitis C virus antibody statuses of only 11 (21.5%) of 51 initially reactive samples from volunteer blood donors could be confirmed by using additional screening and confirmatory assays; 23 (45%) were negative by all subsequent assays. Seventeen samples (33.3%) gave variable results in the different assays. The core and NS5 antigens were most immunogenic. An algorithm for serological screening of volunteer blood donors in blood banks of developing countries is suggested.
The high rate of chronicity and the debilitating cost of treatment for hepatitis C virus (HCV) infection make the use of highly sensitive immunoassays in blood banks imperative in preventing its transmission. Serological assays designed to detect antibody to HCV (HCV-Ab) are associated with a high degree of false positivity in regions with low prevalence and in low-risk populations such as nonremunerated blood donors (7). The low specificity of these assays results in the loss of apparently infected blood units.
Though the phenomenon of the occurrence of false positives during HCV-Ab testing is well known (2), in developing countries like India where only 39% of blood donations are voluntary (5), every effort should be made to prevent false labeling of volunteer blood donors (VBDs). Based on this pilot study, which examined the rate of confirmation of HCV-Ab positivity in a blood bank setting, we propose a simple algorithm for on-site counseling and follow-up for donors in blood banks.
Of 14,128 VBDs tested at the Blood Bank at the Christian Medical College in south India between February 1998 and December 1998, 110 (0.77%) tested positive for HCV-Ab. For the purposes of this study, blood samples from VBDs that tested positive were sent for further evaluation to the clinical virology laboratory. Of the 110 serum samples, 51 with adequate volume were used for further testing. The blood bank currently uses Abbott HCV enzyme immunoassay (EIA) 3.0 (Abbott Laboratories, Abbott Park, Ill.). This is a third-generation assay for the qualitative detection of antibodies to the core, NS3, NS4, and NS5 antigens of HCV. Currently, in this blood bank, all units are tested singly and HCV-Ab-positive units are summarily rejected with no further repeat testing. For this study, such HCV-Ab-positive samples were sent to the clinical virology laboratory for further testing. In the laboratory, serum samples were aliquoted and stored at 4°C for testing in a second EIA, UBI HCV EIA 3.0 (United Biomedicals, Inc.), and a microparticle EIA (MEIA), Axsym HCV version 3.0 (Abbott Laboratories). Additional aliquots were stored at −20°C for immunoblot or line immunoassay (LIA) testing. As samples collected from the blood bank were not appropriate for RNA testing, reverse transcriptase PCR (RT-PCR) testing could not be performed. One of the two kinds of assays, LIA (INNO-LIA HCV Ab III; Innogenetics, Zwijndrecht, Belgium) or immunoblot (HCV BLOT 3.0; Genelabs Diagnostics Pte. Ltd., Singapore), was used as the confirmatory antibody test.
All UBI EIAs, Axsym MEIAs, immunoblotting, and LIAs were performed and interpreted strictly according to the manufacturers' instructions. HCV-Ab reactivity in the Axsym MEIA is expressed as a ratio of the sample absorbance value to the calculated cutoff value for each sample. For purposes of comparison, a similar ratio, which we call the index of reactivity (IR), was computed for the UBI EIA: IR = sample absorbance value/cutoff absorbance value.
The cutoff absorbance value for the UBI EIA was calculated by using a formula supplied by the kit's manufacturer. For UBI EIA, an IR value was calculated and when this value was ≥2, we took it to indicate a high positive. An IR value of ≥10 was taken to indicate the same in an Axsym MEIA.
Of the 51 positive samples from the blood bank, 11 (21.5%) were positive both in UBI EIA or Axsym MEIA and in the confirmatory immunoblot test or LIA. All 11 samples could be classified as high positives based on their IR values. Twenty-three samples (45%) tested negative both in the UBI EIA or Axsym MEIA and in the immunoblot test or LIA. An additional five samples were negative by immunoblotting or LIA but positive in UBI EIA (n = 1) or Axsym MEIA (n = 4). Among these five samples, the IR was >2 for the single UBI EIA positive sample but the IR values were always <10 for the four samples positive by Axsym MEIA. Twelve samples (23.5%) gave indeterminate results with immunoblotting or LIA. Nine of these 12 indeterminate samples (75%) were negative by both UBI EIA and Axsym MEIA (Table 1). Among the three remaining samples that tested positive by either UBI EIA (n = 2) or Axsym MEIA (n = 1), the IR values were >2 for both of the UBI EIA positive samples but <10 for the Axsym MEIA positive sample.
TABLE 1.
Comparison of the performance of UBI EIA and Axsym MEIA with that of immunoblotting and LIA for 51 volunteer blood donor samples positive for HCV-Ab by Abbott HCV EIA 3.0
| Immunoblot or LIA reactivity (no. of samples) | No. of samples reactive in:
|
|
|---|---|---|
| UBI HCV EIA 3.0 | Axsym HCV MEIA | |
| Positive (11) | 11 | 11 |
| Indeterminate (12) | 2 | 1 |
| Negative (28) | 1 | 4a |
The IR was <10 for all four samples.
HCV is poised to become a major health problem, with over 1% of the world's population having been infected (7). Given the high chronicity rate, with ensuing complications, associated with HCV and the expensive treatment modalities, the implementation of sensitive screening methods for HCV in blood banks has become an urgent need. However, the specificity of HCV-Ab assays is an equally important concern in blood banks.
Forty-five percent of the positive donor samples from the blood bank tested negative both in UBI EIA or Axsym MEIA and in the confirmatory immunoblotting or LIA. This could possibly be explained by the multiplicity of intercurrent infections seen in tropical countries that lead to hyperglobulinemia, contributing to false positivity in certain HCV-Ab assays, as has been suggested previously (8). The majority (75%) of samples showing indeterminate results with LIA or immunoblotting were negative in both the UBI EIA and the Axsym MEIA. Since no “gold standard” exists for HCV-Ab testing, the issue of infectivity of these samples could have been resolved only with longitudinal follow-up of these donors by using additional tests such as HCV RT-PCR and liver enzyme estimations.
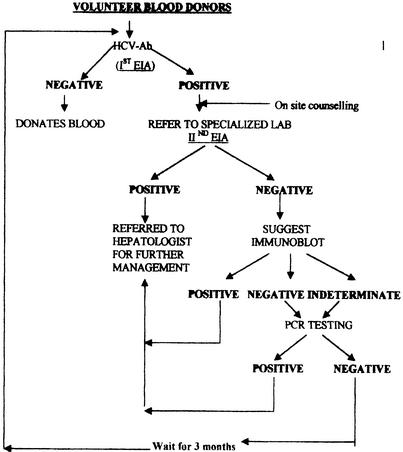
Based on these findings, we suggest the following algorithm for HCV-Ab screening in blood banks (Fig. 1). If a VBD tests positive in a screening assay, he or she should be appropriately counseled in the blood bank about (i) the high rates of false positivity seen in such situations and (ii) the importance of an alternate screening and confirmatory testing in a referral or specialized laboratory. In the event of a donor testing positive in the second EIA, he or she should be directed to a hepatologist for further management, including supplementary HCV-Ab testing and HCV RNA testing.
FIG. 1.
Recommended algorithm for screening of VBDs in blood banks.
If the donor tests negative in the second EIA, he or she should be offered immunoblot testing. Negative or indeterminate results obtained by immunoblotting would necessitate RNA testing. A decision on the exclusion of the VBD or the return of the VBD to the donor pool can be made based on the RNA test results. Donors testing positive in the first EIA, with negative or indeterminate immunoblot results and negative RNA results, can donate only after going through the entire screening process again. The costs for such repeat testing should be met by the blood banks themselves based on the requirement for blood units. This algorithm will help in lowering the HCV burden in the general population, as well as increasing the safety of blood units and encouraging the return of “safe donors.”
It is known, however, that an infected individual may test negative for HCV RNA because of either low viral load (4) or suboptimal amplification in the RT-PCR due to PCR inhibitors (1). In such a scenario, it is recommended that the donor wait for a minimum period of 3 months before attempting to donate again.
For samples showing discrepancy between the results from immunoblotting or LIA and those from UBI EIA or Axsym MEIA, the IR values were high (>2) for the UBI EIA but always <10 for the Axsym MEIA. This is in contrast to a previous report (3) which suggested that a high positive in an EIA would very likely be confirmed in the recombinant immunoblot assay.
Among samples that tested positive both in the UBI EIA or Axsym-MEIA and in the immunoblot test or LIA (n = 11), antibody reactivities to the core antigen were found to be the highest, as has been reported previously (6, 9). NS5 antigen lines were the next highest in intensity. The strong immunogenicity of the NS5 antigen reinforces the importance of using third-generation assays for screening.
In conclusion, this study reveals a high degree of false positivity associated with screening for HCV-Ab among VBDs in an Indian blood bank. The number of samples that tested positive in confirmatory immunoblot tests or LIAs was small. To minimize wastage of otherwise safe units of blood, especially those belonging to rare blood groups, and anxiety to VBDs, the importance of a second EIA and confirmatory testing by immunoblotting or LIA and/or RT-PCR is reiterated. Additionally, counseling of all VBDs who test positive for HCV-Ab at blood banks must be advocated.
REFERENCES
- 1.Al-Soud, W. A., J. L. Jönsson, and P. Rådström. 2000. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 38:345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver. 1999. EASL International Consensus Conference on hepatitis C, Paris, 26-28 February 1999: consensus statement. J. Hepatol. 30:956-961. [PubMed] [Google Scholar]
- 3.Gretch, D., W. Lee, and L. Corey. 1992. Use of aminotransferase, hepatitis C antibody, and hepatitis C polymerase chain reaction RNA assays to establish the diagnosis of hepatitis C virus infection in a diagnostic virology laboratory. J. Clin. Microbiol. 30:2145-2149.1323578 [Google Scholar]
- 4.Haydon, G. H., L. M. Jarvis, C. S. Blair, P. Simmonds, D. J. Harrison, K. J. Simpson, and P. C. Hayes. 1998. Clinical significance of intrahepatic hepatitis C virus levels in patients with chronic HCV infection. Gut 42:570-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapoor, D., R. Saxena, B. Sood, and S. K. Sarin. 2000. Blood transfusion practices in India: results of a national survey. Indian J. Gastroenterol. 19:64-67. [PubMed] [Google Scholar]
- 6.Leroux-Roels, G., C. A. Esquivel, R. DeLeys, L. Stuyver, A. Elewaut, J. Philippe, I. Desombere, J. Paradijs, and G. Maertens. 1996. Lymphoproliferative responses to hepatitis C virus core, E1, E2, and NS3 in patients with chronic hepatitis C infection treated with interferon alfa. Hepatology 23:8-16. [DOI] [PubMed] [Google Scholar]
- 7.Quer, J., and J. I. Esteban. 1998. Hepatitis C virus epidemiology, p 271-283. In A. J. Zuckerman and Howard C. Thomas (ed.), Viral hepatitis, 2nd ed. Churchill Livingstone, London, United Kingdom.
- 8.Trepo, C., F. Zoulin, C. Alonso, M.-A. Petit, C. Pichoud, and L. Vitvitski. 1993. Diagnostic markers of viral hepatitis B and C. Gut 34(Suppl.):S20-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson, H. G., C. A. Ludlam, S. Rebus, L. Q. Zhang, J. F. Peutherer, and P. Simmonds. 1992. Use of several second generation serological assays to determine the true prevalence of hepatitis C virus infection in haemophiliacs treated with non-virus inactivated factor VIII and IX concentrates. Br. J. Haematol. 80:514-518. [DOI] [PubMed] [Google Scholar]