Abstract
The Hox genes have been implicated as central to the evolution of animal body plan diversity. Regulatory changes both in Hox expression domains and in Hox-regulated gene networks have arisen during the evolution of related taxa, but there is little knowledge of whether functional changes in Hox proteins have also contributed to morphological evolution. For example, the evolution of greater numbers of differentiated segments and body parts in insects, compared with the simpler body plans of arthropod ancestors, may have involved an increase in the spectrum of biochemical interactions of individual Hox proteins. Here, we compare the in vivo functions of orthologous Ultrabithorax (Ubx) proteins from the insect Drosophila melanogaster and from an onychophoran, a member of a sister phylum with a more primitive and homonomous body plan. These Ubx proteins, which have been diverging in sequence for over 540 million years, can generate many of the same gain-of-function tissue transformations and can activate and repress many of the same target genes when expressed during Drosophila development. However, the onychophora Ubx (OUbx) protein does not transform the segmental identity of the embryonic ectoderm or repress the Distal-less target gene. This functional divergence is due to sequence changes outside the conserved homeodomain region. The inability of OUbx to function like Drosophila Ubx (DUbx) in the embryonic ectoderm indicates that the Ubx protein may have acquired new cofactors or activity modifiers since the divergence of the onychophoran and insect lineages.
Keywords: Hox genes, molecular evolution, onychophora, morphological diversity
The identification of highly conserved developmental regulatory genes across metazoan taxa has provided many new insights into the genetic basis of body plan evolution. One of the best-characterized gene families is the Hox gene family, which plays a conserved role in patterning regional identities along the antero–posterior (A/P) axis. These genes are best understood in insects and vertebrates (1) but have been described in many animal phyla (2, 3). All Hox genes encode a conserved DNA-binding domain, the homeodomain, and regulate the expression of an extensive set of target genes during development.
Comparative gene expression studies have revealed that Hox genes are deployed in similar body regions of animals with comparable body plans (4–8). However, there are clearly morphological differences between these regions, even when they express the same Hox gene. For example, the Hox gene Ultrabithorax (Ubx) is expressed in the hindwing of all insects, yet the size, shape, pattern, and function of insect hindwings differ markedly among species (6, 9, 10). Moreover, the origin of the Ubx gene predates the evolution of insects and their derived body plan (11, 12), and Ubx is expressed in other arthropods (4, 11–13) and in onychophora (12) in homonomous trunk segments. How, then, are the same (orthologous) Hox genes used in different lineages to regulate morphologically diverse body patterns?
Several models of Hox gene evolution have been proposed to explain changes in body plans. First, duplication and divergence of Hox genes expands the number of genetic regulators available for developmental diversification. This clearly occurred during chordate evolution, as the entire Hox complex was duplicated to create at least four clusters (14–16). However, the Hox gene family did not expand greatly in protostomes, as comparisons of several phyla indicate that most Hox genes predate the early common bilaterian ancestors (2). Second, changes in the regulation of Hox genes can underlie the evolution of body plan diversity (1, 10–13). The expression patterns of Hox genes along the body axis have shifted among related taxa. Both small and large alterations in the spatial and temporal dynamics of Hox gene expression patterns are correlated with morphological change (4, 11–13, 17). Third, regulatory evolution downstream of the Hox genes contributes to morphological evolution, as the sets of target genes regulated by a given Hox gene evolve independently in different lineages (10). Finally, sequence changes in Hox proteins and consequent alterations in biochemical function could also underlie the diversification of body patterns. Changes in DNA-binding specificity, in Hox protein interactions with cofactors, or in the posttranslational regulation of Hox protein activity could evolve in concert with more complex developmental roles for Hox proteins. There is, however, no evidence to date for the functional diversification of Hox orthologs.
The high sequence conservation of the homeodomain and the broad sets of target genes that are under Hox regulation suggest that Hox homeodomain sequence and function are highly constrained. Hox protein specificity has been localized primarily to residues within the homeodomain (18–24), and orthologous Hox proteins from different animals exhibit certain functional similarities to Drosophila Hox genes in vivo (24–30). However, if Hox proteins have evolved more complex functions as their developmental roles have diversified, a detailed assessment of orthologous Hox protein function requires the analysis of the effects of Hox protein expression in multiple tissue types and on multiple target genes. Also, the functional divergence of orthologs may be best identified between taxa that have comparable but qualitatively different body plans and similar sets of Hox genes (such that orthologs can be clearly identified and new gene duplications have not influenced ortholog function). These considerations led us to compare the activity of the onychophoran Ubx (OUbx) protein (12) with its ortholog in Drosophila (DUbx). The onychophora are a sister phylum of the arthropods, with a simpler body plan and less segmental diversity.
We have found that ectopic expression of OUbx in Drosophila can generate several similar tissue transformations and regulate some of the same target genes as DUbx, despite over 540 million years of sequence divergence. However, OUbx is not capable of generating one characteristic DUbx gain-of-function phenotype, the transformation of the embryonic thoracic ectoderm toward abdominal identity. We have mapped this functional difference to sequences outside the conserved homeodomain region. The divergence of Ubx protein function indicates that DUbx may have evolved new cofactor interactions or activity modifiers during the course of insect evolution.
Methods
Generation of Ubx Gain-of-Function Phenotypes.
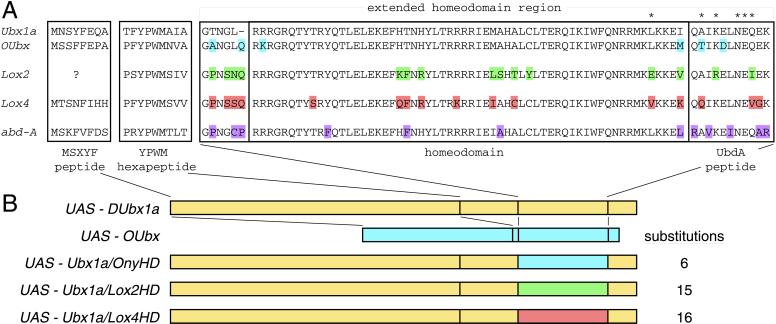
UAS-DUbx1a was previously described (31). The UAS-OUbx construct was made by splicing together the two predicted exons of onychophora Ubx (12) and cloning the predicted ORF into pUAST (32). The chimeric UAS-Ubx1a constructs were made by replacing the UAS-DUbx1a nucleotide sequence with the homologous sequence of OUbx, Lox2 (33), or Lox4 (34) within the conserved homeodomain region (see Fig. 1).
Figure 1.
Sequence similarity of Ubx-like Hox genes. (A) Alignment of regions with sequence similarity of Drosophila Ubx (GenBank accession no. P02834), onychophora Ubx (Acanthokara kaputensis, AAB92412), leech Lox2 (Helobdella robusta, A49130), leech Lox4 (Hirudo medicinalis, AAB35067), and Drosophila abd-A (P29555). Note that the amino-terminal peptide of Lox4 differs from previously published sequence (34), as independent sequencing of the Lox4 cDNA indicated an early frameshift and a different predicted protein sequence. Amino acid differences between DUbx and the other sequences are highlighted within the extended homeodomain. Asterisks mark the residues in the carboxyl-terminal tail of DUbx1a that have been mapped as the epitope of the cross-reactive monoclonal antibody FP6.87 (11). (B) Constructs used to overexpress Ubx orthologs and chimeras in Drosophila. Full-length constructs: UAS-DUbx1a (Drosophila melanogaster; ref. 31), UAS-OUbx (onychophora). Chimeric constructs: UAS-Ubx1a/OnyHD, UAS-Ubx1a/Lox2HD, UAS-Ubx1a/Lox4HD. In the chimeric constructs the extended homeodomain region of DUbx1a is replaced by the homologous sequences of OUbx, Lox2, and Lox4, respectively. The number of incorporated amino acid substitutions is indicated for each chimeric construct.
The Gal4 drivers used to overexpress Ubx orthologs and chimeric genes can be obtained from the Bloomington stock center. The following lines were used: dpp4A3 (antenna), MS1096 (wing), 24B (embryonic visceral mesoderm), and arm-Gal411 (embryonic ectoderm). To observe the activity of the target gene enhancers dpp674 (35) and Dll304 (36), stocks carrying dpp674; 24B and arm-Gal411; Dll304 were constructed. All crosses to generate ectopic expression phenotypes were conducted using virgin females carrying the Gal4 driver and males carrying an autosomal UAS responder.
Assays of Ubx Target Gene Expression and Tissue Transformation.
All antibody stainings were conducted as previously reported for embryos (4) and imaginal discs (9), by using a mouse monoclonal anti-β-galactosidase antibody (Promega), a rabbit anti-Dll antibody (4), a mouse monoclonal anti-Ubx/abd-A antibody (37), and a rat anti-serum response factor (SRF) antibody (38). Embryonic cuticles were prepared as described in ref. 39, and adult structures were mounted in Canada balsam.
Relative Levels of DUbx1a and OUbx Protein Overexpression.
One consideration in these experiments is the relative level of protein expression from the different UAS constructs. To assay OUbx activity, several independent UAS-OUbx P-element lines were tested for each phenotype. For the strongest OUbx lines, the levels of OUbx protein overexpression driven by the MS1096 driver in wing imaginal discs were compared with DUbx1a by using the cross-reactive monoclonal antibody FP6.87 (37). However, this antibody may have lower affinity for the OUbx protein, as there are amino acid substitutions in the region of the epitope recognized by FP6.87 (refs. 11 and 12; see Fig. 1A). This comparison may therefore underestimate the level of OUbx protein expression. Male larvae hemizygous for the MS1096 driver and carrying one copy of either the DUbx or OUbx construct were compared to control for the level of Gal4 activation.
Results and Discussion
OUbx Can Transform the Identity of Drosophila Appendages.
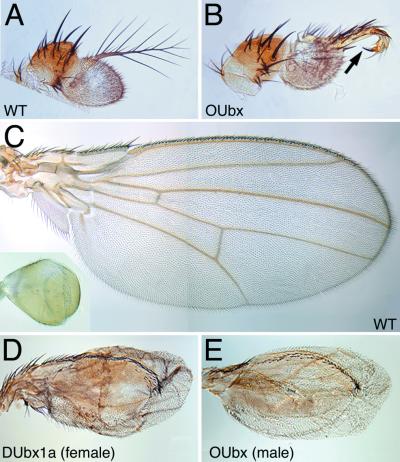
The OUbx and DUbx proteins share little sequence similarity outside the conserved homeodomain region (Fig. 1A). To compare the biochemical activities of these two proteins in vivo, we have used the Gal4-UAS system (32) to overexpress OUbx in Drosophila (Fig. 1B). It has been shown (18, 40) that ectopic expression of DUbx1a, an isoform of DUbx, can transform anterior appendages toward the identity of the corresponding (homologous) appendage in the third thoracic segment (i.e., the T3 leg and the haltere). These phenotypes represent a change in fate of multiple cell types, coordinated through the regulation of many Ubx target genes. We have found that overexpression of OUbx in the antenna can transform this appendage toward leg identity, as seen by the transformation of the distal antennal segment, the arista, into tarsae and claw (Fig. 2 A and B). This transformation is presumably achieved in part by means of the repression of homothorax in the antenna (41). Overexpression of OUbx in the wing imaginal disc can transform the adult wing toward a haltere, characterized by the reduction in size, the induction of a balloon-like shape, and the loss of veins. The degree of transformation observed is similar to the effect of overexpressing DUbx1a (see Fig. 2 C–E).
Figure 2.
Transformation of adult tissues by overexpression of Ubx orthologs. (A) Wild-type adult Drosophila antenna. (B) Antenna transformed toward a thoracic leg by ectopic expression of OUbx, under the control of the dpp4A3-Gal4 driver. The distal antennal segment is transformed into distal leg, ending in a claw (arrow). (C) Wild-type adult Drosophila wing and haltere (Inset). (D) Adult wing transformed toward a haltere by ectopic expression of DUbx1a under the control of the X-linked MS1096-Gal4 driver. This wing is from a female; male siblings hemizygous for MS1096 do not survive larval stages. (E) Adult wing transformed toward a haltere by ectopic expression of OUbx under the control of MS1096. This wing is from a male; female siblings show a less severe transformation. Thus, a higher level of Gal4 is required to generate the strong OUbx transformation. The transformations in D and E are stronger on the dorsal surface of the wing, consistent with higher levels of MS1096-Gal4 expression dorsally (40, 52), and do not seem to interfere with the development of the wing margin bristles.
DUbx regulation of haltere development in Drosophila depends on the repression of several genes required for wing development (42), including the Drosophila ortholog of the SRF gene. This gene is required for proper development of intervein tissue in the wing and the apposition of dorsal and ventral surfaces to generate a flat wing blade. Expression of SRF is repressed by DUbx in the haltere pouch, and ectopic expression of DUbx in the wing pouch is sufficient to repress SRF (42). We have tested whether the ability of OUbx to transform the Drosophila wing toward haltere fate is accompanied by repression of SRF in the wing imaginal disc. We find that the level of SRF protein expression is reduced, particularly in the dorsal compartment, where Gal4 expression is highest (Fig. 3 A–C). Thus, OUbx can transform a field of tissue in the wing disc in a manner similar to DUbx1a, including the repression of a gene known to be downstream of DUbx.
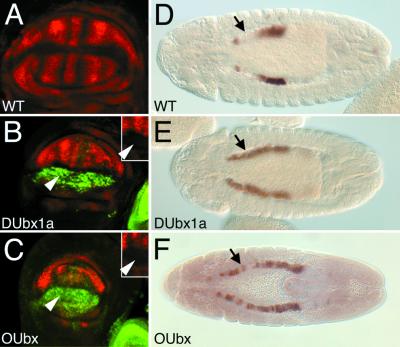
Figure 3.
OUbx regulates target genes in the Drosophila wing and in the embryonic midgut. (A) Wild-type expression of SRF in the pouch region of the wing imaginal disc. (B and C) Expression of SRF (red) and Ubx (green) in the wing pouch where ectopic expression of DUbx1a (B) or OUbx (C) is driven by MS1096-Gal4. SRF expression is repressed, particularly in the dorsal compartment where ectopic DUbx1a (B) or OUbx (C) expression is high (arrowheads). Insets show the single channel of SRF expression in the center of the wing pouch, in the region of the arrowhead in the larger panel. In the lower right corner of B and C the edge of another imaginal disc from the third thoracic segment of the same animal is visible, indicating that the levels of ectopic protein expression in B and C are comparable to each other and to endogenous DUbx protein expression. (D) Wild-type expression of the dpp674 embryonic midgut enhancer in PS7 of the visceral mesoderm in a stage-13 embryo. dpp674 also drives expression in the gastric caeca at the anterior of the midgut independent of Ubx regulation. The gap between these expression domains is indicated by an arrow. Expression of DUbx1a (E) or OUbx (F) driven by 24B-Gal4 causes ectopic expression of dpp674 anterior to PS7 (arrow). The OUbx phenotype is typically less strong than DUbx1a, though the ectopic activation of dpp674 generated by either Ubx protein is often patchy.
OUbx Can Activate dpp, a Specific Target of DUbx in the Embryo.
The adult homeotic transformations of antenna toward leg and of wing toward haltere are not unique to Ubx, as other trunk Hox genes in Drosophila are able to generate similar tissue transformations (19, 26, 40). However, there are gain-of-function phenotypes that are specific to Ubx function in the embryo. To test whether the OUbx protein can generate a Ubx-specific ectopic expression phenotype, we characterized the ability of OUbx to activate a known embryonic DUbx target gene, the signaling molecule decapentaplegic (dpp), in the visceral mesoderm of the developing midgut (43). The cis-regulatory enhancer that controls dpp expression in the midgut has been well characterized as a direct target of DUbx (35, 44). The dpp674 enhancer is activated by DUbx in parasegment 7, is repressed by Abdominal-A (Abd-A) in parasegments 8–12, and does not respond to other trunk Hox genes (35). Overexpression of DUbx1a throughout the visceral mesoderm is sufficient to activate the dpp674 enhancer anterior to its endogenous domain in parasegment 7, but not to override the repression by Abd-A in parasegments 8–12 (ref. 35, see Fig. 3 D and E).
Ectopic expression of OUbx in the visceral mesoderm also activates dpp674, as seen by expansion anterior to parasegment 7 (Fig. 3F). OUbx activation of dpp674 is not dependent on the endogenous DUbx locus, as this phenotype arises in homozygous Ubx6.28 null mutant embryos (data not shown). Ubx activation of dpp674 requires the Hox cofactor Extradenticle (Exd) (45), and the ability of OUbx to generate this phenotype in a DUbx-null background indicates that OUbx can interact with the Drosophila Exd protein.
OUbx Does Not Transform Embryonic Ectodermal Segmental Identity.
Another Drosophila gain-of-function homeotic phenotype specific to Ubx is the transformation of embryonic thoracic cuticle toward abdominal fate. The ventral denticle belts of the cuticle represent a read-out of the development of the underlying epidermal cells of the embryo (Fig. 4A), and the denticle belt pattern of the first abdominal segment (A1) is unique and is regulated by DUbx. Ectopic expression of DUbx1a in the ectoderm transforms the denticle belts of thoracic segments toward A1 identity (refs. 18 and 31; see Fig. 4 A–C). Also, the development of Keilin's organs and ventral pits, which arise in bilateral pairs in each of the thoracic segments of the cuticle, is repressed by ectopic expression of DUbx1a (ref. 18; Fig. 4C).
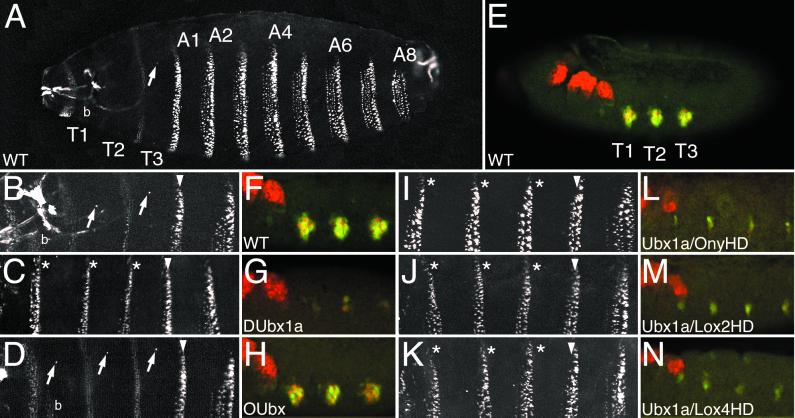
Figure 4.
OUbx does not transform embryonic ectoderm segmental identity or repress the Dll target gene. (A and B) Wild-type embryonic cuticle (A) and higher magnification of thorax and anterior abdomen (B). (C) Cuticle transformation of thoracic segments toward A1 fate when ectopic DUbx1a is expressed under the control of armadillo-Gal4. Transformed denticle belts are indicated with asterisks. (D) Ectopic expression of OUbx does not transform the thoracic denticle belts toward A1 fate, nor does it repress the development of Keilin's organs or ventral pits (arrows). (E–H) Expression of Dll protein (red) and the Dll304 early thoracic limb enhancer (green) in wild-type (E and F) and transgenic (G and H) embryos. F–H show higher magnification of thoracic segments. (G) Ectopic DUbx1a represses expression of Dll protein and the Dll304 enhancer in the thorax. Some residual expression is typically seen when DUbx is overexpressed (31). (H) Ectopic expression of OUbx does not affect Dll protein or Dll304 expression. (I–K) Ectopic expression of Ubx1a chimeric genes strongly transforms head and thoracic segments toward A1 identity. In I, J, and K, respectively, Ubx1a/OnyHD, Ubx1a/Lox2HD, and Ubx1a/Lox4HD transformed denticle belts are indicated with asterisks. (L–N) Ectopic expression of Ubx1a chimeric genes represses expression of Dll protein (red) and the Dll304 early limb enhancer (green) in the thorax. (L) Ubx1a/OnyHD. (M) Ubx1a/Lox2HD. (N) Ubx1a/Lox4HD. In A and E, T1, T2, T3 are thoracic segments; A1–A8 are abdominal segments; in A, B, and D, b indicates the T1 denticle beard. In B–D and I–K, the A1 segment is indicated with an arrowhead, transformed thoracic segments are marked with asterisks, and arrows point to ventral pits (when present).
We were surprised to find that expression of OUbx in the embryonic ectoderm does not transform thoracic segmental identity in the cuticle. The thoracic denticle belts appear wild-type and Keilin's organs and ventral pits are present (Fig. 4D). One DUbx target gene that is affected by the transformation of thoracic segment identity is Distal-less (Dll). Dll expression in thoracic limb primordia is required for the development of Keilin's organs (46), and Dll expression is normally repressed by DUbx (and Abd-A) in the Drosophila abdomen (36). An early thoracic limb enhancer of Dll, Dll304, has been shown to be a direct target of DUbx (36). Dll304 activity and Dll protein expression are reduced or eliminated by ectopic expression of DUbx1a, consistent with the absence of Keilin's organs in the cuticle of these embryos (see Fig. 4 E–G). The activity of this enhancer and the expression of Dll protein are not altered in embryos in which OUbx is ectopically expressed (Fig. 4H).
The Functional Divergence of DUbx1a and OUbx Maps Outside the Homeodomain.
The difference in the activity of DUbx1a and OUbx proteins in the embryonic ectoderm could reflect functional divergence of these orthologs. The sequence similarity of these proteins is limited to a few short peptide motifs and the region including the homeodomain and flanking residues (Fig. 1A). There may be other functional elements of DUbx that are not conserved in OUbx, such as protein–protein interaction domains that are required for DUbx cofactors or regions that modify the ability of DUbx to act as a repressor or an activator. Alternatively, the few sequence changes within the conserved regions may be responsible for the functional divergence of DUbx1a and OUbx. In the conserved homeodomain region, there are six differences in the amino acid sequence of DUbx and OUbx (Fig. 1A). To test whether these changes are responsible for the functional divergence of DUbx1a and OUbx, we created a chimeric Ubx gene based on DUbx1a but with the onychophoran sequence in and near the homeodomain. We also built similar chimeras that have more substitutions in the conserved homeodomain region, based on the sequence of the Ubx-like leech genes Lox2 (33) and Lox4 (34) (Fig. 1).
We repeated all of the adult ectopic Ubx gain-of-function assays with each of the Ubx1a chimeras, and in all cases these chimeras generate the same phenotypes as DUbx1a (Table 1). When ectopically expressed in the embryonic ectoderm, all of the Ubx1a chimeras can transform the cuticular identity of thoracic segments, including transformation of the denticle belts to A1 identity and repression of Keilin's organs and ventral pits (Fig. 4 I–K). Expression of both Dll protein and the Dll304 early limb enhancer in these embryos is reduced or absent (Fig. 4 L–N). The strong transforming activity of each of the Ubx1a chimeras in the embryonic ectoderm indicates that the amino acid differences between DUbx and OUbx within the conserved homeodomain region are not solely responsible for the different activities of DUbx1a and OUbx proteins. Thus, the functional divergence of DUbx1a and OUbx resides in sequences outside the homeodomain.
Table 1.
Ectopic Ubx expression phenotypes
| UAS construct | Tissue transformations
|
Target genes
|
||||
|---|---|---|---|---|---|---|
| Antenna to leg | Wing to haltere | Thorax to A1 | Repress SRF | Activate dpp | Repress Dll | |
| DUbx1a | +++ | +++ | +++ | +++ | +++ | +++ |
| OUbx | +++ | ++ | − | ++ | ++ | − |
| abd-A* | +++ | +++ | (To A2–A8) | ND | (Repress) | +++ |
| Ubx1a/OnyHD | +++ | +++ | +++ | ND | +++ | +++ |
| ubx1a/Lox2HD | +++ | +++ | +++ | ND | +++ | +++ |
| Ubx1a/Lox4HD | +++ | +++ | +++ | ND | +++ | +++ |
Evolution of Ubx Protein Activity.
We have shown that OUbx can mimic DUbx gain-of-function adult tissue transformations, can both activate and repress downstream genes, and can apparently interact with the Hox cofactor Exd. These shared properties of OUbx and DUbx reflect biochemical activities of Ubx that have been conserved since the common ancestor of onychophora and Drosophila. However, we have found one tissue where DUbx and OUbx have different activities, as ectopic OUbx does not transform embryonic segmental identity in Drosophila. Since the amino acid differences between OUbx and DUbx in the conserved homeodomain region are not responsible for the different behavior of these proteins, we suggest that a new functional element may have evolved in the DUbx protein that is required for interaction with a cofactor or for activity regulation.
In addition to the homeodomain, regions of the DUbx amino-terminal exon that contain conserved peptide motifs (Fig. 1A) contribute to the functional specificity of DUbx (18). Much of the remainder of the DUbx protein is not required for transformation of the ectoderm, including the DUbx microexons, most of the amino-terminal exon, and the carboxyl-terminal tail (18, 22). While OUbx does contain these conserved motifs (Fig. 1A), sequence divergence in or near these regions may be responsible for the difference in OUbx function in Drosophila. Interestingly, the behavior of chimeric (19, 22) or altered (47) Hox proteins indicates that Ubx regulation of denticle belt patterning is separable from repression of Dll expression and subsequent development of Keilin's organs, and may require different regions of the DUbx protein. Thus, OUbx may lack more than one key functional element required for Ubx transformation of the Drosophila ectoderm.
This comparison of Ubx protein activity highlights the importance of understanding the biochemical mechanisms by which DUbx regulates target genes during development. While the specificity of homeodomain binding is key to Hox protein function, Hox activity is also modulated by the regulation of activation states (47–49) and by cofactor interactions (50). Exd is the only Ubx cofactor that has been well studied, but it is not required in all cells and structures that are patterned by DUbx (51). Our study suggests that additional DUbx cofactors remain to be discovered.
Ubx Evolution Contributes to the Segmental Diversification of the Arthropod Body Plan.
Functional differences between Ubx orthologs may reflect the evolving role of Ubx in regulating segmental diversity in arthropods. The common ancestor of arthropods and onychophora likely had a homonomous trunk with little segmental diversity. The subsequent course of arthropod evolution is marked by a pronounced increase in the number of distinct segment types. In the insects, Ubx regulates several differences between trunk segments, such as changes in the morphology and placement of appendages. Some of these new developmental roles have evolved through downstream changes in Ubx regulation of target genes, and they required no apparent modification to Ubx protein function. For example, morphological differences between the Drosophila wing and haltere may primarily reflect the evolution of Ubx binding sites in the cis-regulatory enhancers of wing patterning genes (10).
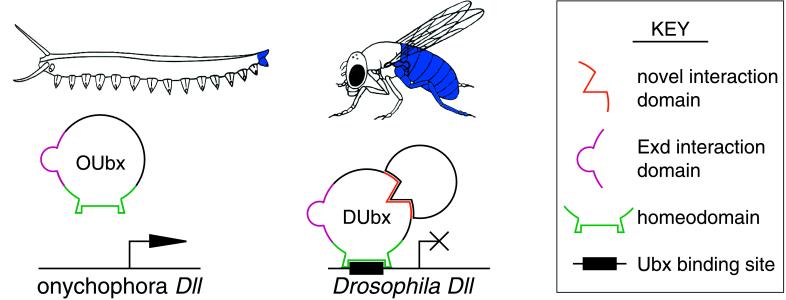
In contrast, other developmental roles for Ubx may have evolved in concert with functional evolution of the Ubx protein. Segmental diversification of the embryonic ectoderm may reflect the evolution of novel functional elements in the DUbx protein that are required for Ubx regulation of target genes in this tissue. The evolution of DUbx regulation of Dll in insects provides one example. Ubx repression of Dll expression likely evolved during insect evolution, as this interaction is not observed in other arthropods or onychophora, where Ubx and Dll proteins are coexpressed in trunk limbs during early stages of development (4, 11, 12). We propose that the evolution of Dll repression in insects involved changes both in the biochemical activity of the Ubx protein and in the Dll cis-regulatory limb enhancer (Fig. 5). Thus, in addition to evolutionary changes in the Hox-regulated network of target genes, changes in Hox protein sequence and function have contributed to morphological evolution.
Figure 5.
Evolution of Ubx regulation of Dll in the insect lineage. (Upper) The expression domains of Ubx are different in the onychophoran (Left) and Drosophila (Right). OUbx is expressed on the extreme posterior of the onychophora, overlapping the expression of Dll in the terminal limb of the homonomous trunk. In contrast, DUbx is expressed in the third thoracic segment of Drosophila, where it patterns the haltere and the T3 leg, and throughout much of the Drosophila abdomen, where it represses Dll expression and limb development. (Lower) Ubx repression of Dll in the insect abdomen may have evolved because of changes in Ubx protein function and in cis-regulatory sequences of the Dll target gene. The OUbx protein (Left) shares some functional elements with DUbx (Right), including the homeodomain and the Exd interaction domain. However, evolutionary modifications to Ubx protein function in the insect lineage may have created a novel cofactor interaction domain (shown in orange) or a novel posttranslational regulation of Ubx activity required for Ubx repression of Dll. Ubx regulation of Dll in the insect lineage also involved the evolution of Ubx binding sites in the Dll early limb enhancer.
Acknowledgments
We thank Michael Akam, Juan Botas, Maria Capovilla, Georg Halder, and Grace Panganiban for Drosophila stocks and reagents, Victoria Wong and Marty Shankland for leech Hox cDNAs, Georg Halder and Craig Nelson for their comments on the manuscript, and Victoria Kassner, Kathy Vacarro, and Jamie Wilson for excellent technical support. J.K.G. is a Predoctoral Fellow and S.B.C. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- OUbx
onychophoran Ubx
- DUbx
Drosophila Ubx
- SRF
serum response factor
- dpp, decapentaplegic
Dll, Distal-less
References
- 1.Carroll S. Nature (London) 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 2.de Rosa R, Grenier J K, Andreeva T, Cook C, Adoutte A, Akam M, Carroll S B, Balavoine G. Nature (London) 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- 3.Finnerty J R, Martindale M Q. Curr Opin Genet Dev. 1998;8:681–687. doi: 10.1016/s0959-437x(98)80037-3. [DOI] [PubMed] [Google Scholar]
- 4.Panganiban G, Sebring A, Nagy L, Carroll S B. Science. 1995;270:1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- 5.Palopoli M F, Patel N H. Curr Biol. 1998;8:587–590. doi: 10.1016/s0960-9822(98)70228-3. [DOI] [PubMed] [Google Scholar]
- 6.Warren R, Nagy L, Selegue J, Gates J, Carroll S. Nature (London) 1994;372:458–461. doi: 10.1038/372458a0. [DOI] [PubMed] [Google Scholar]
- 7.Burke A C, Nelson C E, Morgan B A, Tabin C. Development (Cambridge, UK) 1995;121:333–346. doi: 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- 8.Cohn M J, Tickle C. Nature (London) 1999;399:474–479. doi: 10.1038/20944. [DOI] [PubMed] [Google Scholar]
- 9.Carroll S, Weatherbee S, Langeland J. Nature (London) 1995;375:58–61. doi: 10.1038/375058a0. [DOI] [PubMed] [Google Scholar]
- 10.Weatherbee S D, Nijhout H F, Grunert L W, Halder G, Galant R, Selegue J, Carroll S. Curr Biol. 1999;9:109–115. doi: 10.1016/s0960-9822(99)80064-5. [DOI] [PubMed] [Google Scholar]
- 11.Averof M, Akam M. Nature (London) 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- 12.Grenier J, Garber T, Warren R, Whitington P, Carroll S. Curr Biol. 1997;7:547–553. doi: 10.1016/s0960-9822(06)00253-3. [DOI] [PubMed] [Google Scholar]
- 13.Averof M, Patel N H. Nature (London) 1997;388:682–686. doi: 10.1038/41786. [DOI] [PubMed] [Google Scholar]
- 14.Pendleton J, Nagai B, Murtha M, Ruddle F. Proc Natl Acad Sci USA. 1993;90:6300–6304. doi: 10.1073/pnas.90.13.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holland P W, Garcia-Fernandez J. Dev Biol. 1996;173:382–395. doi: 10.1006/dbio.1996.0034. [DOI] [PubMed] [Google Scholar]
- 16.Amores A, Force A, Yan Y L, Joly L, Amemiya C, Fritz A, Ho R K, Langeland J, Prince V, Wang Y L, et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 17.Stern D L. Nature (London) 1998;396:463–466. doi: 10.1038/24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mann R S, Hogness D S. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- 19.Gibson G, Schier A, LeMotte P, Gehring W J. Cell. 1990;62:1087–1103. doi: 10.1016/0092-8674(90)90386-s. [DOI] [PubMed] [Google Scholar]
- 20.Lin L, McGinnis W. Genes Dev. 1992;6:1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- 21.Zeng W, Andrew D J, Mathies L D, Horner M A, Scott M P. Development (Cambridge, UK) 1993;118:339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]
- 22.Chan S K, Mann R S. Genes Dev. 1993;7:796–811. doi: 10.1101/gad.7.5.796. [DOI] [PubMed] [Google Scholar]
- 23.Furukubo-Tokunaga K, Flister S, Gehring W J. Proc Natl Acad Sci USA. 1993;90:6360–6364. doi: 10.1073/pnas.90.13.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann R. BioEssays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- 25.Malicki J, Schughart K, McGinnis W. Cell. 1990;63:961–967. doi: 10.1016/0092-8674(90)90499-5. [DOI] [PubMed] [Google Scholar]
- 26.McGinnis N, Kuziora M A, McGinnis W. Cell. 1990;63:969–976. doi: 10.1016/0092-8674(90)90500-e. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J J, Lazzarini R A, Pick L. Genes Dev. 1993;7:343–354. doi: 10.1101/gad.7.3.343. [DOI] [PubMed] [Google Scholar]
- 28.Hunter D P, Kenyon C. Nature (London) 1995;377:229–232. doi: 10.1038/377229a0. [DOI] [PubMed] [Google Scholar]
- 29.Zhao J J, Lazzarini R A, Pick L. EMBO J. 1996;15:1313–1322. [PMC free article] [PubMed] [Google Scholar]
- 30.Brown S, Holtzman S, Kaufman T, Denell R. Dev Genes Evol. 1999;209:389–398. doi: 10.1007/s004270050269. [DOI] [PubMed] [Google Scholar]
- 31.Castelli-Gair J, Greig S, Micklem G, Akam M. Development (Cambridge, UK) 1994;120:1983–1985. doi: 10.1242/dev.120.7.1983. [DOI] [PubMed] [Google Scholar]
- 32.Brand A, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 33.Nardelli-Haefliger D, Shankland M. Development (Cambridge, UK) 1992;116:697–710. doi: 10.1242/dev.116.3.697. [DOI] [PubMed] [Google Scholar]
- 34.Wong V, Aisemberg G, Wen-Biao G, Macagno E. J Neurosci. 1995;15:5551–5559. doi: 10.1523/JNEUROSCI.15-08-05551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capovilla M, Brandt M, Botas F. Cell. 1994;76:461–475. doi: 10.1016/0092-8674(94)90111-2. [DOI] [PubMed] [Google Scholar]
- 36.Vachon G, Cohen B, Pfeifle C, McGuffin M, Botas J, Cohen S. Cell. 1992;71:437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]
- 37.Kelsh R, Weinzierl R, White R, Akam M. Dev Genet. 1994;15:19–31. doi: 10.1002/dvg.1020150104. [DOI] [PubMed] [Google Scholar]
- 38.Affolter M, Montagne J, Walldorf U, Groppe G, Kloter U, LaRosa M, Gehring W J. Development (Cambridge, UK) 1994;120:743–753. doi: 10.1242/dev.120.4.743. [DOI] [PubMed] [Google Scholar]
- 39.Roberts D B. Drosophila: A Practical Approach. New York: Oxford Univ. Press; 1998. [Google Scholar]
- 40.Casares F, Calleja M, Sanchez-Herrero E. EMBO J. 1996;15:3934–3942. [PMC free article] [PubMed] [Google Scholar]
- 41.Yao L C, Liaw G J, Pai C Y, Sun Y H. Dev Biol. 1999;211:268–276. doi: 10.1006/dbio.1999.9309. [DOI] [PubMed] [Google Scholar]
- 42.Weatherbee S, Halder G, Hudson A, Kim J, Carroll S. Genes Dev. 1998;10:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuter R, Panganiban G E, Hoffmann F M, Scott M P. Development (Cambridge, UK) 1990;110:1031–1040. doi: 10.1242/dev.110.4.1031. [DOI] [PubMed] [Google Scholar]
- 44.Manak J R, Mathies L S, Scott M P. Development (Cambridge, UK) 1994;120:3605–3619. doi: 10.1242/dev.120.12.3605. [DOI] [PubMed] [Google Scholar]
- 45.Rauskolb C, Wieschaus E. EMBO J. 1994;13:3561–3569. doi: 10.1002/j.1460-2075.1994.tb06663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen S M, Jurgens G. EMBO J. 1989;8:2045–2055. doi: 10.1002/j.1460-2075.1989.tb03613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, McGinnis W. Proc Natl Acad Sci USA. 1999;96:6802–6807. doi: 10.1073/pnas.96.12.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Murre C, McGinnis W. EMBO J. 1999;18:198–211. doi: 10.1093/emboj/18.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaffe L, Ryoo H D, Mann R S. Genes Dev. 1997;11:1327–1340. doi: 10.1101/gad.11.10.1327. [DOI] [PubMed] [Google Scholar]
- 50.Johnson F B, Parker E, Krasnow M A. Proc Natl Acad Sci USA. 1995;92:739–743. doi: 10.1073/pnas.92.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.González-Crespo S, Morata G. Development (Cambridge, UK) 1995;121:2117–2125. doi: 10.1242/dev.121.7.2117. [DOI] [PubMed] [Google Scholar]
- 52.Capdevilla J, Guerrero I. EMBO J. 1994;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]