Abstract
Nocardia veterana is a newly described species named after the veteran's hospital where it was first isolated. This initial type strain was not thought to be clinically significant. We describe three cases of pulmonary disease attributable to N. veterana: two cases in patients presenting with multiple pulmonary nodules in a setting of immunocompromise and one case of exacerbation of chronic pulmonary disease. The isolates were susceptible to ampicillin, imipenem, gentamicin, amikacin, and trimethoprim-sulfamethoxazole and had reduced susceptibilities to ceftriaxone, cefotaxime, minocycline, and ciprofloxacin. The MICs of amoxicillin-clavulanate were higher than that of ampicillin alone, and the bacteria produced a β-lactamase detectable only after induction with clavulanic acid. Phenotypically, the isolates could not be characterized beyond the Nocardia genus level. All three isolates were definitively identified as N. veterana by PCR and sequencing of the 16S rRNA gene. On the basis of their susceptibility and restriction enzyme analysis profiles, our findings indicate that they could potentially be misidentified as N. nova. These cases illustrate the pathogenic potential of this newly described species and emphasize the importance of accurate identification of Nocardia isolates to the species level by integrated use of phenotypic and genotypic methods.
Nocardia spp. are gram-positive, nonmotile aerobic actinomycetes that are ubiquitous environmental saprophytes. They cause a variety of suppurative and granulomatous infections in humans, ranging from localized cutaneous mycetomas to disseminated systemic disease (2, 19). While Nocardia spp. can cause disease in an immunocompetent host (16), they are often considered opportunistic pathogens, causing disease in settings of compromised immunity, such as organ transplantation or lymphoreticular neoplasia (2, 18).
Identification of Nocardia isolates to the species level is important to define the spectrum of diseases caused by each species and to predict antimicrobial susceptibility (16). While the number of species that make up the genus Nocardia is rapidly expanding, the majority (∼80%) of pulmonary and disseminated infections are caused by the members of the Nocardia asteroides complex (2, 18). Less frequently, other Nocardia species, like N. braziliensis, N. transvalensis, and N. otitidiscavarium, have also been implicated (2). Recently, Hamid et al. reported the isolation of a new pathogenic species, N. africana, from sputum samples of patients with chronic pulmonary infections in Africa (9). N. veterana is a newly described species whose pathogenicity is poorly appreciated (8). Kano et al. reported the first isolation of N. veterana from a patient with a case of mycetoma (11). Here we report the first case of severe pulmonary infection caused by this newly described Nocardia species in an immunocompromised host. We have subsequently identified N. veterana by molecular methods in two other cases of pulmonary disease.
Case 1.
A 47-year-old male with end stage liver disease secondary to hepatitis C underwent an orthotopic liver transplant. Postoperative immunosuppression included cyclosporine, azathioprine, and prednisone. Because of a history of possible allergy to trimethoprim-sulfamethoxazole, dapsone was used for Pneumocystis carinii pneumonia prophylaxis.
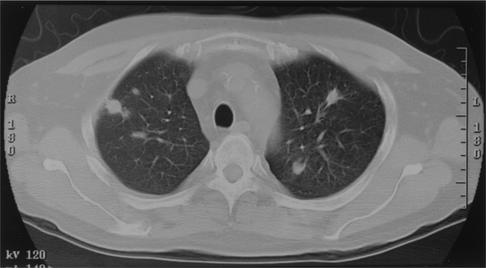
Four months after receiving his transplant, he presented with a 4-day history of increasing temperatures associated with chills and diaphoresis, nasal discharge, and a nonproductive cough. Leukocytosis was absent. A chest X-ray demonstrated a small right upper lobe and a left upper lobe nodule. A computed tomography (CT) scan of his chest demonstrated numerous nodules in his upper lobes bilaterally, which were distributed both peripherally and centrally (Fig. 1). He underwent a CT-guided needle biopsy of a left lung nodule. Histopathological examination of the biopsy tissue showed evidence of cytomegalovirus (CMV) infection with enlarged lung parenchymal cells containing cytoplasmic and nuclear inclusions. Infection with CMV was further confirmed by immunohistochemical staining of the tissue and ultimately by viral culture. There was no histopathological evidence of fungal, bacterial, or mycobacterial infection on special stained sections. Bacterial and fungal cultures of this needle biopsy tissue were also negative, and intravenous (i.v.) ganciclovir therapy was begun.
FIG. 1.
CT scan showing multiple pulmonary nodules bilaterally, distributed both peripherally and centrally.
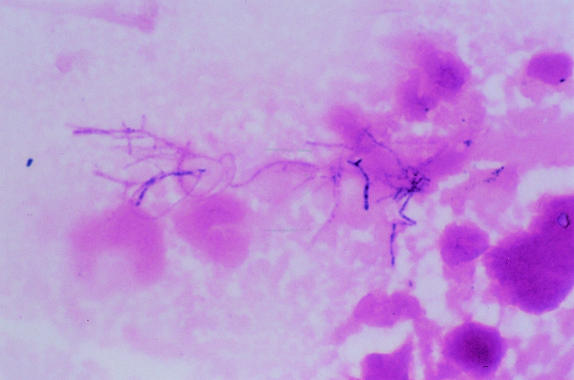
Given the progression of his pulmonary infiltrates despite i.v. ganciclovir and because CMV is often found as a copathogen (22), he underwent a thoracoscopy and a wedge biopsy of his left upper lobe nodule. Histopathological examination of the tissue obtained showed evidence of organizing and active pneumonia with filamentous variably gram-positive bacterial organisms, which also stained positive with Gomori ammoniacal silver stain. No CMV inclusions were seen. Gram staining of the open lung biopsy tissue revealed branching, beady-staining, gram-positive rods amid a moderate inflammatory response (Fig. 2). The cultures produced a pure growth of nocardiae. The tissue was negative for staining for P. carinii. Specific cultures for mycobacteria, fungi, legionellae, and mycoplasmas were negative. A CT scan of his head was normal. Blood cultures remained negative after 6 weeks of incubation. Despite his possible history of allergy, he was treated with i.v. trimethoprim-sulfamethoxazole, which he tolerated well. He was discharged on home infusion of i.v. ganciclovir and trimethoprim-sulfamethoxazole. Six months after initiation of therapy, follow-up imaging demonstrated resolution of the pulmonary nodules.
FIG. 2.
Gram staining of open-lung biopsy tissue showing branching, beady-staining, gram-positive rods amid a moderate inflammatory response.
Case 2.
A 43-year-old female with long-standing systemic lupus erythematosus (SLE) presented with fevers, chills, and a dry cough of several days duration. She had received multiple courses of cyclophosphamide as treatment for her SLE and was chronically receiving prednisone (10 mg/day). A chest radiograph revealed bilateral infiltrates, and chest CT revealed multiple pulmonary nodules, some with evidence of cavitation. Bronchoscopy with bronchoalveolar lavage (BAL) was performed. Culture of the BAL fluid produced a pure growth of Nocardia sp. No other potential pathogens were isolated from cultures of blood, urine, or BAL fluid. She was treated with high-dose trimethoprim-sulfamethoxazole (initially i.v.) with a good clinical response and resolution of the nodules. Her therapy was subsequently changed to oral minocycline, and she was treated for 6 months. No evidence of recurrence was noted during follow-up for 1 year.
Case 3.
A 67-year-old female with a history of recurrent pneumonia was diagnosed with bronchiectasis in the middle and lower lobes bilaterally, which was confirmed by a CT scan. She presented with an exacerbation of her symptoms with a severe cough productive of purulent sputum but no fever, chills, or sweats. Symptoms persisted despite routine antibiotic therapy, which consisted of 1-week courses of azithromycin, doxycycline, and levofloxacin in the preceding 6-week period. Her sputum culture grew Nocardia sp. along with Paecilomyces sp. and a black mold identified as Exophiala sp. Given her clinical presentation and poor response to 7-day courses of routine antibiotic therapy, the Nocardia sp. was thought to be clinically significant. She was initially treated with minocycline for 2 weeks and later with amoxicillin for 2 weeks, which she tolerated poorly. Her therapy was changed to azithromycin, which she tolerated well with symptomatic improvement.
Microbiological identification.
The three isolates were recovered over a period of 16 months, from December 2000 to March 2002. They had identical biochemical and morphological characteristics. They grew well on blood agar after 3 to 5 days of incubation at 35°C in a CO2-enriched atmosphere. Equivalent growth was noted at 35, 42, and 45°C. They formed dry, powdery colonies that were very adherent to the agar, with white to light coral pigmentation and an earthy odor. The isolates were resistant to lysozyme. Typical Gram stain morphology for Nocardia spp., with branching gram-positive rods with beaded staining, was noted, and the isolates were modified Ziehl-Neelsen acid-fast stain positive. Typical tap water agar morphology for Nocardia spp., with very fine dichotomously branched filaments of substrate and surface hyphae, along with the presence of aerial hyphae, was observed (14). All three isolates were positive for catalase and hydrolyzed esculin and urea.
There was no growth on MacConkey agar devoid of crystal violet. They were also negative for decomposition of tyrosine, xanthine, hypoxanthine, and casein, and the 14-day arylsulfatase test result was negative. Growth without acid production was present in heart infusion broth with either adonitol, glucose, inositol, rhamnose, trehalose, arabinose, mannitol, or sorbitol. Similarly, the isolates grew without acid production in peptone water containing either dextrose, lactose, maltose, mannitol, sorbitol, sucrose, or lactose. Hydrolysis of acetamide, reduction of nitrate, and utilization of citrate as the sole carbon source were negative. On the basis of the results of the biochemical tests, along with their morphology and staining characteristics, the isolates were identified as belonging to the genus Nocardia but species identification was not possible.
Antimicrobial susceptibility testing.
MICs were obtained for the three isolates by the broth microdilution method and the E test (AB Biodisk, Solna, Sweden) performed in accordance with published methods and interpreted on the basis of the tentative NCCLS guidelines (Table 1) (1, 17). With the E test, the MICs of all of the drugs tested correlated, within 1 dilution difference, with those obtained by the broth microdilution method, except for ceftriaxone, cefotaxime, trimethoprim-sulfamethoxazole, and ciprofloxacin. This led to minor interpretative errors only for ceftriaxone and cefotaxime when the E test was used. All three isolates had a very characteristic growth pattern around the cefotaxime and ceftriaxone E-test strips. They exhibited a distinct ellipse but had inner colonies extending right up to the highest concentration on the E-test strip, which was interpreted as a MIC of >32 μg/ml. This was unlike the broth microdilution method, where the endpoint was well defined. The MIC of amoxicillin-clavulanic acid was higher than that of ampicillin. In all three isolates, β-lactamase production was detectable by the nitrocefin disk method only after induction with clavulanic acid.
TABLE 1.
Susceptibilities of the four N. veterana isolates to 10 antimicrobials
| Antimicrobial | MIC (μg/ml)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Broth microdilution
|
E test
|
|||||||||
| Case 1 | Case 2 | Case 3 | Type strain DSM 44445T | Range | Case 1 | Case 2 | Case 3 | Type strain DSM 44445T | Range | |
| Ampicillin | 2 | 2 | 2 | 2 | 2 | 2 | 0.5 | 1 | 2 | 0.5-2 |
| Amoxicillin-clavulanic acida | 32 | 16 | 16 | 32 | 16-32 | 16 | 16 | 16 | 8 | 8-16 |
| Cefotaxime | 16 | 4 | 4 | 16 | 4-16 | >32 | >32 | >32 | >32 | >32 |
| Ceftriaxone | 16 | 4 | 8 | 16 | 8-16 | >32 | >32 | >32 | >32 | >32 |
| Imipenem | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | 0.032 | 0.064 | 0.064 | 0.016 | 0.016-0.064 |
| Ciprofloxacin | 8 | 4 | 8 | 8 | 8 | >32 | >32 | >32 | >32 | >32 |
| Gentamicin | 4 | 8 | 4 | 8 | 4-8 | 2 | 8 | 2 | 4 | 2-8 |
| Amikacin | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | 0.25 | 0.25 | 0.125 | 0.25 | 0.125-0.25 |
| Minocycline | 4 | 4 | 2 | 4 | 2-4 | 4 | 4 | 2 | 4 | 2-4 |
| Trimethoprim-sulfamethoxazole | 1/19 | 0.5/9.5 | 1/19 | 0.5/9.5 | 0.5/9.5-1/19 | 0.12/2.3 | 0.25/4.75 | 0.12/2.3 | 0.064/1.1 | 0.064/1.1-0.25/4.75 |
All four isolates were positive for beta-lactamase, which was only detectable after induction with clavulanic acid, by the nitrocefin disk method.
Molecular identification.
DNA was extracted with a QIAamp DNA Mini Kit. The entire 1,510-bp 16S rRNA gene was PCR amplified with primers P1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and P10 (5′-AGGA GGTGATCCACCGCA-3′). The 100-μl DNA amplification reaction mixture contained each primer at a concentration of 1 μM, 1× PCR buffer, 3 mM MgCl2, each deoxynucleotide at a concentration of 0.08 mM, and 1 U of AmpliTaq Gold (Perkin-Elmer, Norwalk, Conn.). The DNA Thermal Cycler (model 9700; PE Applied Biosystems) was set to the following parameters: 95°C for 10 min, followed by 30 cycles of 95°C for 30 s, 68°C for 30 s, and 72°C for 45 s, followed by one extension at 72°C for 10 min. The product was visualized by 1% agarose gel electrophoresis. The PCR products were filtered with a Microcon column (YM-100; Amicon, Inc., Beverly, Mass.) and resuspended in 50 μl of sterile pharmacy water.
To sequence the whole gene, the following four sets of primers were used, P1 and P3 (5′-TTACCGCGGCTGCTGGCA-3′), P2 (5′-TGCCAGCAGC CGCGGTAA-3′) and P5 (5′-GGACTACCAGGGTATCTAAT-3′), P4 (5′-ATTAGATACCCTGGTAGTCC-3′) and P7 (5′-ACGTCATCCCCACCTTCCTC-3′), and P6 (5′-GAGAAGGTGGGGATGACGT-3′) and P10. Cycle sequencing was performed with the Ready-Reaction mixture (ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit; PE Applied Biosystems) in accordance with the manufacturer's instructions on a model 9700 PE thermocycler by using preprogrammed BigDye cycling parameters on the instrument. Each sequencing product was concentrated to dryness with a SpeedVac after removal of excess DyeDeoxyR terminators with CENTRI-SEP columns (Princeton Separations, Adelphia, N.J.). The sequencing products were purified with Centri-Sep columns, and gel analysis of the sequences was performed on an ABI BioPRISM377. Sequences derived from both DNA strands were assembled and edited with the Sequencher program (version 4.1). The sequence was compared with the reported sequences in the GenBank database (National Center for Biotechnology Information). The 16S rRNA sequence of the isolate from case 1 was identical at 1,339 of 1,341 positions to the sequence of the N. veterana 16S rRNA gene (accession no. AF278572): insertion of a C at position 1329 in our isolate and a C in our isolate versus an N in the type strain at position 1335 were the only differences.
Comparison with N. veterana type strain DSM 44445T (8).
Because of the two-nucleotide difference between our patient isolate and the type strain, we obtained the type strain and compared the two by phenotypic, susceptibility testing, and molecular methods. DSM 44445T had the same phenotypic pattern as our patient isolates described above. Within a twofold dilution difference, there was 100% agreement in the MICs for the isolates by both the broth microdilution and E-test methods (Table 1). Sequencing of the 16S rRNA gene revealed 100% nucleotide identity at 1,510 positions between our isolates and DSM 44445T.
For the isolates from cases 2 and 3, the first 500 bases of the 16S rRNA gene were sequenced. The first 500 base pairs of the sequences of the isolates from cases 1 and 3 and the type strain were identical. The sequence of the isolate from case 2 had one base pair mismatch from the other three strains. Restriction endonuclease analysis of the PCR-amplified hsp65 gene of these four isolates with MspI, HinfI, and BsaHI showed it to be indistinguishable from those of N. nova and N. veterana (data not shown; 20, 21).
Discussion.
N. veterana is a new species named after the veteran's hospital (Heidelberg, Victoria, Australia) where the organism was first isolated. On the basis of morphological, physiological, chemotaxonomic, and molecular methods, the strain was described as a unique species of the genus Nocardia (8). This initial type strain was isolated from a bronchoscopic lavage fluid of a 78-year-old patient with a history of tuberculous pleurisy. Further clinical details were not provided, except that this isolate was thought not to be clinically significant. Consistent with this idea, Nocardia sp., when isolated from respiratory specimens, may occasionally represent either transient colonization or a spontaneously resolving subclinical infection (7, 16, 19). Respiratory colonization is said to occur in individuals with underlying pulmonary disorders that do not progress to invasive infections in the absence of steroid therapy (7, 19).
Several lines of evidence that N. veterana was the cause of the invasive pulmonary infection in our first case include (i) febrile illness, (ii) clinical progression without specific treatment, (iii) radiological evidence of multiple pulmonary nodules, (iv) positive tissue Gram staining and pure growth in culture, and (v) histopathological evidence of inflammation. In case 2, infection occurred in a setting of chronic immunosuppression. It was associated with multiple pulmonary nodules with cavitation, a typical presentation for Nocardia infection (7, 19). N. veterana was the sole pathogen isolated, and treatment was associated with a good clinical response. In case 3, N. veterana was isolated from a sputum culture from an immunocompetent patient with underlying chronic pulmonary disease who was not on steroid therapy. The organism was not isolated in pure culture and could be thought to represent either transient colonization or a subclinical infection (7). Because of persistent symptoms despite routine week-long courses of antimicrobial therapy (which included a quinolone the isolate was resistant to) and given the severity of her underlying lung disease, the clinical decision was made to administer specific treatment against Nocardia sp.
In our first patient, pulmonary nocardiosis with coexistent CMV infection developed 4 months after a liver transplant without any evidence of dissemination. In a review of nocardiosis in liver transplant patients, coexistent bacterial or viral disease was present in four patients, with two cases having coexistent CMV infection (6). The presence of coexistent infections was associated with 75% mortality. This emphasizes the importance of being aware of uncommon infections like nocardiosis, which can often be overlooked when other pathogens have already been isolated. Patel et al. also stressed that the most important approach to infection in a solid-organ transplant recipient is prevention; failing this, prompt diagnosis and aggressive therapy are essential (18). Trimethoprim-sulfamethoxazole administration for prophylaxis of P. carinii pneumonia is said to play a role in preventing nocardiosis in transplant recipients (22). However, this regimen was not used for our patient because of a history of possible allergy.
Phenotypically, the isolates exhibited all of the characteristic features of the genus Nocardia. However, species identification based on the characteristics of the other well-described Nocardia species was not possible (3, 10). Phenotypic characteristics are often used in conjunction with antimicrobial susceptibility patterns to help identify some Nocardia isolates (23, 24). On the basis of the NCCLS guidelines, the isolates were susceptible to ampicillin, trimethoprim-sulfamethoxazole, gentamicin, imipenem, and amikacin (17). Susceptibility to ceftriaxone, cefotaxime, minocycline, and ciprofloxacin was reduced. Except for the elevated MICs of the two expanded-spectrum cephalosporins obtained by the E test, the rest of the MICs of the various antibiotics were within the ranges reported for N. nova isolates (23). Also similar to those for N. nova, the MICs of amoxicillin-clavulanate for the four isolates were higher than that of ampicillin alone (1, 23). Wallace et al. reported that this unusual resistance pattern of N. nova isolates was due to an inducible membrane-bound β-lactamase with penicillinase activity and that the enzyme was induced by clavulanic acid and not ampicillin (23). While β-lactamase was detectable only after induction with clavulanic acid in all 4 of our isolates, in a study reported by Ambaye et al., β-lactamase was detectable before induction in 9 of 10 N. asteroides sensu stricto and 5 of 7 N. nova isolates (1). Phenotypically, these nocardiae could be differentiated from N. nova by their very adherent growth, equivalent growth at 35 and 45°C, lack of production of acid from glucose, and 14-day arylsulfatase test negativity (3, 23).
Because traditional phenotypic methods of identification of Nocardia species are not only time consuming and laborious but can also be challenging and inconclusive, various molecular methods including gene probes, gene sequencing, and restriction endonuclease (RE) analysis after PCR have been developed (5, 15). Restriction fragment length polymorphism of an amplified 439-bp segment of the 65-kDA heat shock protein has been shown to be a rapid and sensitive method for identification of clinically significant aerobic actinomycetes (20, 21). However, we found, by RE analysis of the PCR-amplified hsp65 gene with MspI, HinfI, and BsaHI, that N. nova and N. veterana were indistinguishable. Chun and Goodfellow performed phylogenetic analyses of 16S rRNA sequences derived from nine species of the genus Nocardia to establish the relationships among the species (4). They found 16S rRNA gene sequencing to be a powerful tool for defining the different Nocardia species. In the phylogenetic tree derived from aligned 16S rRNA sequences, Gurtler et al. found that N. veterana is most closely related to N. vaccinnii, with a similarity of 98.6% (8). The next most closely related strain is N. nova, with a similarity of 98.1%. Kano et al. identified N. veterana from a case of mycetoma by PCR and sequencing of the 16S rRNA gene (11). While our patient isolates were not clearly distinguishable from N. nova on the basis of their susceptibility pattern or RE analysis, they were clearly different on the basis of their phenotypic characteristics and production of inducible β-lactamase, and their unique identity as N. veterana was confirmed by 16S rRNA PCR and sequencing.
Our three cases illustrate the spectrum of pulmonary disease that is attributable to N. veterana, ranging from severe progressive pulmonary disease leading to multiple pulmonary nodules with or without cavitation in a setting of immunocompromise secondary to either transplantation or treatment for SLE to isolation from the sputum of an immunocompetent patient presenting with exacerbation of underlying chronic pulmonary disease. This is the first report on the pathogenic potential of N. veterana in pulmonary disease. Our findings also illustrate the potential for misidentification of this species as N. nova. We therefore conclude that accurate identification of Nocardia isolates by integrated use of phenotypic and genotypic methods will help determine the virulence, epidemiology, and pathogenic potential of this and other newly identified species (12, 13).
Nucleotide sequence accession number.
The additional 161-bp sequence identified in this report has been submitted to the GenBank database and assigned accession no. AY171039.
REFERENCES
- 1.Ambaye, A., P. C. Kohner, P. C. Wollan, K. L. Roberts, G. D. Roberts, and F. R. Cockerill III. 1997. Comparison of agar dilution, broth microdilution, disk diffusion, E-test, and BACTEC radiometric methods for antimicrobial susceptibility testing of clinical isolates of Nocardia asteroides complex. J. Clin. Microbiol. 35:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaman, B. L., and L. Beaman. 1994. Nocardia species: host-parasite relationships. Clin. Microbiol. Rev. 7:213-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, J. M., M. M. McNeil, and E. P. Desmond. 1999. Nocardia, Rhodococcus, Gordona, Actinomadura, Streptomyces, and other actinomycetes of medical importance, p. 370-398. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 4.Chun, J., and M. Goodfellow. 1995. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 45:240-245. [DOI] [PubMed] [Google Scholar]
- 5.Conville, P. S., S. H. Fischer, C. P. Cartwright, and F. G. Witebsky. 2000. Identification of Nocardia species by restriction endonuclease analysis of an amplified portion of the 16S rRNA gene. J. Clin. Microbiol. 38:158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forbes, G. M., F. A. H. Harvey, J. N. Philpott-Howard, J. G. O'Grady, R. D. Jensen, M. Sahathevan, M. W. Casewell, and R. Williams. 1990. Nocardiosis in liver transplantation: variation in presentation, diagnosis and therapy. J. Infect. 20:11-19. [DOI] [PubMed] [Google Scholar]
- 7.Georghiou, P. R., and Z. M. Blacklock. 1992. Infection with Nocardia species in Queensland: a review of 102 clinical isolates. Med. J. Aust. 156:692-697. [DOI] [PubMed] [Google Scholar]
- 8.Gurtler, V., R. Smith, B. C. Mayall, G. Potter-Reinmann, E. Stackebrandt, and R. M. Kroppenstadt. 2001. Nocardia veterana sp. nov., isolated from human bronchial lavage. Int. J. Syst. E vol. Microbiol. 51:933-936. [DOI] [PubMed] [Google Scholar]
- 9.Hamid, M. E., L. Maldonado, G. S. Sharaf Eldin, M. F. Mohamed, N. S. Saeed, and M. Goodfellow. 2001. Nocardia africana sp. nov., a new pathogen isolated from patients with pulmonary infections. J. Clin. Microbiol. 39: 625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed., p. 625-650. The Williams & Wilkins Co., Baltimore, Md.
- 11.Kano, R., Y. Hattori, N. Murakami, N. Mine, M. Kashima, R. M. Kroppenstedt, M. Mizoguchi, and A. Hasegawa. 2002. The first isolation of Nocardia veterana from a human mycetoma. Microbiol. Immunol. 46:409-412. [DOI] [PubMed] [Google Scholar]
- 12.Kattar, M. M., B. T. Cookson, L. C. Carlson, S. K. Stiglich, M. A. Schwartz, T. T. Nguyen, R. Daza, C. K. Wallis, S. L. Yarfitz, and M. B. Coyle. 2001. Tsukamurella strandjordae sp. nov., a proposed new species causing sepsis. J. Clin. Microbiol. 39:1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kattar, M. M., J. F. Chavez, A. P. Limaye, S. L. Rassoulian-Barrett, S. L. Yarfitz, L. C. Carlson, Y. Houze, S. Swanzy, B. L. Wood, and B. T. Cookson. 2000. Application of 16S rRNA gene sequencing to identify Bordetella hinzii as a causative agent of fatal septicemia. J. Clin. Microbiol. 38:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Land, G., M. R. McGinnis, J. Staneck, and A. Gatson. 1991. Aerobic pathogenic Actinomycetales, p. 340-360. In A. Balows, W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 15.Laurent, F. J., F. Provost, and P. Boiron. 1999. Rapid identification of clinically relevant Nocardia species to genus level by 16S rRNA gene PCR. J. Clin. Microbiol. 37:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNeil, M. M., and J. M. Brown. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin. Microbiol. Rev. 7:357-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NCCLS. 2000. Susceptibility testing of mycobacteria, nocardia and other aerobic actinomycetes. Tentative standard, second edition. NCCLS document M24-T2. NCCLS, Wayne, Pa. [PubMed]
- 18.Patel, R., and C. V. Paya. 1997. Infections in solid organ transplant recipients. Clin. Microbiol. Rev. 10:86-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorrell, T. C., J. R. Iredell, and D. H. Mitchell. 2000. Nocardia species, p. 2637-2645. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.) Mandell, Douglas and Bennett's principles and practice of infectious diseases. Churchill Livingstone, Philadelphia, Pa.
- 20.Steingrube, V. A., B. A. Brown, J. L. Gibson, R. W. Wilson, J. Brown, Z. Blacklock, K. Jost, S. Locke, R. F. Ulrich, and R. J. Wallace, Jr. 1995. DNA amplification and restriction endonuclease analysis for differentiation of 12 species and taxa of Nocardia, including recognition of four new taxa within the Nocardia asteroides complex. J. Clin. Microbiol. 33:3096-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steingrube, V. A., R. W. Wilson, B. A. Brown, K. C. Jost, Jr., Z. Blacklock, J. L. Gibson, and R. J. Wallace, Jr. 1997. Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella isolates, by DNA amplification and restriction endonuclease analysis. J. Clin. Microbiol. 35:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Burik, J. A., R. C. Hackman, S. Q. Nadeem, J. W. Heimenz, M. H. White, M. E. Flowers, and R. A. Bowden. 1997. Nocardiosis after bone marrow transplantation: a retrospective study. Clin. Infect. Dis. 24:1154-1160. [DOI] [PubMed] [Google Scholar]
- 23.Wallace, R. J., B. A. Brown, M. Tsukamura, J. M. Brown, and G. O. Onyi. 1991. Clinical and laboratory features of Nocardia nova. J. Clin. Microbiol. 29:2407-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace, R. J., Jr., L. C. Steele, G. Sumter, and J. M. Smith. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob. Agents Chemother. 32:1776-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]