Abstract
The classical Mycobacterium tuberculosis complex (MtbC) subspecies include Mycobacterium tuberculosis, Mycobacterium africanum (subtypes I and II), Mycobacterium bovis (along with the attenuated M. bovis bacillus Calmette-Guérin [BCG]), and Mycobacterium microti; increasingly recognized MtbC groupings include Mycobacterium bovis subsp. caprae and “Mycobacterium tuberculosis subsp. canettii.” Previous investigations have documented each MtbC subspecies as a source of animal and/or human tuberculosis. However, study of these organisms is hindered by the lack of a single protocol that quickly and easily differentiates all of the MtbC groupings. Towards this end we have developed a rapid, simple, and reliable PCR-based MtbC typing method that makes use of MtbC chromosomal region-of-difference deletion loci. Here, seven primer pairs (which amplify within the loci 16S rRNA, Rv0577, IS1561′, Rv1510, Rv1970, Rv3877/8, and Rv3120) were run in separate but simultaneous reactions. Each primer pair either specifically amplified a DNA fragment of a unique size or failed, depending upon the source mycobacterial DNA. The pattern of amplification products from all of the reactions, visualized by agarose gel electrophoresis, allowed immediate identification either as MtbC composed of M. tuberculosis (or M. africanum subtype II), M. africanum subtype I, M. bovis, M. bovis BCG, M. caprae, M. microti, or “M. canettii” or as a Mycobacterium other than MtbC (MOTT). This MtbC PCR typing panel provides an advanced approach to determine the subspecies of MtbC isolates and to differentiate them from clinically important MOTT species. It has proven beneficial in the management of Mycobacterium collections and may be applied for practical clinical and epidemiological use.
Mycobacteria that cause tuberculosis in mammals form the Mycobacterium tuberculosis complex (MtbC) and include Mycobacterium tuberculosis, Mycobacterium africanum (divided into subtype I [group A/West African/“M. bovis-like”] and subtype II [group B/East African/“M. tuberculosis-like”]), Mycobacterium bovis (along with the M. bovis-derived bacillus Calmette-Guérin [BCG] vaccine strains), Mycobacterium microti, Mycobacterium bovis subsp. caprae (M. caprae), and “Mycobacterium tuberculosis subsp. canettii” (“M. canettii”) (7, 29) (the name “M. canettii” is in quotation marks since it does not appear on the official List of Bacterial Names with Standing in Nomenclature [http://www.bacterio.cict.fr]). Members of the MtbC are highly related mycobacteria exhibiting remarkable nucleotide sequence level homogeneity despite varying in pathogenicity, geographic range, certain physiological features (such as colony morphology as well as profiles of resistance and susceptibility to inhibitors), epidemiology, and host preference (10, 11, 39). Notably, M. bovis has a wide host range but is primarily a bovid pathogen, goats are the natural host of M. caprae, and M. microti is most often isolated from small rodents, while M. tuberculosis is the predominant cause of human tuberculosis (2, 3, 39, 44, 45). However, each of the MtbC subspecies is known to infect humans (16, 17, 20, 22, 27, 33, 43-45), and since most laboratories do not fully identify MtbC isolates, the true cause of tuberculosis in these patients and its source often remain undiscovered. An important health concern is the zoonotic transmission of some MtbC subspecies from animals to humans and vice versa. Of particular significance is the transmission of M. bovis to humans from cattle and unpasteurized milk as well as M. bovis BCG infection of immunocompromised individuals (1, 21, 27, 33). M. bovis is naturally resistant to pyrazidamide, a first-line antituberculosis drug (31, 36). Therefore, complete identification of MtbC isolates at the subspecies level is required in order to collect information on their epidemiology and also to enable appropriate patient treatment and public health measures.
Various biological and molecular mycobacterial characteristics have been utilized to identify MtbC isolates but have limited applicability as MtbC taxonomical tools. Although certain Mycobacterium species-specific gene sequence differences work well to differentiate mycobacteria other than MtbC (MOTT) from each other and from the MtbC, to date none can discriminate the individual MtbC subspecies due to genetic invariance in the target loci (8, 11, 26, 28, 34, 37, 39, 41). In contrast, a series of classical tests based upon growth, phenotypic, and biochemical properties have been traditionally used to segregate members of the MtbC (17, 30). However, together these tests can be slow, cumbersome, imprecise, nonreproducible, and time-consuming, and they may not give an unambiguous result in every case and may not be performed by every laboratory. To complement the classical tests for determination of MtbC species, well-defined MtbC lineage- and subspecies-restricted single-nucleotide polymorphisms (SNPs) in the gyrA, katG, pncA, oxyR′, hsp65, and gyrB genes have been used to specify certain MtbC groupings through sequence analysis and/or digestion of PCR products followed by restriction fragment length polymorphism (PCR-RFLP) analysis (12, 13, 18, 30, 31, 36, 38, 39). However, these loci on their own are unable to differentiate all of the MtbC subspecies. Likewise, molecular genetic MtbC typing assays (e.g., variable numbers of tandem repeat analysis, mixed linker PCR, and IS6110 RFLP) that have been designed to reveal interstrain relationships (12, 17, 25, 39) cannot be used as efficient taxonomic tools to unambiguously classify individual MtbC strains. Spacer oligonucleotide typing (spoligotyping) is the only DNA-based methodology for which most MtbC members are believed to have signature features (3, 16, 23, 25, 31, 42-45). However, spoligotypes, the numerical output of spoligotyping, are not necessarily exclusive to one MtbC member, nor are they restricted, as strains can waver from the expected minimal consensus spoligopattern for their MtbC subspecies. Hence, an improved protocol for MtbC species determination with greater discriminatory power is needed.
Comparative genomics studies employing several different genetic hybridization strategies revealed regions of difference (RD) representing the loss of genetic material in M. bovis BCG compared to M. tuberculosis H37Rv (4, 6, 14). One of these deletions is believed to have been the primary attenuation event in the derivation of M. bovis BCG from M. bovis, since all M. bovis BCG isolates bear this deletion (4). PCR analysis for these long sequence polymorphisms (LSPs) found some RD loci to be restricted to one MtbC strain or subspecies, while others appeared to be differentially distributed among the MtbC groupings (7, 14, 29). These data suggested a sequential accumulation of LSPs and was used to construct a phylogenetic map for the evolution of the MtbC (7). Recently, PCR analysis of certain LSPs, in combination with phenotypic testing, was shown to accurately differentiate several MtbC groupings and was used to evaluate an large collection of clinical isolates (35). Unfortunately, some of the targeted RD loci do not have restricted profiles for particular MtbC subspecies, nor was the protocol evaluated for the identification of M. caprae, “M. canettii,” and MOTT isolates.
In this study we investigated the RD profile of the MtbC groupings, and based upon this information we developed a solely PCR-based system using seven PCR primer pairs specific to the loci 16S rRNA, Rv0577, IS1561′, Rv1510, Rv1970, Rv3877/8, and Rv3120, which together form a MtbC PCR typing panel (Table 1). The final pattern of amplification products of all reactions, given by failure or success, clearly segregated the tested strains from MOTT isolates and by MtbC subspecies identity.
TABLE 1.
Primers used in this study
| Primer type and target locus | Primer name | Nucleotide sequence | Locationa | Size (bp) | Programb |
|---|---|---|---|---|---|
| Main MtbC PCR typing panel primer pairs | |||||
| 16S rRNA | 16SRNAF | 5′ ACG GTG GGT ACT AGG TGT GGG TTT C 3′ | 1472650-674 | 543 | 1 |
| 16SRNAR | 5′ TCT GCG ATT ACT AGC GAC TCC GAC TTC A 3′ | 1473192-165 | |||
| Rv0577 | Rv0577F | 5′ ATG CCC AAG AGA AGC GAA TAC AGG CAA 3′ | 671164-190 | 786 | 1 |
| Rv0577R | 5′ CTA TTG CTG CGG TGC GGG CTT CAA 3′ | 671949-926 | |||
| IS1561′ (Rv3349c) | IS1561F | 5′ GCT GGG TGG GCC CTG GAA TAC GTG AAC TCT 3′ | 3733670-699 | 943 | 1 |
| IS1561R | 5′ AAC TGC TCA CCC TGG CCA CCA CCA TTG ACT 3′ | 3754583-612 | |||
| Rv1510 (RD4) | Rv1510F | 5′ GTG CGC TCC ACC CAA ATA GTT GC 3′ | 1701578-600 | 1,033 | 1 |
| Rv1510R | 5′ TGT CGA CCT GGG GCA CAA ATC AGT C 3′ | 1702610-586 | |||
| Rv1970 (RD7) | Rv1970F | 5′ GCG CAG CTG CCG GAT GTC AAC 3′ | 2214244-264 | 1,116 | 1 |
| Rv1970R | 5′ CGC CGG CAG CCT CAC GAA ATG 3′ | 2215359-339 | |||
| Rv3877/8 (RD1) | Rv3877/8F | 5′ CGA CGG GTC TGA CGG CCA AAC TCA TC 3′ | 4356295-320 | 999 | 1 |
| Rv3877/8R | 5′ CTT GCT CGG TGG CCG GTT TTT CAG C 3′ | 4357269-293 | |||
| Rv3120 (RD12) | Rv3120F | 5′ GTC GGC GAT AGA CCA TGA GTC CGT CTC CAT 3′ | 3485555-584 | 404 | 1 |
| Rv3120R | 5′ GCG AAA AGT GGG CGG ATG CCA GAA TAG T 3′ | 3485958-931 | |||
| Additional PCR primer pairs | |||||
| rpoB | rpoBF | 5′ TCA AGG AGA AGC GCT ACG A 3′ | 760688-706 | 359 | 1 |
| rpoBR | 5′ GGA TGT TGA TCA GGG TCT GC 3′ | 761047-028 | |||
| IS1081 | 1081F | 5′ TCG CGT GAT CCT TCG AAA CG 3′ | 1169369-388c | 238 | 1 |
| 1081R | 5′ GCC GTT GCG CTG ATT GGA CC 3′ | 1169369-587c | |||
| MPB70 (Rv2875) | MB1 | 5′ GGC GAT CTG GTG GGC CCG 3′ | 3187117-134 | 489 | 2 |
| MB2 | 5′ CGC CGG AGG CAT TAG CAC GCT 3′ | 3187605-585 | |||
| IS1561′-2 | 2 IS1561F | 5′ GAC CTG ACG CCG CTG ACA C 3′ | 3754127-145 | 530 | 1 |
| 2 IS1561R | 5′ CAC CTA CAC CGC TTC CTG CC 3′ | 3754638-657 | |||
| Rv3879c (RD1) | Rv3879cF | 5′ TCT CCG GAA TGT CAT CTG GCT CCA GCA CAA 3′ | 4357958-987 | 1,000d | 1 |
| Rv3879cR | 5′ GCA ACC CCG GCC ACG CCC GTT ACC 3′ | 4358957-934 | |||
| Rv1510-2 (RD4) | Y277-32F | 5′ GAC ATG TAC GAG AGA CGG CAT GAG 3′ | 1701290-313 | 1,031 | 1 |
| Y277-32R | 5′ AAT CCA ACA CGC AGC AAC CAG 3′ | 1702320-300 | |||
| RD8 (5′ junction) | RD8-5′jnF | 5′ ATG CGC CAA CCG CCG TGT AGG 3′ | 4056590-610 | 956 | 1 |
| RD8-5′jnR | 5′ TGC CGG CCA GGT CCA GTT CAA AT 3′ | 4057545-523 | |||
| Rv2073c (RD9) | Rv2073cF | 5′ TCG CCG CTG CCA GAT GAG TC 3′ | 2330577-596 | 600 | 1 |
| Rv2073cR | 5′ TTT GGG AGC CGC CGG TGG TGA TGA 3′ | 2331171-148 | |||
| Rv0222 (RD10) | RD10iF | 5′ AGG GAT TCG GCG TTC GGG ATT CCT G 3′ | 265886-910 | 666 | 2 |
| RD10iR | 5′ GAT CGC GAT CGC CAA CGA TTC AGT GTA TG 3′ | 266551-523 | |||
| Rv1257c (RD13) | Rv1257cF | 5′ GGT GGC GAG CTG GAA TTC GTG AGA CAT TAC 3′ | 1404563-592 | 456 | 2 |
| Rv1257cR | 5′ CAT CGC CGA GGA GCG GAA TCT GAT GAT 3′ | 1405018-4992 | |||
| Primers for gene sequencing and/or PCR-RFLP | |||||
| oxyR285 | oxyRF | 5′ CTA TGC GAT CAG GCG TAC TTG 3′ | 2725558-578 | 556 | 3 |
| oxyRR | 5′ GGT GAT ATA TCA CAC CAT A 3′ | 2726113-095 | |||
| pncA57 | pncAF | 5′ CAG GAG CTG CAA ACC AAC TCG 3′ | 2288680-700 | 664 | 1 |
| pncAR | 5′ GCT GGT CAT GTT CGC GAT CG 3′ | 2289343-324 | |||
| hsp65631 | hsp65F | 5′ ACC AAC GAT GGT GTG TCC AT 3′ | 528750-769 | 441 | 1 |
| hsp65R | 5′ CTT GTC GAA CCG CAT ACC CT 3′ | 529190-171 | |||
| gyrBΔ | gyrBF | 5′ TCG GAC GCG TAT GCG ATA TC 3′ | 5570-589 | 1,040 | 1 |
| gyrBR | 5′ ACA TAC AGT TCG GAC TTG CG 3′ | 6609-590 | |||
| gyrA95 | gyrAF | 5′ GGA GGT GCG CGA CGG GCT CAA G 3′ | 7427-448 | 354 | 2 |
| gyrAR | 5′ ACC CGG CCG TCG TAG TTA GGG ATG AAA TC 3′ | 7780-752 | |||
| katG203 | katG203F | 5′ GCC GGC GCC ATG GGT CTT ACC GAA AGT GTG 3′ | 2155273-302 | 370 | 2 |
| katG203R | 5′ CAA GAA GCT CTC ATG GGC GGA CCT GAT TGT 3′ | 2155642-613 | |||
| katG463 | katG463F | 5′ GAC GAG GTC GGC GAA GGA CAC TTT GA 3′ | 2154478-503 | 351 | 2 |
| katG463R | 5′ GGG CCG CTG GTC CCC AAG CAG AC 3′ | 2154828-806 | |||
| 16S rRNA (long) | 16SLongF | 5′ TAA CAC ATG CAA GTC GAA CGG AAA GG 3′ | 1471895-920 | 1,436 | 1 |
| 16SLongR | 5′ ACT TCG TCC CAA TCG CCG ATC CCA CC 3′ | 1473331-305 | |||
| Additional sequencing primers | |||||
| 16S rRNA Seq | 16S seqR | 5′ TTC ACG AAC AAC GCG ACA AAC CAC 3′ | 1472442-419 | ||
| gyrB Seq | gyrB seqR | 5′ GCG GTT CGC TGA CCT TCA CCG AGA TCA C 3′ | 6335-307 |
(This study contributed to the fulfillment of the Ph.D. degree requirements by R. C. Huard.)
MATERIALS AND METHODS
Strains analyzed.
A total of 71 MtbC strains and 44 MOTT isolates were tested (Table 2). These strains were characterized by various typing methods, i.e., numerical analysis of phenotypic and biochemical characteristics, 16S rRNA sequencing, IS6110-RFLP analysis, and spoligotyping. The country of origin of these strains (where available), as well as references to prior publications that utilized certain strains, is given in Table 2. A list associating mycobacterial strains with the contributing laboratory is available upon request.
TABLE 2.
PCR analysis of mycobacterial genetic elements
| Straina | Previous publication (reference) | MtbC subspecies or MOTT species | Host | Location | Resultb for:
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA | rpoB | Rv0577 | IS1081 | MPB70 | IS1561′ | Rv3877/8 (RD1) | Rv1510 (RD4) | Rv3120 (RD12) | Rv1257c (RD13) | RV1970 (RD7) | RD8 | Rv2073c (RD9) | Rv0222 (RD10) | ||||||||||||||||||
| 217-94 | 13 | “M. canettii” | Human | Zurich | + | + | + | + | + | + | + | + | −c | + | + | + | + | + | |||||||||||||
| 17727 (116) | 25 | “M. canettii” | Human | Somalia | + | + | + | + | + | + | + | + | −c | + | + | + | + | + | |||||||||||||
| 96-46 | “M. canettii” | Human | France | + | + | + | + | + | + | + | + | −c | + | + | + | + | + | ||||||||||||||
| 97-488 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 97-742 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 97-803 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 97-818 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 97-1177 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 97-1289 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 97-1438 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| W | M. tuberculosis | Human | United States | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| CA-43 (43) | 25 | M. tuberculosis | Human | China | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| CA-65 (65) | 25 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| CDC1551 | M. tuberculosis | Human | United States | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| Cb3.3 | M. tuberculosis | Human | United States | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 2001-1225 | M. tuberculosis or M. africanum subtype II | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| CA-56 (56) | 25 | M. tuberculosis | Human | Curacao | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| 97-66 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 97-279 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 97-1503 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| AH TN13475 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| Mtb21 | M. tuberculosis | Human | United States | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| Mtb22 | M. tuberculosis | Human | United States | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| 970623 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| ATCC 27294 | M. tuberculosis | H37Rv | United States | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| ATCC 25177 | M. tuberculosis | H37Ra | United States | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| ATCC 51910 | M. tuberculosis | Human | United States | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| NY iso | M. tuberculosis | Human | United States | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| CA-105 (105) | 25 | M. tuberculosis | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| 13876 (100) | 25 | M. africanum subtype II | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| 15082 (92) | 25 | M. africanum subtype II | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| 15199 (47) | 25 | M. africanum subtype II | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| 133/92 | 17 | M. africanum subtype II | Human | Uganda | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| 178/92 | 17 | M. africanum subtype II | Human | Uganda | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| 290/92 | 17 | M. africanum subtype II | Human | Uganda | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| 1167/93 | 17 | M. africanum subtype II | Human | Uganda | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| ATCC 25420 | 12 | M. africanum subtype I | Human | Senegal | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||||||||||||
| ATCC 35711 | 12 | M. africanum subtype I | Human | NDd | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||||||||||||
| 1255/93 | 17 | M. africanum subtype I | Human | Sierra Leone | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||||||||||||
| 1457/93 | 17 | M. africanum subtype I | Human | Sierra Leone | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||||||||||||
| 1565/93 | 17 | M. africanum subtype I | Human | Sierra Leone | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||||||||||||
| 1567/93 | 17 | M. africanum subtype I | Human | Sierra Leone | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||||||||||||
| 17902 (85) | 25 | M. africanum subtype I | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||||||||||||
| 17316 (6) | 25 | M. africanum subtype I | Human | The Netherlands | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||||||||||||
| ATCC 19422 | 44 | M. microti | Vole | United Kingdom | + | + | + | + | + | − | + | + | + | + | − | − | − | − | |||||||||||||
| ATCC 35782 | M. microti | Vole | ND | + | + | + | + | + | − | + | + | + | + | − | − | − | − | ||||||||||||||
| ATCC 11152 | 44 | M. microti | Vole | United Kingdom | + | + | + | + | + | − | + | + | + | + | − | − | − | − | |||||||||||||
| 97-2272 | 44 | M. microti | Human | The Netherlands | + | + | + | + | + | − | + | + | + | + | − | − | − | − | |||||||||||||
| 97-2257 | 44 | M. microti | Human | The Netherlands | + | + | + | + | + | − | + | + | + | + | − | − | − | − | |||||||||||||
| 15496 | 44 | M. microti | Vole | United Kingdom | + | + | + | + | + | − | + | + | + | + | − | − | − | − | |||||||||||||
| 15498 | 44 | M. microti | Vole | United Kingdom | + | + | + | + | + | − | + | + | + | + | − | − | − | − | |||||||||||||
| 16420 | 44 | M. microti | Vole | United Kingdom | + | + | + | + | + | − | + | + | + | + | − | − | − | − | |||||||||||||
| 15912 | 44 | M. microti | Llama | Belgium | + | + | + | + | + | − | + | + | + | + | − | − | − | − | |||||||||||||
| 97-1297 | 44 | M. microti | Feret | The Netherlands | + | + | + | + | + | − | + | + | + | + | − | − | − | − | |||||||||||||
| Cip 105776 | 2 | M. caproe | Goat | Spain | + | + | + | + | + | + | + | + | − | − | − | − | − | − | |||||||||||||
| ATCC 19210 | M. bovis | Cow | United States | + | + | + | + | + | + | + | − | − | − | − | − | − | − | ||||||||||||||
| TN5022 | M. bovis | Human | United States | + | + | + | + | + | + | + | − | − | − | − | − | − | − | ||||||||||||||
| ATCC 35725 | M. bovis | ND | ND | + | + | + | + | + | + | + | − | − | − | − | − | − | − | ||||||||||||||
| ATCC 35726 | M. bovis | Cow | ND | + | + | + | + | + | + | + | − | − | − | − | − | − | − | ||||||||||||||
| ATCC 35730 | M. bovis | Cow | ND | + | + | + | + | + | + | + | − | − | − | − | − | − | − | ||||||||||||||
| 2001-831 | M. bovis | Human | The Netherlands | + | + | + | + | + | + | + | − | − | − | − | − | − | − | ||||||||||||||
| CA-73 (73) | 25 | M. bovis | Cow | The Netherlands | + | + | + | + | + | + | + | − | − | − | − | − | − | − | |||||||||||||
| CA-117 (117) | 25 | M. bovis | Cow | Argentina | + | + | + | + | + | + | + | − | − | − | − | − | − | − | |||||||||||||
| CA-126 (126) | 25 | M. bovis | Cow | Argentina | + | + | + | + | + | + | + | − | − | − | − | − | − | − | |||||||||||||
| CA-130 (130) | 25 | M. bovis | Cow | The Netherlands | + | + | + | + | + | + | + | − | − | − | − | − | − | − | |||||||||||||
| BCGt | M. bovis BCG | Vaccine | Pasteur-Tunis | + | + | + | + | + | + | − | − | − | − | − | − | − | − | ||||||||||||||
| r4-93 | M. bovis BCG | ND | ND | + | + | + | + | + | + | − | − | − | − | − | − | − | − | ||||||||||||||
| ATCC 27290 | M. bovis BCG | Vaccine | Copenhagen | + | + | + | + | + | + | − | − | − | − | − | − | − | − | ||||||||||||||
| ATCC 35736 | M. bovis BCG | Vaccine | Brazil | + | + | + | + | + | + | − | − | − | − | − | − | − | − | ||||||||||||||
| ATCC 35737 | M. bovis BCG | Vaccine | Japan | + | + | + | + | + | + | − | − | − | − | − | − | − | − | ||||||||||||||
| TN10130 | M. bovis BCG | Human | United States | + | + | + | + | + | + | − | − | − | − | − | − | − | − | ||||||||||||||
| NYS 9976 | M. abscessus | Ref.e | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| H 15882 | M. abscessus | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| H 21479 | M. abscessus | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| T 61715 | M. abscessus | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| MAC (BJ) | M. avium complex | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| MAC (FL) | M. avium complex | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| ATCC 19074 | M. avium subsp. avium | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| ATCC 25291 | M. avium subsp. avium | Chicken | ND | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| ATCC 35781 | M. avium subsp. avium | ND | ND | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 9800847 | M. avium subsp. avium | Goose | Belgium | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 9601138 | M. avium subsp. avium | Human | The Netherlands | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| ATCC 19077 | M. avium subsp. intracellulare | Human | ND | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| ATCC 13950 | M. avium subsp. intracellulare | ND | ND | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 9601103 | M. avium subsp. intracellulare | Human | The Netherlands | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| EG | M. chelonae | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| PB | M. chelonae | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| NYS IS0136 | M. fortuitum | Ref. | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| W 37396 | M. fortuitum | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| H 49581 | M. fortuitum | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| NYS IS9775 | M. fortuitum | Ref. | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| ML | M. fortuitum | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| YG | M. fortuitum | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| BD | M. gordonae | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| GM | M. gordonae | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| Straina | Previous publication (reference) | MtbC subspecies or MOTT species | Host | Location | Resultb for:
|
||||||||||||||||||||||||||
| 16S rRNA | rpoB | Rv0577 | IS1081 | MPB70 | IS1561′ | Rv3877/8 (RD1) | Rv1510 (RD4) | Rv3120 (RD12) | Rv1257c (RD13) | RV1970 (RD7) | RD8 | Rv2073c (RD9) | Rv0222 (RD10) - | ||||||||||||||||||
| 900419 | M. kansasii | Human | The Netherlands | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 2000-1049 | M. kansasii | Human | The Netherlands | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 2000-1454 | M. kansasii | Tap water | Belgium | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 2000-1457 | M. kansasii | Biofilm | Germany | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 2000-1458 | M. kansasii | Tap water | Germany | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 2000-1459 | M. kansasii | Tap water | Belgium | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 2000-1461 | M. kansasii | Toilet | Belgium | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 2000-1471 | M. kansasii | Environment | Italy | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 2000-1493 | M. kansasii | Water | Germany | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 2000-1495 | M. kansasii | Hot water | Czech Republic | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| J18698 | M. malmoense | Human | The Netherlands | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| ATCC 927 | M. marinum | Fish | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 9801810 | M. marinum | Human | The Netherlands | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| 990036 | M. marinum | Human | The Netherlands | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| xsim | M. simiae | ND | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| ATCC 23038 | M. smegmatis | ND | ND | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| mc2 | M. smegmatis | mc2 155 | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| SCC 74/31 | M. szulgai | Human | The Netherlands | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| myc 941 | M. xenopi | Human | The Netherlands | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
| SR-BL | M. xenopi | Human | United States | + | + | − | − | − | − | − | − | − | − | − | |||||||||||||||||
Underlined strains were reclassified in this study and were originally designated M. africanum [no subtype] (13876 and 15082), M. bovis (2001-1225, 15199, 17902, 17316, and TN10130), or M. microti (ATCC 35781 and xsim). Identification numbers from reference 25 are in boldface.
−, negative for a strong band of the correct size; +, positive for such a band.
RDcan partially overlaps with RD12 (see reference 6).
ND, no data.
Ref., reference strain.
DNA preparation for PCR.
DNA from mycobacterial cultures was purified as follows. Cultured bacteria were spun down, resuspended in 1 ml of Tris-EDTA buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA), and transferred to a 2-ml Eppendorf tube. Lysozyme solution (100 μl; 10 mg/ml in Tris-EDTA buffer) and five 3-mm-diameter glass beads were then added, vortexed, sonicated for 10 min, vortexed again, and then incubated at 37°C for 2 h with brief vortexing every 30 min. To the resultant suspension, after water bath sonication for 10 min and division into two 1.5-ml Eppendorf tubes, was added 70 μl of 10% sodium dodecyl sulfate and 10 μl of proteinase K (10 mg/ml). The mixture was then vortexed and incubated for 2 h at 65°C, with brief vortexing every 30 min. Afterwards, 100 μl of 5 M NaCl was added and vortexed, and following the addition of 80 μl of 10% hexadecyltrimethyl ammonium bromide (Sigma, St. Louis, Mo.) in pure water, the mixture was incubated at 65°C for 30 min. For DNA extraction 750 μl of chloroform was added, mixed well, and centrifuged at 14,000 rpm for 5 min in a Microfuge. The resultant upper phase was transferred to a clean tube with 420 μl of isopropanol and mixed gently. The tubes were then cooled on ice and spun in a Microfuge for 30 min at 14,000 rpm and 4°C. Following removal of the supernatant, the DNA pellet was washed with 75% ethanol and air dried. The DNA was then resuspended in RNase- and DNase-free water, quantified, diluted to 50 to 500 μg/ml, and used for PCR.
PCR amplification primers and conditions.
The target gene loci and their primer names, primer sequences, primer locations in the M. tuberculosis H37Rv genome (accession no. NC_000962) (9), and amplification product sizes and the programs used to amplify are listed in Table 1. Primer pairs suited to evaluate the chosen genetic elements were created by using the DNASTAR program (DNASTAR, Inc., Madison, Wis.) and sequence database information (http//:www.ncbi.nlm.nih.gov and http//:www.sanger.ac.uk/Projects/M_tuberculosis). Each PCR mixture was prepared with 25 μl of PCR Master (Roche Diagnostics, Indianapolis, Ind.), 20 μl of water, 2.5 μl of dimethyl sulfoxide, 1 μl of each primer at 20 μM, and 0.5 μl of DNA (equaling 25 to 250 ng). The sole difference was with the (additional) Rv3879c primer pair, which required 5 μl of dimethyl sulfoxide for good amplification (final volume, 52.5 μl). PCR amplifications were performed in a Gene Amp PCR system 9700 (PE Applied Biosystems, Foster City, Calif.), using either program 1 (with an initial denaturation step of 5 min at 94°C followed by 25 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, and ending with a final elongation step for 10 min at 72°C), program 2 (program 1 but with an annealing temperature of 65°C), or program 3 (program 1 but with an annealing temperature of 50°C). All of the main MtbC PCR typing panel amplifications used program 1. When DNA samples were limited or of low concentration, the number of PCR cycles was increased to 35. PCR products and a 100-bp ladder (Gibco BRL, Grand Island, N.Y.) were visualized by agarose (1.5%) gel electrophoresis and ethidium bromide staining. Images were captured with the Nighthawk imaging system (PDI Inc., Huntington Station, N.Y.) and Quality One software package (PDI Inc.). For this work all of the described PCR results from mycobacteria included a positive control for amplification of the target locus as well as a positive control for the test DNA. Furthermore, all negative and unexpected positive PCR results, as given in Table 2, were repeated and confirmed at least once again.
DNA sequencing and PCR product restriction enzyme digest analysis.
Direct sequencing of PCR fragments was performed by the Cornell University BioResource Center (Ithaca, N.Y.) (http://www.brc.cornell.edu), using a BigDye Terminator kit (PE Applied Biosystems), and the output was analyzed with an ABI 3700 DNA sequencer. The PCR amplification primers were also used as sequencing primers, and a minimal single overlap from two directions for each was usually achieved. Additional sequencing primers for gyrB and the 16S rRNA long fragment are listed in Table 1. The DNASTAR program was used to compare the derived sequence data with DNA sequence information taken from the GenBank database. Comparative analysis of polymorphisms in the gyrA codon 95 (gyrA95; G→C at nucleotide 7585 [7585G→C]) (nucleotide numberings are relative to the plus strand of the M. tuberculosis H37Rv chromosome, accession no. NC_000962 [9]) and katG codons 203 and 463 (katG203 and katG463; 2155501G→A and 2154722C→A, respectively) (7, 12, 17, 39) allows the segregation of the MtbC subspecies into four distinct genotypic groups: 1a, 1b, 2, and 3. M. tuberculosis strains partition into either group 1b, 2, or 3; M. africanum subtype II strains fall into either MtbC group 1b or 2; “M. canettii” strains are MtbC group 1b; and M. bovis, M. bovis BCG, M. caprae, M. microti, and M. africanum subtype I strains fall into group 1a. For the strains indicated in Table 3, the determination of MtbC grouping was done by sequence analysis of the gyrA95 PCR fragment and PCR-RFLP of the katG203 and katG463 amplicons with the restriction enzyme BstNI (New England Biolabs, Beverly, Mass.). BstNI digestion of the katG203 PCR product amplified from MtbC group 1b, 2, and 3 isolates produces two DNA fragments (230 and 140 bp), while that from MtbC group 1a strains remains uncut. Similarly, BstNI digestion of the katG463 PCR product produces three DNA fragments (12, 61, and 278 bp) from MtbC group 2 and 3 isolates, while that from MtbC group 1a and 1b strains is cut into four DNA fragments (12, 61, 104, and 174 bp). Digest products were visualized by agarose (4%) gel electrophoresis with ethidium bromide staining. Nucleotide substitutions in pncA, oxyR′, hsp65, and gyrB have also been used to identify specific MtbC members. The pncA codon 57 SNP (pncA57; 2289071G→C) segregates M. bovis (strict sense) and M. bovis BCG from the other MtbC subspecies (36), while the oxyR′ pseudogene bears a position 285 SNP (oxyR285; 2725801C→T) that differentiates M. bovis (strict sense), M. bovis BCG, and M. caprae isolates from the other MtbC members (31, 38). As a result, the pncA57 nucleotide usage relative to that of oxyR285 singles out M. caprae strains. Similarly, a position 631 SNP in the hsp65 gene (hsp65631; 529004C→T) is restricted to “M. canettii” strains, differentiating them from the other MtbC subspecies (13), while unique SNPs in a >1-kb section of the gyrB gene (gyrBΔ [Δ represents the polymorphic amplicon]) allow M. africanum subtype I (gyrBAf, 6446G→T), M. microti (gyrBMic, 5617C→T and 6446G→T), M. bovis (strict sense) (gyrBBv, 5752G→A, 6406C→T, and 6446G→T), and M. caprae (gyrBCp, 5752G→A, 6307T→G, and 6446G→T), to be discriminated from each other and from M. tuberculosis or M. africanum subtype II (gyrBTb) (24, 30). PCR-RFLP of the oxyR285, hsp65631, and gyrBΔ PCR fragments was done to suggest subspecies identity, for the strains indicated in Table 3, as previously described (13, 31, 38). Likewise, PCR-RFLP of the pncA57 PCR product was done with BstEII (New England Biolabs), a restriction enzyme that cuts the M. bovis (strict sense) pncA57 amplicon into two fragments (170 and 494 bp) and cuts pncA57 from most other MtbC strains into three fragments (103, 170, and 391 bp) (Table 3). Additional digestion of the gyrBΔ PCR fragment (to probe for the 6406C→T SNP) with the MaeIII restriction enzyme (Roche Diagnostics) was done to cut the gyrBBv amplicon into three fragments (306, 331, and 403 bp) and to cut the gyrBΔ from the remaining MtbC into four DNA fragments (123, 208, 306, and 403 bp) (Table 3). Direct sequencing of katG203, katG463, oxyR285, pncA57, hsp65631, and gyrBΔ PCR products was done to confirm the respective PCR-RFLP results from a sample of MtbC strains.
TABLE 3.
Single-nucleotide polymorphism analysis of MtbC isolatesa
| Strainb | MtbC subspecies | MtbC grouping | katG203 (2155501) | katG463 (2154722) | gyrA95 (7585) |
gyrBΔ
|
hsp65631 (529004) | oxyR285 (2725801) | pncA57 (2289071) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5671 | 5752 | 6307 | 6406 | 6446 | |||||||||
| 217-94 | “M. canettii” | 1b | G | A | C | C | G | T | C | G | T | C | G |
| 17727 | “M. canettii” | 1b | G | A | C | C | G | T | C | G | T | C | G |
| 96-46 | “M. canettii” | 1b | G | A | C | C | G | T | C | G | T | C | G |
| 97-488 | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| 97-742 | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| 97-803 | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| 97-818 | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| 97-1177 | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| 97-1289 | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| 97-1438 | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| W | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| CA-43 | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| CA-65 | M. tuberculosis | 1b | G | A | C | C | G | T | C | G | C | C | G |
| CDC1551 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| Cb3.3 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| 2001-1225 | M. tuberculosis or M. africanum subtype II | 2 | G | C | C | C | G | T | C | G | C | C | G |
| 97-56 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| 97-66 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| 97-279 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| 97-1503 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| AH TN13475 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| Mtb21 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| Mtb22 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| 970623 | M. tuberculosis | 2 | G | C | C | C | G | T | C | G | C | C | G |
| H37Rv | M. tuberculosis | 3 | G | C | G | C | G | T | C | G | C | C | G |
| H37Ra | M. tuberculosis | 3 | G | C | G | C | G | T | C | G | C | C | G |
| ATCC 51910 | M. tuberculosis | 3 | G | C | G | C | G | T | C | G | C | C | G |
| NY iso | M. tuberculosis | 3 | G | C | G | C | G | T | C | G | C | C | G |
| CA-105 | M. tuberculosis | 3 | G | C | G | C | G | T | C | G | C | C | G |
| 13876 | M. africanum subtype II | 1b | G | A | C | C | G | T | C | G | C | C | G |
| 15082 | M. africanum subtype II | 1b | G | A | C | C | G | T | C | T | C | C | G |
| 15199 | M. africanum subtype II | 1b | G | A | C | C | G | T | C | G | C | C | G |
| 133/92 | M. africanum subtype II | 2 | G | C | C | C | G | T | C | G | C | C | G |
| 178/92 | M. africanum subtype II | 2 | G | C | C | C | G | T | C | G | C | C | G |
| 290/92 | M. africanum subtype II | 2 | G | C | C | C | G | T | C | G | C | C | G |
| 1167/93 | M. africanum subtype II | 2 | G | C | C | C | G | T | C | G | C | C | G |
| ATCC 25420 | M. africanum subtype I | 1a | A | A | C | C | G | T | C | T | C | C | G |
| ATCC 35711 | M. africanum subtype I | 1a | A | A | C | C | G | T | C | T | C | C | G |
| 1255/93 | M. africanum subtype I | 1a | A | A | C | C | G | T | C | T | C | C | G |
| 1457/93 | M. africanum subtype I | 1a | A | A | C | C | G | T | C | T | C | C | G |
| 1565/93 | M. africanum subtype I | 1a | A | A | C | C | G | T | C | T | C | C | G |
| 1567/93 | M. africanum subtype I | 1a | A | A | C | C | G | T | C | T | C | C | G |
| 17902 | M. africanum subtype I | 1a | A | A | C | C | G | T | C | T | C | C | G |
| 17316 | M. africanum subtype I | 1a | A | A | C | C | G | T | C | T | C | C | G |
| ATCC 19422 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| ATCC 35782 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| ATCC 11152 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| 9402272 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| 9702257 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| 15496 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| 15498 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| 16420 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| 15912 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| 97-1297 | M. microti | 1a | A | A | C | T | G | T | C | T | C | C | G |
| Cip 105776 | M. caprae | 1a | A | A | C | C | A | G | C | T | C | T | G |
| ATCC 19210 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| TN5022 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| ATCC 35725 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| ATCC 35726 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| ATCC 35730 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| 2001-831 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| CA-73 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| CA-117 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| CA-126 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| CA-130 | M. bovis | 1a | A | A | C | C | A | T | T | T | C | T | C |
| BCGt | M. bovis BCG | 1a | A | A | C | C | A | T | T | T | C | T | C |
| r4-93 | M. bovis BCG | 1a | A | A | C | C | A | T | T | T | C | T | C |
| ATCC 27290 | M. bovis BCG | 1a | A | A | C | C | A | T | T | T | C | T | C |
| ATCC 35736 | M. bovis BCG | 1a | A | A | C | C | A | T | T | T | C | T | C |
| ATCC 35737 | M. bovis BCG | 1a | A | A | C | C | A | T | T | T | C | T | C |
| TN10130 | M. bovis BCG | 1a | A | A | C | C | A | T | T | T | C | T | C |
RESULTS
Various chromosomal loci were evaluated to ascertain their usefulness as part of an MtbC PCR typing panel.
Selective amplification of the mycobacterial 16S rRNA gene.
For the present work it was necessary to have a means of confirming the presence of mycobacterial DNA. Previously, species-specific nucleotide polymorphisms in the 16S rRNA gene have been used to determine Mycobacterium species identity (37, 41). For pan-genus PCR amplification of the mycobacterial 16S rRNA, a primer pair was designed to consensus nonpolymorphic segments of the 16S rRNA GenBank gene sequences from two MtbC subspecies and several MOTT species. These primers were successful in amplifying a DNA fragment from all mycobacterial isolates (n = 115) in the collection (Table 2). Likewise, an alternative PCR primer pair to the rpoB gene, designed in the same manner as the 16S rRNA primers, amplified DNA from all tested mycobacteria (n = 115) (Table 2). Importantly, the evaluated MOTT species included the nontuberculous organisms most commonly encountered in clinical practice (19). To further validate species identity, the PCR amplification products from several MtbC strains (n = 21) and all MOTT strains (n = 44) were sequenced and confirmed to have the 16S rRNA nucleotide sequence reported for that species in GenBank (data not shown). Amplification for the 16S rRNA was chosen to provide the positive control when evaluating mycobacteria by PCR.
The Rv0577 gene is restricted to the MtbC subspecies.
Ongoing tuberculosis-related projects revealed the potential of Rv0577 as an MtbC-restricted gene. Primers were generated to amplify the full coding region of Rv0577 and were then used to test the collected mycobacteria (n = 115). Rv0577 was specifically and consistently amplified from all MtbC subspecies and strains tested (n = 71), whereas none of the MOTT strains (n = 44) showed a PCR fragment (Table 2). To establish the appropriate amplification of Rv0577, the PCR product from M. tuberculosis H37Ra was sequenced (data not shown). Moreover, PCR amplification results for IS1081 and MPB70, genetic elements believed to be absent from most or all MOTT species (27), paralleled the MtbC-restricted pattern of Rv0577 (n = 115 mycobacteria tested) (Table 2). These results support that Rv0577 is a genotypic marker for the MtbC and that it can be used to distinguish the MtbC subspecies from MOTT species.
Absence of the IS1561′ element differentiates M. microti from the other MtbC subspecies.
Previously, Gordon et al. (15) noted that PCR amplification for the transposase pseudogene fragment IS1561′ (Rv3349c) was positive for all MtbC isolates tested except for their single M. microti (OV254) strain, and they postulated that it might prove to be a M. microti-specific marker. For this study, primers to the IS1561′ element were generated and the collected MtbC strains were evaluated. The IS1561′ PCR fragment was found to be absent from all of the M. microti strains (n = 10) but was present in the other evaluated MtbC isolates (n = 61) (Table 2). The identity of the IS1561′ amplicon was verified by sequence analysis of the PCR fragment from M. tuberculosis H37Ra. In addition, amplification with a previously described IS1561′ primer pair (15) confirmed the lack of IS1561′ amplicons from the M. microti strains (n = 71 MtbC strains tested) (data not shown). The M. microti strains in this collection were isolated from diverse sources (including llama, ferret, and vole, as well as human) and locations (43). Moreover, a very recent paper describing new M. microti LSPs also found the IS1561′ locus to be deleted (5). Therefore, these data support that the absence of IS1561′ is a good genotypic indicator for M. microti.
Absence of the Rv1510 gene differentiates M. bovis and M. bovis BCG from the other MtbC subspecies.
Previous studies indicated that PCR primers generated to the RD4 locus could distinguish M. bovis (strict sense) and M. bovis BCG from the rest of the MtbC subspecies (4, 5, 14). In order to confirm M. bovis subspecies identity, primers were generated to Rv1510, a gene located internal to the RD4 deletion. Amplification experiments confirmed that Rv1510 was absent in all of the evaluated M. bovis strains (n = 10) and M. bovis BCG strains (n = 6) but was present in the other MtbC isolates of the collection (n = 55) (Table 2). The identity of the Rv1510 amplification product generated from M. tuberculosis H37Ra was proven by sequence analysis. Furthermore, using alternative Rv1510 primers described previously (14), M. bovis and M. bovis BCG were also segregated from the remaining MtbC strains (n = 71 MtbC isolates tested) (data not shown). Therefore, these results support that RD4 is a genotypic marker for M. bovis (strict sense) and M. bovis BCG and that the absence of Rv1510 can be used to differentiate them from the other MtbC subspecies.
The Rv1970 gene differentiates M. tuberculosis, M. africanum subtype II, and “M. canettii” from the other MtbC subspecies.
Rv1970 (lprM) is a gene located internal to RD7 (4, 14). In prior studies, PCR analysis revealed RD7 to be present in all of the tested M. tuberculosis and “M. canettii” strains, as well as some M. africanum isolates, but not in the evaluated M. microti, M. bovis, M. bovis BCG, and M. caprae strains (7, 14, 29). Primers were generated to amplify Rv1970, and the PCR results were positive for M. tuberculosis (n = 26), M. africanum subtype II (n = 7), and “M. canettii” (n = 3) strains, while the gene from MtbC group 1a subspecies (n = 35) failed to amplify (Table 2). To ensure that Rv1970 was correctly amplified, the PCR product generated from M. tuberculosis H37Ra was sequenced and shown to be Rv1970 (data not shown). In addition, the RD7 LSP is absent concomitantly with the RD8 and RD10 locus deletions and is often, but not in all cases, associated with the loss of the RD9 locus (7, 29). Primer pairs that amplify either the RD9-internal gene Rv2073c or within the RD10 deletion Rv0222 gene or the RD8 deletion intergenic region of Rv3616c and Rv3617 each segregated the evaluated MtbC isolates identically to the Rv1970 PCR (n = 71 MtbC strains evaluated) (Table 2). Therefore, amplification for these loci supported the determined Rv1970 MtbC subspecies segregation, and amplification for Rv1970 was chosen for inclusion in the MtbC PCR typing panel.
Absence of the Rv3877 and Rv3878 genes of the RD1 locus differentiates M. bovis BCG from the other MtbC subspecies.
The RD1 locus was found previously to be selectively absent in all M. bovis BCG isolates (4, 14), and the presence of RD1 has been shown to be a good negative marker for M. bovis BCG (35, 40). For this study, a PCR primer pair was designed to amplify within the RD1 deletion genes Rv3877 and Rv3878 (Rv3877/8). PCR testing of these primers revealed that, with the exception of the M. bovis BCG isolates (n = 6), all of the tested MtbC strains (n = 65) amplified a specific DNA fragment (Table 2). Sequence analysis of this PCR product generated from M. tuberculosis H37Ra confirmed it as Rv3877/8 (data not shown). As a result, this element appeared to provide a good target for M. bovis BCG identification. A second RD1 primer pair that amplifies a portion of Rv3879c was also evaluated. These primers failed to amplify from all M. bovis BCG isolates, as expected, but also failed to amplify from M. tuberculosis strain CA-56, an isolate that has been described to have an undefined LSP that partially overlaps RD1 (reference 7 and data not shown). In addition, the M. tuberculosis strains 97-803, 97-117, and 97-1438 were each found to have a novel and identical 57-bp deletion within Rv3879c (nucleotides 4368661 to 4368707 relative to the M. tuberculosis H37Rv genome sequence [9]) that truncates the predicted protein by 19 amino acids while maintaining the codon frame (data not shown).
The Rv3120 gene differentiates M. bovis, M. bovis BCG, M. caprae, and “M. canettii” from the other MtbC members.
RDcan is a unique “M. canettii”-specific deletion that partially overlaps with RD12, both of which include the gene Rv3120 (7). To further segregate members of the MtbC, PCR primers were generated to amplify within Rv3120. A PCR fragment of the correct size was detected from the evaluated M. tuberculosis (n = 26), M. africanum subtype I (n = 8), M. africanum subtype II (n = 7), and M. microti (n = 10) strains but not from the M. bovis (n = 10), M. bovis BCG (n = 6), M. caprae (n = 1), and “M. canettii” (n = 3) strains (Table 2). Sequence analysis of the Rv3120 PCR product from M. tuberculosis H37Ra further confirmed it as Rv3120 (data not shown). In the process of building the MtbC PCR typing panel, a primer pair to the RD13 deletion gene Rv1257c was also tested. The Rv1257c primers exhibited the same MtbC amplification profile as Rv3120 but with the distinct ability to amplify for “M. canettii” (n = 71 MtbC isolates tested) (Table 2), thereby supporting the selectivity of the Rv3120 amplification results.
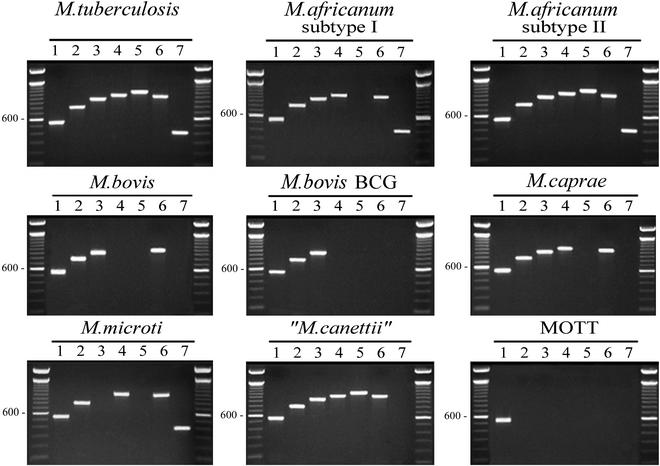
The composite PCR panel provides a unique pattern to type individual MtbC subspecies.
The optimized and thoroughly evaluated primers to the loci 16S rRNA, Rv0577, IS1561′, Rv1510, Rv1970, Rv3877/8, and Rv3120 (chosen for their respective amplification profiles from the evaluated mycobacteria) form the MtbC PCR typing panel (Table 1). Figure 1 illustrates the representative PCR amplification pattern for each MtbC grouping and a single MOTT (M. avium subsp. avium) with this set of seven primer pairs. With the exception of M. tuberculosis and M. africanum subtype II (which were identical), this panel gave a unique pattern of DNA fragments for each MtbC subspecies. When the MtbC PCR typing panel was applied to the DNAs from the collected MOTT strains (n = 44) (Table 2), only the 16S rRNA primer pairs amplified, thereby giving the MOTT samples a single pattern. Table 4 summarizes the distinct patterns whereby the amplification result (positive or negative) for each MtbC subspecies and the tested MOTT species is indicated. Based upon the presence of the expected PCR fragment, for each numerically assigned locus, an algorithm is proposed for a MOTT and each MtbC subspecies. As examples, a MOTT which amplifies only for the 16S rRNA in lane 1 has a profile of 1, while M. tuberculosis and M. africanum subtype II, which amplify for all loci in the MtbC PCR typing panel, have a profile of 1234567.
FIG. 1.
The composite MtbC PCR typing panel. Illustrated are the typical MtbC PCR panel typing results for a single representative of each MtbC subspecies as well as MOTT (M. avium subsp. avium is shown). A total of 71 MtbC isolates and 44 MOTT isolates were tested. PCR products and a 100-bp ladder were visualized by agarose gel electrophoresis and ethidium bromide staining. Images were captured with the Nighthawk imaging system (PDI Inc.) and Quality One software package (PDI Inc.). Lanes: 1, 16S rRNA; 2, Rv0577; 3, IS1561′; 4, Rv1510; 5, Rv1970; 6, Rv3877/8; 7, Rv3120. Unlabeled lanes in each panel contain the 100-bp ladder.
TABLE 4.
An algorithm to identify individual MtbC subspeciesa
| Organism(s) | Result for the following locus (lane in Fig. 1):
|
Profile | ||||||
|---|---|---|---|---|---|---|---|---|
| 16S rRNA (1) | Rv0577 (2) | IS1561′ (3) | Rv1510 (4) | Rv1970 (5) | Rv3877/8 (6) | Rv3120 (7) | ||
| M. tuberculosis | + | + | + | + | + | + | + | 1234567 |
| M. africanum subtype II | + | + | + | + | + | + | + | 1234567 |
| “M. canettii” | + | + | + | + | + | + | − | 123456 |
| M. africanum subtype I | + | + | + | + | − | + | + | 123467 |
| M. caprae | + | + | + | + | − | + | − | 12346 |
| M. bovis | + | + | + | − | − | + | − | 1236 |
| M. bovis BCG | + | + | + | − | − | − | − | 123 |
| M. microti | + | + | − | + | − | + | + | 12467 |
| MOTT | + | − | − | − | − | − | − | 1 |
Based on the results shown in Fig. 1.
MtbC strains which did not give the expected MtbC PCR typing panel pattern.
Throughout the course of this study, a few mycobacterial isolates as designated conflicted with the expected results with the MtbC PCR typing panel as well as one or more of the following: PCR-RFLP analysis, 16S rRNA sequencing, amplification using the corroborative primer pairs, and/or published spoligotype data (25, 31, 45). Each of these isolates was reclassified in the final synopsis and indicated by an underlined name (with reference to the prior designation) in Tables 2 and 3. For instance, a single “M. microti” isolate (ATCC 35781) in the collection had the MtbC PCR typing panel profile of 1, the MOTT profile. This isolate failed to amplify for the additional MtbC-specific IS1081 and MPB70 elements, and sequence determination of a 1,436-bp fragment of the 16S rRNA from this isolate found it to match perfectly to that of M. avium subsp. avium (accession no. AF306455). This “M. microti” strain was purchased directly at a time that no M. avium subsp. avium strains were present in the testing laboratory and so was reclassified as M. avium subsp. avium (strain ATCC 35781 in Table 2). As further examples, two strains determined phenotypically and given as “M. bovis” human isolates did not yield the expected M. bovis profile 1236 (strains 15199 and 2001-1225 in Tables 2 and 3) when tested by the PCR panel but gave the profile 1234567 and were determined to be either M. africanum subtype II or M. tuberculosis. Support for reassignment came from their wild-type nucleotide usage in oxyR285 and pncA57 and for having gyrBTb. Furthermore, sequence determination of katG203, katG463, and gyrA95 revealed strain 15199 to be MtbC group 1b, while strain 2001-1225 was MtbC group 2. Both groups are known to include M. tuberculosis and M. africanum subtype II strains but not M. bovis (12, 17, 39). The spoligotype of strain 15199 (25 [shown as strain 47]) corresponded to the M. africanum subtype II spoligotype patterns illustrated by Niemann et al. (31). The spoligotype of strain 2001-1225 was unavailable. Therefore, for this study strain 15199 was designated M. africanum subtype II, while strain 2001-1225 could not be definitively identified and so is given as M. tuberculosis or M. africanum subtype II. The MtbC PCR typing panel also identified a probable cross-contamination and confirmed its correction. Here, one M. bovis BCG strain (BCGt) maintained in culture gave the expected 123 profile from an earlier DNA sample, but a later DNA preparation gave the 1234567 profile of M. tuberculosis or M. africanum subtype II. Restarting the culture from stored stocks recapitulated the 123 profile of M. bovis BCG (data not shown). These examples illustrate the usefulness of the MtbC PCR typing panel for intralaboratory control practices and the management of MtbC cultures. Importantly, there was near-complete concordance between the MtbC PCR panel-determined MtbC subspecies identities (Table 2) and the known MtbC subspecies-defining SNPs (Table 3). To our knowledge, this study provides the most comprehensive comparative analysis and validation of the known MtbC-defining SNPs. However, M. africanum subtype II strain 15082 (25 [shown as strain 92]), which is otherwise typical by all other criteria (Tables 2 and 3), was not gyrBTb but rather was gyrBAf by both PCR-RFLP and sequence analysis and is a noteworthy exception.
DISCUSSION
This work describes the development of a simple, straightforward, rapid, and specific PCR-based typing method that unambiguously differentiates individual subspecies of the MtbC and segregates them from various clinically important MOTT species. This MtbC PCR typing panel evaluates independent loci that vary due to unidirectional chromosomal region deletions, and, as a result, our data were in close concordance with recent independently derived RD data (5, 7, 29). As such, this protocol is an improvement over current phenotypic and molecular-based MtbC subspecies-determining strategies. However, an inherent concern of RD locus PCR targeting strategies for MtbC subspecies determination is the potential for independent strain-specific deletions that overlap other subspecies-specific or lineage-specific LSPs. Emerging RD loci may result in uncommon isolates with novel strain-specific MtbC PCR typing panel profiles or rare outliers that give the MtbC PCR typing panel profile of another MtbC subspecies. In both cases, alternative PCR target amplifications (such as those provided in Table 1), SNP-based subspecies determination, spoligotyping, or phenotypic characterization may be warranted. Alternatively, to compensate for such variants or novel MtbC subspecies, the MtbC PCR typing panel may be expanded to incorporate new target loci so as to increase its discriminatory power.
The major limitation of our MtbC PCR typing panel is that it cannot differentiate M. tuberculosis from M. africanum subtype II. Compared to M. tuberculosis, M. africanum subtype II is known to have certain unique phenotypic and biogeographical features (17, 22, 30, 32), and yet we were unable to identify any chromosomal differences to set these two apart. In fact, to this point no single DNA-based protocol has been able to positively identify all M. africanum subtype II strains on its own. Indeed, there is a growing consensus that M. africanum subtype II strains are really simply clustered biovariants of M. tuberculosis with divergent properties in phenotypic analyses (7, 35). The facts that all M. africanum subtype II strains tested to date also lacked the “modern” M. tuberculosis deletion TbD1 (7) and that M. africanum subtype II isolates do not form a single lineage but can fall into either MtbC genotypic group 1b or group 2 by katG203, katG463, and gyrA95 determinations (Table 3) support this view. If previously characterized M. africanum subtype II isolates actually represent atypical M. tuberculosis strains, then the MtbC PCR typing panel grouped these isolates appropriately, reiterating the advantages of this typing approach.
The MtbC PCR typing panel was originally intended for use in the confirmation of MtbC subspecies identity and as an intralaboratory control for cross-species contamination and other laboratory errors. For these uses the MtbC typing panel has been of great value, revealing several erroneously designated MtbC strains. In the future, the MtbC PCR typing panel may be applied to determine subspecies of stored clinical and agricultural isolates in retrospective epidemiological studies that may unveil previously unknown correlates of infection, at-risk populations, animal reservoirs, and nuances of clinical course and treatment response, as well as pathogen geographic range and spread over time. The MtbC PCR typing panel may also be useful for the typing of new clinical isolates, especially in areas where the geographic distributions of various MtbC subspecies may overlap, as is potentially the case with M. tuberculosis, M. africanum subtype II, M. africanum subtype I, M. bovis, and “M. canettii” in Africa. This simple and rapid typing protocol could also help to determine the optimal therapeutic strategy for individual cases in areas of high tuberculosis incidence where the diagnosis of tuberculosis is made more difficult by human immunodeficiency virus coinfection and/or where the distinction from MOTT species is of the most importance. The MtbC PCR typing panel may also aid the diagnosis of M. bovis BCG dissemination as a complication of BCG vaccination or immunostimulation against bladder cancer (1, 21). Likewise, the panel could contribute to important public health investigations, as with M. bovis transmission from cattle to humans (27, 33). Since PCR is increasingly used, it is anticipated that both reference institutions and clinical diagnostic laboratories may be able to adopt our strategy for MtbC subspecies determination.
In addition to providing a streamlined means to evaluate MtbC strain identity, this work expands upon the present knowledge of genotypic features of the MtbC. First, Rv0577 was evaluated and identified as an MtbC-restricted gene. The commonly used MtbC-defining element IS6110 has been shown to be absent from some M. tuberculosis strains (39) and to be present in some MOTT strains (27). Hence, Rv0577 may be a preferable MtbC PCR marker. Second, a novel 57-bp deletion within Rv3879c was identified in three M. tuberculosis isolates. This new RD locus joins the previously described RvD1 through -5 and TbD1 LSPs of numerous M. tuberculosis strains (7) and suggests that an epidemiological relationship exists among these three isolates. Together, the collective RD locus data support the theory that the accumulation of LSPs is a significant adaptive strategy of the MtbC subspecies that serves to generate genetic and biological diversity. Therefore, it is expected that other M. tuberculosis LSPs will be found and that such LSPs will offer an additional means of assigning genetic relatedness to M. tuberculosis strains. Third, the loci RD7 through RD10 were confirmed to be present in M. tuberculosis, M. africanum subtype II, and “M. canettii” and confirmed absent in the evaluated MtbC group 1a subspecies. Importantly, PCR amplification testing for these LSPs allowed the segregation of M. africanum subtype I from M. africanum subtype II, and together they offer a genotypic basis for the classically described M. bovis-like versus M. tuberculosis-like phenotypic differences of M. africanum isolates (17). Recent studies have also described additional M. africanum subtype I-like strains that possess the RD7 locus but lack the RD9 locus and are unlike any strains evaluated in this report (7, 29, 35). These strains likely represent phylogenetic precursors of the more common M. africanum subtype I isolates but may also prove to be a novel MtbC subspecies. However, without exclusive genetic markers for M. africanum subtype I, the taxonomic status of these unique strains containing RD7 but lacking RD9 remains unclear. As a potential aid to type these strains, an RD9 primer pair was evaluated in this study and is included in the list of primers in Table 1 (running at 600 bp, this amplicon fits well between lanes 6 and 7 in the PCR fragment cascade presented in Fig. 1). Finally, the previously identified MtbC subspecies- or lineage-specific SNPs of gyrA95, katG203, katG463, oxyR285, pncA57, hsp65631, and gyrBΔ were used collectively here for the first time and correlated perfectly with one another and with the RD locus data for all tested strains except one. This M. africanum subtype II strain, 15082, is of particular interest because it had the appropriate 1234567 profile, contained RD9, and was MtbC genotypic group 1b (by katG203, katG463, and gyrA95), but it bore the gyrBAf (6446G→T SNP, M. africanum subtype I) sequence for this amplicon. Until now, a gyrBΔ with the 6446G→T SNP has been described only for MtbC genotypic group 1a strains (24, 30), thereby suggesting that strain 15082 (if the gyrBΔ 6446G→T here is not simply a chance homologous mutation) is a remnant of the strain(s) that diverged from the preexisting MtbC lineage to become the group 1a MtbC subspecies (7). If so, then the gyrBΔ 6446G→T SNP arose prior to the fixation of the katG203 SNP and also before the accumulation of the known LSPs of the MtbC group 1a subspecies.
ADDENDUM
After submission of this paper, M. africanum subtype II strain 15082 was PCR tested for the TbD1 locus. As described recently, this locus is believed to be absent from “modern” M. tuberculosis strains but present in both “ancient” M. tuberculosis strains and MtbC genotypic group 1a strains (6). Strain 15082 yielded a DNA fragment for TbD1 by PCR that was determined to be of the appropriate nucleotide sequence. These data are in keeping with our hypothesis that this strain is positioned along the MtbC phylogenetic tree intermediate to the MtbC genotypic groups 1a and 1b.
Acknowledgments
We are indebted to Lee W. Riley, Sabine Ehrt, Walter Haas, Gaby Pfyffer, Barry Kreiswirth, Davise Larone, and Timothy Kiehn for providing vital bacterial isolates. We also thank Hongxia Zhu, Petra de Haas, Krishna Menon, Benjamin See, and Susan Mossarella for their time and effort in maintaining and preparing isolates for our use. We are also grateful to Barry Kreiswirth for critical appraisal of the manuscript and to Warren D. Johnson for trust and encouragement.
Funding support was provided by NIH grants R0-1 AI39606 and R0-1 HL61960 (to J.L.H.), an NIH Fogarty International Center Training grant (FICTG) (D43 TW00018), a grant from the Coordenação de Aperfeicoamento de Pessoal de Nivel Superior (CAPES) (Ministry of Education-Brazil), and a grant from the Laura Cook Hull Trust Fund (LCHTF) (Warren D. Johnson, principal investigator). R.C.H. was supported by the LCHTF, and L.C.D.O.L. was a FICTG and CAPES trainee. D.V.S. received funding from EC grant 200-0630 for the Molecular Epidemiology of Tuberculosis.
REFERENCES
- 1.Anonymous. 1998. Case records of the Massachusetts General Hospital. Weekly clinicopathological exercises. Case 29-1998. A 57-year-old man with fever and jaundice after intravesical instillation of bacille Calmette-Guérin for bladder cancer. N. Engl. J. Med. 339:831-837. [DOI] [PubMed] [Google Scholar]
- 2.Aranaz, A., E. Liebana, E. Gomez-Mampaso, J. C. Galan, D. Cousins, A. Ortega, J. Blazquez, F. Baquero, A. Mateos, G. Suarez, and L. Dominguez. 1999. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int. J. Syst. Bacteriol. 49:1263-1273. [DOI] [PubMed] [Google Scholar]
- 3.Aranaz, A., E. Liebana, A. Mateos, L. Dominguez, D. Vidal, M. Domingo, O. Gonzolez, E. F. Rodriguez-Ferri, A. E. Bunschoten, J. D. Van Embden, and D. Cousins. 1996. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J. Clin. Microbiol. 34:2734-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 5.Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann, S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito, B. G. Barrell, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunello, F., M. Ligozzi, E. Cristelli, S. Bonora, E. Tortoli, and R. Fontana. 2001. Identification of 54 mycobacterial species by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J. Clin. Microbiol. 39:2799-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentiles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jegels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Eisenach, K. D., J. T. Crawford, and J. H. Bates. 1986. Genetic relatedness among strains of the Mycobacterium tuberculosis complex. Analysis of restriction fragment heterogeneity using cloned DNA probes. Am. Rev. Respir. Dis. 133:1065-1068. [DOI] [PubMed] [Google Scholar]
- 11.Frothingham, R., H. G. Hills, and K. H. Wilson. 1994. Extensive DNA sequence conservation throughout the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 32:1639-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frothingham, R., P. L. Strickland, G. Bretzel, S. Ramaswamy, J. M. Musser, and D. L. Williams. 1999. Phenotypic and genotypic characterization of Mycobacterium africanum isolates from West Africa. J. Clin. Microbiol. 37:1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goh, K. S., E. Legrand, C. Sola, and N. Rastogi. 2001. Rapid differentiation of “Mycobacterium canettii” from other Mycobacterium tuberculosis complex organisms by PCR-restriction analysis of the hsp65 gene. J. Clin. Microbiol. 39:3705-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, S. V., B. Heym, J. Parkhill, B. Barrell, and S. T. Cole. 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology 145:881-892. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez, M., S. Samper, M. S. Jimenez, J. D. van Embden, J. F. Marin, and C. Martin. 1997. Identification by spoligotyping of a caprine genotype in Mycobacterium bovis strains causing human tuberculosis. J. Clin. Microbiol. 35:3328-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas, W. H., G. Bretzel, B. Amthor, K. Schilke, G. Krommes, S. Rusch-Gerdes, V. Sticht-Groh, and H. J. Bremer. 1997. Comparison of DNA fingerprint patterns of isolates of Mycobacterium africanum from east and west Africa. J. Clin. Microbiol. 35:663-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas, W. H., K. Schilke, J. Brand, B. Amthor, K. Weyer, P. B. Fourie, G. Bretzel, V. Sticht-Groh, and H. J. Bremer. 1997. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob. Agents Chemother. 41:1601-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland, S. M. 2001. Nontuberculous mycobacteria. Am. J. Med. Sci. 321:49-55. [DOI] [PubMed] [Google Scholar]
- 20.Horstkotte, M. A., I. Sobottka, C. K. Schewe, P. Schafer, R. Laufs, S. Rusch-Gerdes, and S. Niemann. 2001. Mycobacterium microti llama-type infection presenting as pulmonary tuberculosis in a human immunodeficiency virus-positive patient. J. Clin. Microbiol. 39:406-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jouanguy, E., F. Altare, S. Lamhamedi, P. Revy, J. F. Emile, M. Newport, M. Levin, S. Blanche, E. Seboun, A. Fischer, and J. L. Casanova. 1996. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N. Engl. J. Med. 335:1956-1961. [DOI] [PubMed] [Google Scholar]
- 22.Kallenius, G., T. Koivula, S. Ghebremichael, S. E. Hoffner, R. Norberg, E. Svensson, F. Dias, B. I. Marklund, and S. B. Svenson. 1999. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J. Clin. Microbiol. 37:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leclerc, M. C., N. Haddad, R. Moreau, and M. F. Thorel. 2000. Molecular characterization of environmental mycobacterium strains by PCR-restriction fragment length polymorphism of hsp65 and by sequencing of hsp65, and of 16S and ITS1 rDNA. Res. Microbiol. 151:629-638. [DOI] [PubMed] [Google Scholar]
- 27.Liebana, E., A. Aranaz, B. Francis, and D. Cousins. 1996. Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 34:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitarai, S., A. Kurashima, A. Tamura, H. Nagai, and H. Shishido. 2001. Clinical evaluation of Amplicor Mycobacterium detection system for the diagnosis of pulmonary mycobacterial infection using sputum. Tuberculosis 81:319-325. [DOI] [PubMed] [Google Scholar]
- 29.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 30.Niemann, S., D. Harmsen, S. Rusch-Gerdes, and E. Richter. 2000. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niemann, S., E. Richter, and S. Rusch-Gerdes. 2000. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis. J. Clin. Microbiol. 38:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemann, S., S. Rusch-Gerdes, M. L. Joloba, C. C. Whalen, D. Guwatudde, J. J. Ellner, K. Eisenach, N. Fumokong, J. L. Johnson, T. Aisu, R. D. Mugerwa, A. Okwera, and S. K. Schwander. 2002. Mycobacterium africanum subtype II is associated with two distinct genotypes and is a major cause of human tuberculosis in Kampala, Uganda. J. Clin. Microbiol. 40:3398-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Reilly, L. M., and C. J. Daborn. 1995. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber. Lung Dis. 76(Suppl. 1):1-46. [DOI] [PubMed] [Google Scholar]
- 34.Park, H., H. Jang, C. Kim, B. Chung, C. L. Chang, S. K. Park, and S. Song. 2000. Detection and identification of mycobacteria by amplification of the internal transcribed spacer regions with genus- and species-specific PCR primers. J. Clin. Microbiol. 38:4080-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662-667. [DOI] [PubMed] [Google Scholar]
- 37.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Bottger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sreevatsan, S., P. Escalante, X. Pan, D. A. Gillies II, S. Siddiqui, C. N. Khalaf, B. N. Kreiswirth, P. Bifani, L. G. Adams, T. Ficht, V. S. Perumaalla, M. D. Cave, J. D. van Embden, and J. M. Musser. 1996. Identification of a polymorphic nucleotide in oxyR specific for Mycobacterium bovis. J. Clin. Microbiol. 34:2007-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talbot, E. A., D. L. Williams, and R. Frothingham. 1997. PCR identification of Mycobacterium bovis BCG. J. Clin. Microbiol. 35:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. van Der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Soolingen, D., T. Hoogenboezem, P. E. de Haas, P. W. Hermans, M. A. Koedam, K. S. Teppema, P. J. Brennan, G. S. Besra, F. Portaels, J. Top, L. M. Schouls, and J. D. van Embden. 1997. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol. 47:1236-1245. [DOI] [PubMed] [Google Scholar]
- 44.van Soolingen, D., A. G. van der Zanden, P. E. de Haas, G. T. Noordhoek, A. Kiers, N. A. Foudraine, F. Portaels, A. H. Kolk, K. Kremer, and J. D. van Embden. 1998. Diagnosis of Mycobacterium microti infections among hu-mans by using novel genetic markers. J. Clin. Microbiol. 36:1840-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viana-Niero, C., C. Gutierrez, C. Sola, I. Filliol, F. Boulahbal, V. Vincent, and N. Rastogi. 2001. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J. Clin. Microbiol. 39:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]