Abstract
The human immunodeficiency virus type 1 initially assembles and buds as an immature particle that is organized by the viral Gag polyprotein. Gag is then proteolyzed to produce the smaller capsid protein CA, which forms the central conical capsid that surrounds the RNA genome in the mature, infectious virus. To define CA surfaces that function at different stages of the viral life cycle, a total of 48 different alanine-scanning surface mutations in CA were tested for their effects on Gag protein expression, processing, particle production and morphology, capsid assembly, and infectivity. The 27 detrimental mutations fall into three classes: 13 mutations significantly diminished or altered particle production, 9 mutations failed to assemble normal capsids, and 5 mutations supported normal viral assembly but were nevertheless reduced more than 20-fold in infectivity. The locations of the assembly-defective mutations implicate three different CA surfaces in immature particle assembly: one surface encompasses helices 4 to 6 in the CA N-terminal domain (NTD), a second surrounds the crystallographically defined CA dimer interface in the C-terminal domain (CTD), and a third surrounds the loop preceding helix 8 at the base of the CTD. Mature capsid formation required a distinct surface encompassing helices 1 to 3 in the NTD, in good agreement with a recent structural model for the viral capsid. Finally, the identification of replication-defective mutants with normal viral assembly phenotypes indicates that CA also performs important nonstructural functions at early stages of the viral life cycle.
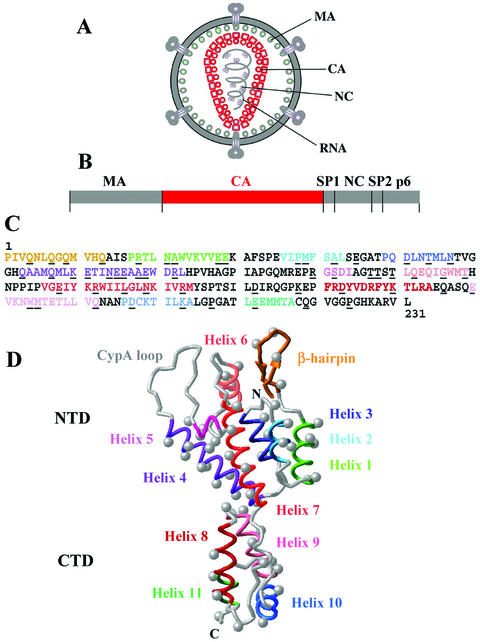
Several stages of the retroviral life cycle, including virion assembly, budding, maturation, entry, uncoating, and replication, occur within large, multicomponent particles that are organized by the structural Gag polyprotein or its proteolytic products (see references 31, 41, 54, 79, 85, 90 and 97 for reviews). Late in the infectious cycle, unprocessed human immunodeficiency virus type 1 (HIV-1) Gag molecules assemble beneath the plasma membrane and then bud from the cell as immature, enveloped virions. These immature particles are roughly spherical but lack global symmetry elements and exhibit a range of diameters (∼100 to 140 nm), indicating that they are not strictly regular objects (33, 96, 99, 101). The rod-shaped Gag molecules associate on the membrane with their long axes projecting radially toward the center of the immature virion. Each HIV-1 Gag molecule is composed of a series of distinct domains (denoted MA, CA, NC, and p6, from the N to C terminus), as well as two small spacer peptides (SP1 and SP2) (Fig. 1B). The myristoylated MA domain associates with the membrane, and the CA and NC polypeptides form shells of successively smaller radii (96). Although the lateral interactions between adjacent Gag molecules in the immature particle lattice are not yet understood in detail, there are indications that the lattice may be constructed from Gag trimers (46, 66, 74).
FIG. 1.
Structure and assembly of the HIV-1 CA protein. (A) Locations of the Gag-derived proteins in the mature HIV-1 virion. The conical capsid is highlighted in red. (B) The domain structure of the HIV-1 Gag polyprotein, with the CA polypeptide highlighted in red. (C) HIV-1NL4-3 CA amino acid sequence. Secondary structures are color-coded as shown in Fig. 1D and described in the corresponding legend. Residues mutated in this study and in reference 92 are underlined. (D) Structural model for the HIV-1 CA protein based upon crystal structures of the protein's NTD and CTD. Mutated residues are shown as gray balls (at the Cβ positions) and secondary structures are coded as follows. Orange, β-hairpin; light green, helix 1; turquoise, helix 2; dark blue, helix 3; purple, helix 4; gray, cyclophilin A-binding loop (CypA loop); pink, helix 5; rose, helix 6; red, helix 7; dark red, helix 8; salmon, helix 9; blue, helix 10; and green, helix 11. A short 310 helix in the CTD (CA residues 149 to 152) is not highlighted, and the disordered C-terminal 11 amino acids (including P224) are not shown.
As the virus buds, HIV-1 Gag is sequentially processed at five different sites by the viral protease, releasing the MA, CA, and NC polypeptides as discrete proteins. This triggers a major structural rearrangement (maturation) in which NC and the RNA genome condense into a complex at the center of the virion, CA assembles into a conical shell (the capsid) that surrounds the NC/RNA complex, and MA remains associated with the viral membrane (Fig. 1A). Maturation is required for infectivity and presumably converts the virion from a particle that can assemble and bud into a particle that can disassemble and replicate in a new host cell (94).
Previous studies of retroviral Gag proteins have revealed the involvement of several distinct elements at different stages of immature particle production (31, 41, 54, 79, 85, 90, 97). The major signal(s) for trafficking and retention of HIV-1 Gag at the plasma membrane (the M domain), reside within MA (26, 68, 70, 80, 104), and signal(s) required for late stages of particle release (the L domain) are located within the p6 polypeptide (14, 32, 42, 60, 72). In contrast, the regions of Gag that participate in spherical particle assembly appear to be larger and more complex, presumably because Gag proteins associate along their lengths and may therefore make multiple contacts with neighboring molecules. Nevertheless, genetic analyses have revealed that the most important determinant(s) for Gag assembly span the C-terminal domain of CA, SP1, and NC (which contains the I, or interaction, domain) (31, 41). The I domain within NC appears to facilitate assembly primarily by tethering Gag molecules together via nonspecific RNA binding interactions (10, 18, 19, 22, 102).
High-resolution structures of the mature HIV-1 MA, CA, and NC proteins have been determined (reviewed in reference 89), and the structures of these proteins appear to be conserved across retroviruses. In particular, CA proteins from HIV-1 (4, 34, 38, 64), equine infectious anemia virus (47), human T-cell leukemia virus type I (20, 49), and Rous sarcoma virus (13, 51) all adopt similar two-domain structures, with their helical N- and C-terminal domains (NTD and CTD) connected by a short flexible linker. The HIV-1 CA NTD is composed of an N-terminal β-hairpin, seven α-helices, and an extended loop, which create a wedge-shaped molecule with overall dimensions of 45 by 35 by 18 Å (Fig. 1C and D). The cellular protein cyclophilin A binds to the extended loop formed by residues 85 to 93 (34) and is thereby incorporated into the assembling virus. CA residues S146 and P147 act as a flexible linker that connects the NTD with a smaller CTD composed of four α-helices (Fig. 1D, bottom). The CTD dimerizes in solution (35, 78) and in the crystal (35), and the dimer is created by parallel packing of helix 9 against its symmetry-related mate (35, 98). The CTD also contains an essential, highly conserved stretch of 20 amino acids (the major homology region [MHR]) that is found in all retroviruses and in the related yeast transposon Ty3 (8, 21, 61, 69, 82, 97). Finally, the C-terminal 11 residues of the HIV-1 CA CTD are disordered, and equivalent C-terminal tails are also disordered in other mature retroviral CA proteins.
In vitro, purified HIV-1 CA proteins can spontaneously assemble into helical tubes and cones that appear to recapitulate the CA-CA interactions of authentic viral capsids (12, 25, 36, 45, 57). Cryoelectron microscopy reconstructions have revealed that the helical tubes are composed of CA hexamers, and docking studies suggest a model in which the CA NTD forms hexameric rings while the CA CTD makes dimeric contacts that connect each ring to its six nearest neighbors (57). We have proposed that conical lentiviral capsids assemble on analogous hexagonal nets, following the principles of a fullerene cone (24, 36, 47, 57). Thus, these studies have provided a low-resolution model for the body of the HIV-1 capsid that can be tested by other experimental methods.
Several known HIV-1 CA functions have been analyzed by mutagenesis. For example, extensive mutational analyses of the binding site for human cyclophilin A have defined the CA sequences required for cyclophilin A packaging (9, 30, 88, 100). Similarly, mutational analyses of the MHR have demonstrated the importance of this element for both viral assembly and reverse transcription (3, 11, 21, 61, 82). In addition to these directed studies, several global mutational analyses of HIV-1 CA have also been reported (23, 28, 75, 76, 81). These studies have been very important for defining various CA functions and mapping them to the NTD and CTD. However, a limitation of some of the published studies is that they utilized deletion (23, 81), insertion (75, 76), or proline-scanning (28) mutations that may have destabilized secondary and tertiary structural elements. Hence, the mutants generated in those studies cannot reliably be used to localize functional surfaces at high resolution.
We now report a structure-based alanine-scanning mutagenic analysis of the HIV-1 CA surface. The overall goals of this work were (i) to map known CA functions more precisely, (ii) to identify potential new CA functions, (iii) to provide a data set for evaluating emerging structural models of the HIV-1 CA and Gag lattices, and (iv) to identify mutations that will be useful in future studies aimed at dissecting different CA functions.
MATERIALS AND METHODS
Viral constructs.
Point mutations were introduced into the HIVNL4-3 CA protein by oligonucleotide-directed, single-stranded mutagenesis of the phagemid vector pSL1180-gag (92). The complete viral fragments were sequenced to confirm mutations and exclude additional mutations. Mutant BssHII-ApaI fragments were recloned into the full-length HIV expression plasmid R9ΔApa, which is missing the backbone ApaI site (a gift from Didier Trono, University of Geneva [86]).
The primers used to create new CA mutations are shown in Table 1.
TABLE 1.
Primers used to create new CA mutations
| HIV CA mutant | 5′-3′ (forward) sequence (mutated codons underlined) | Altered restriction site |
|---|---|---|
| Q4A | TACCCCTATAGTCGCGAACCTCCAGGG | NruI |
| Q13A | GGCAAATGGTGCATGCGGCCATATCACCTAG | SphI |
| R18A/N21A | CATATCACCTGCAACTTTAGCTGCATGGGTA | NsiI destroyed |
| P38A | CCAGAAGTAATCGCGATGTTTTCAGC | NruI |
| E45A | CATGTTTTCAGCCTTATCGGCAGGAGCCACC | BglI |
| T54A | CACAAGATTTGAATGCCATGCTAAAC | BsmI |
| T54A/N57A | AGATTTAATGCCATGCTAGCCACAGTGGG | NheI |
| Q63A/Q67A | GGGGGGACATGCAGCAGCCATGGCAATGTTAAAAG | DsaI |
| K70A | CCATGCAAATGCTAGCAGAGACCATC | NheI |
| E71A | CATGCAAATGCTTAAGGCGACCATCAAT | AflII |
| N74A | GTTAAAAGAGACGATCGCTGAGGAAGCTG | PvuI |
| E75A/E76A | GACCATCAATGCGGCGGCCGCAGAATGGG | NotI |
| A78D/E79A/R82A | GAGGAAGCTGACGCATGGGATGCATTGCATCCAG | PstI destroyed |
| R100A/S102A | GAGAGAACCAGCGGGAGCGGACATAGCAGG | BsrBI |
| T107A/T108A | CATAGCAGGAGCTGCTAGTACCCTTC | SpeI destroyed |
| T110A/Q112A | GAACTACTAGCGCCCTTGCGGAACAAATAG | SpeI destroyed |
| G116A | GAACAAATAGCATGGATGACA | None |
| T119A | CAAATAGGATGGATGGCACATAATCC | None |
| E128A/R132Aa | CCCAGTAGGAGCAATCTATAAAGCTTGGATAATCC | HindIII |
| L136D | TAAAAGATGGATAATCGATGGATTAAATAAAATAG | ClaI |
| N139A | GATAATCCTGGGACTCGCGAAAATAGTAAG | NruI |
| R143A | AATAAAATAGTGGCCATGTATAGCCC | MscI |
| D152A | TACCAGCATTTTAGCCATAAGACAAG | BsmI destroyed |
| K158A | ACAAGGACCAGCTGAACCCTTTAG | PvuII |
| K158D | ACAAGGACCAGATGAACCCTTTA | None |
| K158R | GACAAGGACCTCGAGAACCCTTTA | XhoI |
| K158Q | ACAAGGACCACAAGAACCCTTA | None |
| D163A | CCCTTTAGAGCCTACGTAGACCGA | SnaBI |
| K170A | CCGATTCTATGCGACGCTAAGAGCC | HgaI |
| Q176A | AAGAGCCGAGGCAGCTTCACAAG | HindIII destroyed |
| E180A | CGAGCAAGCTACACAAGCGGTAAAAAAT | HindIII destroyed |
| W184A | AGGTAAAAAATGCGATGACAGAAACGTTGTTGGTCCAA | Psp 1406I |
| M185A | AGGTAAAAAATTGGGCGACAGAAACGTTGTTGGTCCAA | Psp 1406I |
| Q192A | CTTGTTGGTCGCGAATGCGAACC | NruI |
| D197A | TGCGAACCCAGCATGCAAGACTATTTT | SphI |
| D197N | TGCGAACCCAAACTGTAAGACTAT | XhoI |
| D197E | CAAAATGCGAATCCGGAGTGGAAGACTATTTTAAAAGC | BbsI |
| K203A | GATTGTAAGACAATATTAGCAGCATTGGGA | SspI |
| P207A | AAGCATTGGGCGCCGGAGCGACAC | EheI |
| E212A | CAGGAGCGACGCTAGCAGAAATGATGAC | NheI |
| Q219A | TGACAGCATGCGCAGGAGTGGGGG | FspI |
| P224A | GAGTGGGGGGAGCGGGCCATAAAG | BsrBI |
R132A was also inadvertently made as a single mutant, but it was fully infectious and showed a phenotype similar to that of the double mutant, so it is not listed separately.
HIV-1 production.
Wild-type (WT) and mutant plasmids were transiently transfected by the calcium phosphate method (17) into 293T human embryonic kidney cells carrying the simian virus 40 large T antigen (71). Supernatants were harvested after 40 to 44 h, and virus production was measured in p24 enzyme-linked immunosorbent (Dupont) and reverse transcriptase (RT) assays (40). The Q67A mutation abolished anti-CA antibody binding in the commercial DuPont p24 enzyme-linked immunosorbent assay kit, and the Q63A/Q67A mutant virus was therefore quantitated by RT assay only. During the course of these studies, we found that both methods often overestimated the release of assembly-defective mutants because the CA and RT proteins were also released in nonparticulate forms. Viral particle production was therefore assayed in Western blots of virions purified through sucrose cushions (see below).
Viral infectivity.
Viral infectivities were quantitated in multinucleate activation of galactosidase indicator cell (MAGIC) assays by using P4 HeLa.CD4.LTR.β-gal indicator cells (16), which stain blue with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) after HIV-1 infection and Tat protein expression (50). Infected cells were counted after 48 h, and relative infectivities were quantitated as the number of blue cells per nanogram of p24 of input virus (or units of RT activity for the Q63A/Q67A mutant) and compared with that of WT. Infective titers for the WT virus were typically 2.5 × 105 infectious units/ml. The data in Table 2 are the averaged results of at least three repetitions for each mutant; viral titers reduced more than 100-fold were labeled 0. Growth curves (K158A, K158D, K158Q, M185A, D197A, D197N) were performed for up to 100 days in CEM-ss T cells after normalization for CA protein (92).
TABLE 2.
Viral phenotypes of HIV-1 CA mutations
| Mutation | Location in domaina | Particle productionb | Infectivityc | Presence of conical capsidsd | Comments | Color codee |
|---|---|---|---|---|---|---|
| NTD mutations | ||||||
| Q4A | β-hairpin | + | 1.9 ± 1.4x ↓ | + | Green | |
| Q7A/Q9Af | β-hairpin | + | 10 ± 3.5x ↓ | + | Green | |
| Q13A | β-hairpin | + | 2.5 ± 0.9x ↓ | + | Green | |
| R18A/N21Ag | Helix 1 | + | 0 | ± | Multiple capsids | Yellow |
| A22Df | Helix 1 | + | 0 | − | Yellow | |
| E28A/E29Af | Helix 1 | + | 0 | − | Yellow | |
| P38A | Helix 2 | + | 33 ± 16x ↓ | + | Capsid assembly but reduced infectivity | Blue |
| M39Df | Helix 2 | + | 0 | − | Yellow | |
| A42Df | Helix 2 | + | 0 | − | Yellow | |
| E45A | Helix 2 | + | 29 ± 3.2x ↓ | + | Capsid assembly but reduced infectivity | Blue |
| D51Af | Hx 3 Pro1 salt bridge | + | 0 | − | Yellow | |
| T54A | Helix 3 | + | 10 ± 5x ↓ | + | Green | |
| T54A/N57A | Helix 3 | + | 80 ± 9x ↓ | ± | Few conical capsids | Yellow |
| Q63A/Q67A | Helix 4 | + | 34 ± 14x ↓ | + | Capsid assembly but reduced infectivity | Blue |
| K70A | Helix 4 | + | 21 ± 8x ↓ | ± | Few conical capsids | Yellow |
| E71A | Helix 4 | + | 14 ± 4x ↓ | + | Green | |
| N74A | Helix 4 | + | 5.2 ± 1x ↓ | + | Green | |
| E75A/E76A | Helix 4 | ↓ | 0 | − | Gag assembly defect, aberrant capsids | Red |
| A78D/E79A/R82A | Helix 4 | + | 3.3 ± 0.8x ↓ | + | Green | |
| R100A/S102A | Helix 5 | ↓ | 0 | − | Gag assembly defect | Red |
| T107A/108A | Helix 5/6 | ↓ | 0 | − | Gag assembly defect | Red |
| T110A/Q112A | Helix 6 | ↓ | 0 | − | Gag assembly defect | Red |
| G116A | Helix 6 | + | 1.8 ± 0.1x ↑ | + | Green | |
| T119A | Helix 6 | + | 1.2 ± 0.0x ↑ | + | Green | |
| E128A/R132A | Helix 7 | + | 6.2 ± 0.7x ↓ | + | Green | |
| L136D | Helix 7 | + | 91 ± 13x ↓ | ± | Few cones | Yellow |
| N139A | Helix 7 | + | 1.2 ± 0.2x ↓ | + | Green | |
| R143A | Helix 7 | + | 5.4 ± 4.5x ↓ | + | Green | |
| CTD mutations | ||||||
| D152A | Interdomain linker | + | 6.2 ± 3x ↓ | + | Green | |
| K158A | Turn (MHR) | − | 0 | − | Gag assembly defect | Red |
| K158D | Turn (MHR) | ↓ | 0 | − | Gag assembly defect | Red |
| K158Q | Turn (MHR) | ↓ | 0 | − | Gag assembly defect | Red |
| K158R | Turn (MHR) | + | 3.4 ± 0.5x ↓ | + | Red | |
| D163A | Helix 8 (MHR) | + | 15 ± 7x ↓ | + | Green | |
| K170A | Helix 8 (MHR) | + | 0 | + | Capsid assembly but no infectivity | Blue |
| Q176A | Helix 8/9 | + | 2.7 ± 0.6x ↓ | + | Green | |
| E180A | Helix 9 | + | 13 ± 3x ↓ | + | Green | |
| W184A | Hx 9 dimer interface | ↓ | 0 | − | Gag and capsid assembly defects | Red |
| M185A | Hx 9 dimer interface | ↓ | 0 | − | Gag and capsid assembly defects | Red |
| Q192A | Helix 10 | + | 2.8 ± 0.7x ↓ | + | Green | |
| D197A | Helix 10 | − | 0 | − | Gag assembly defect, aberrant capsids | Red |
| D197E | Helix 10 | ↓ | 0 | ND | Gag assembly defect | Red |
| D197N | Helix 10 | ↓ | 0 | − | Gag assembly defect, aberrant capsids | Red |
| K203A | Helix 10 | + | 0 | + | Capsid assembly but reduced infectivity | Blue |
| P207A | Turn | + | 1.3 ± 0.3x ↓ | + | Green | |
| E212A | Helix 11 | + | 3.1 ± 1.8x ↓ | + | Green | |
| Q219A | Helix 11 | + | 14 ± 12x ↓ | + | Green | |
| P224A | Unstructured | ↓ | 0 | − | Gag assembly defect, aberrant capsids | Red |
Position in the secondary structure of HIV-1 CA, as derived from crystal structures of the protein's NTD and CTD (see Fig. 1).
Detected in Western blots of pelleted virions from the supernatant of transfected 293T cells. +, similar to WT; ↓, consistently lower than WT; −, undetectable.
Infectivity of virus released into the supernatant of transfected cells was analyzed in MAGIC assays and is reported as fold reduced (↓) or fold increased (↑) ± standard deviation relative to that of the WT virus (typically ∼2.5 × 105 infectious units/ml). Any infectivity less than 1/100 that of the WT is called 0.
Conical capsids were assayed by transmission EM of thin sections of pelleted virions as described in Materials and Methods. A minimum of 10 fields of virions from at least two different grids at × 30,000 to 60,000 magnification were screened for each mutant (see Fig. 3A, overview panels). Each field typically contained at least 40 virions, distinguished from microvesicles by their electron-dense cores. For mutants with reduced virus yields, at least 100 identifiable virions with electron-dense cores or immature Gag rings were screened. +, conical capsids frequency of at least 2 cones per field; ±, fewer than 5 normal cones in 10 fields of virions; −, no discernible conical capsids in 10 fields or highly aberrant particles and capsid shapes.
Color codes for the different mutant phenotypes are as described in the legends for Fig. 4B and C.
These six point mutants were described previously (see reference 92) and are included here for completeness.
The primary defect in R18A/N21A assembly appeared to be in capsid formation, but this mutation also reduced particle production modestly and sometimes produced enlarged virions with multiple capsids, suggesting a slight Gag assembly defect as well (see Fig. 3). ND, not done.
Western blotting.
Transfected 293T cells were harvested directly into radioimmunoprecipitation assay buffer (10 mM TrisCl [pH 7.0], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS]) for Western blotting, whereas virus-containing supernatants were pelleted through 20% sucrose and then resuspended in 25 μl of SDS sample buffer. Proteins from 2 μl of pelleted virions were separated by 10 to 15% SDS-polyacrylamide gel electrophoresis, transferred, blotted with antisera, and detected by chemiluminescence (91). The primary antibodies used were rabbit anti-MA at 1:50,000 from Didier Trono, rabbit anti-CA (nos. 40 and 49) at 1:2,000 from Hans-Georg Kräusslich , mouse monoclonal anti-RT mAb21 at 1:500 (obtained from Stephen Hughes through the AIDS Research and Reference Reagent Program) (27), and human monoclonal anti-Env gp41 2F5 at 1:1,000 (obtained from Hermann Katinger through the AIDS Reagent Program, cat. no. 1475) (73).
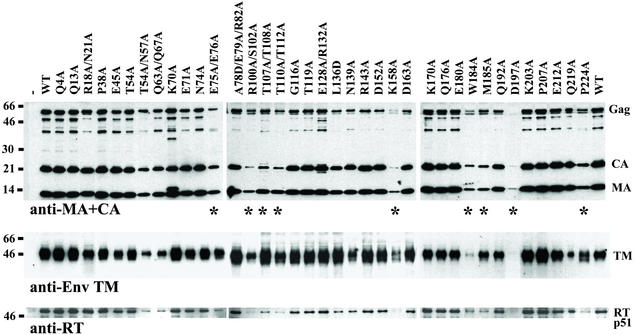
At least three independent Western blots were performed for each mutant. Figure 2 and Table 2 show representative data and were fully reproducible, except that (i) the intensities of the unprocessed p55 Gag bands varied between experiments, (ii) CA degradation products of ∼23 and ∼18 kDa were sometimes observed, particularly in the K70A and E128A/R132A mutants (see Fig. 2), and (iii) the slight reduction in particle production with the T54A/N57A mutation apparent in Fig. 2 was not reproducible.
FIG. 2.
Western blot analysis of HIV-1 particle production. Western blots of sucrose-pelleted virions, incubated with anti-MA and anti-CA (top), anti-Env TM (middle), and anti-RT (bottom) antibodies. −, mock transfected control. Molecular weight markers are shown at left. Mutants with reduced particle yields are marked with an asterisk.
Transmission EM.
Free viral particles were concentrated for electron microscopy (EM) studies either by pelleting through 20% sucrose (as described for Western blotting) or by adjustment of the supernatant to 1× fixative (2.5% glutaraldehyde-1% paraformaldehyde in sodium cacodylate buffer) and 20% fetal calf serum and pelleting for 90 min in a microcentrifuge. Pelleted virions were resuspended in 1× EM fixative and processed for EM as described previously (92). Cellular vesicles present in these preparations (6, 39) served as carriers through this procedure, particularly for mutants with lower particle yields. To visualize cell-associated virions, transfected 293T cells were fixed in 1× EM fixative for 30 min without a rinse and processed the same way, except that dehydration was in EtOH and embedding was in Spurr's plastic. Transfection and preparation of K158A-transfected HeLa cells for EM was performed as described previously (95). Electron micrographs were collected on a Hitachi H-7100 transmission electron microscope at magnifications of ×30,000 to 100,000.
RESULTS
Experimental design.
An ensemble of 48 single and double point mutations, encompassing 54 different amino acids, was designed to sample the entire surface of the monomeric CA molecule (omitting the cyclophilin A binding loop). The side chains of all mutated residues were solvent exposed and made no significant intramolecular interactions in the CA crystal structures, thereby minimizing the chance of structural perturbations beyond the mutation site. Residues were typically mutated to alanine to remove side chain atoms while retaining phi/psi restraints (exceptions are given in Tables 1 and 2). The desired mutations were introduced into the CA gene of proviral HIV-1NL4-3 within the R9 ΔApa plasmid and transfected into 293T producer cells, and viral phenotypes were tested as described in Materials and Methods. A complete list of the CA mutations tested, their locations within the CA structure, and their viral phenotypes is presented in Table 2 and Fig. 1C and D.
Viral infectivity.
Infectivities of mutant viruses were analyzed in single-cycle MAGIC assays (Table 2 and Fig. 4A). Of the 48 mutants tested, 21 retained substantial infectivity (within 20-fold that of the WT), 6 had significantly reduced infectivity (20- to 100-fold), and 21 were noninfectious (>100-fold reduced). Even among the mutants that retained substantial infectivity, however, all but five were reduced at least twofold in infectivity. Reductions in viral replication generally reflected slightly lower particle yields and fewer ideal mature particles (see below).
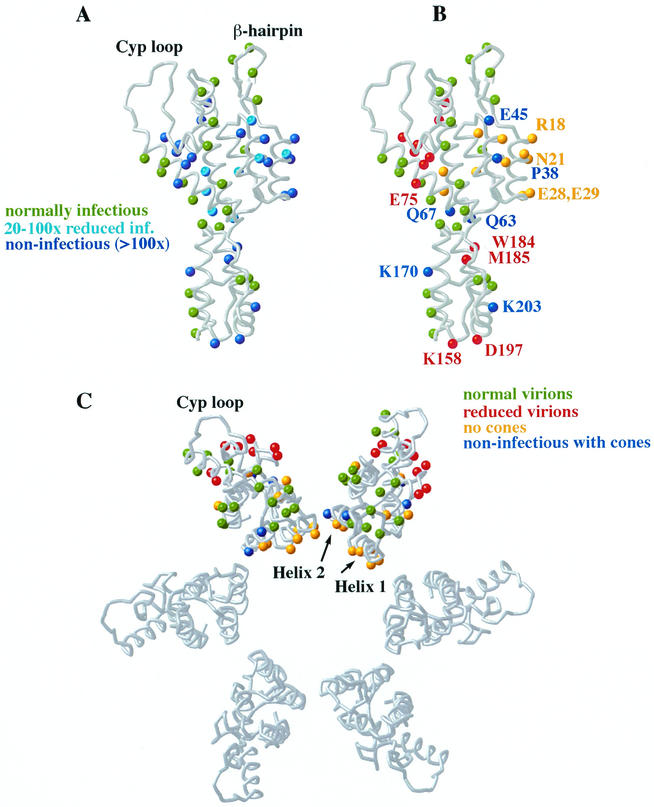
FIG. 4.
Summary of CA mutant infectivity and assembly data. (A) Relative infectivities of the different CA mutants mapped onto the structure of the monomeric CA protein. Green, infectivity reduced less than 20-fold; turquoise, infectivity reduced 20- to 100-fold; blue, noninfectious (infectivity reduced >100-fold). The P224A mutation, which blocks virus assembly and infectivity, lies in the unstructured CA C terminus (not shown). (B) Viral assembly phenotypes of the different CA mutants mapped onto the structure of the monomeric CA protein. Green, infectious virus with conical capsids; blue, noninfectious virus with conical capsids; yellow, normal particle production but no conical capsids; red, reduced particle production and no conical capsids. A subset of the mutations is labeled to help in orientation. (C) Viral assembly phenotypes of the different NTD CA mutants mapped onto a model for the CA hexamer (viewed down the sixfold axis). Color coding is as given in the legend for panel B. Helices 1 and 2, which make the primary contacts predicted to stabilize the hexamer, are labeled, as is the external CypA binding loop.
To confirm the relevance of the MAGIC results to spreading viral infections, we also tested the ability of several representative CA mutants to replicate in cultured T cells. As expected, mutant viruses that were noninfectious in the MAGIC assay (e.g., K158A, K158D, K158Q, M185A, D197A, and D197N) also failed to replicate detectably in cultured CEM cells (not shown). In contrast, a prototypic infectious mutant (Q7A/Q9A) which was reduced only 10-fold in the MAGIC assay replicated to the same peak level as WT virus, but with a delay of 4 days (92). Thus, there is a reasonable correlation between the MAGIC and viral replication assays, although some mutants, particularly those with modest virus assembly defects in the 293T overexpression system, have even more severe phenotypes in peripheral blood mononuclear cells or T cells (29).
Gag expression, processing, and particle production.
Viral protein expression in transfected 293T cells was analyzed in Western blots of cytoplasmic extracts. Steady-state Gag expression levels were not significantly affected by any of the mutations except D197A (not shown). However, a more conservative mutation at the same site (D197N) resulted in normal cytoplasmic Gag protein levels. All CA proteins with detrimental mutations were also tested for expression in Escherichia coli. They were expressed at normal levels as soluble proteins, with the exception of CA E75A/E76A, which was reduced in solubility but nevertheless could be purified and was shown to assemble normally into helical cylinders (B. K. Ganser, U. K. von Schwedler, K. Stray, and W. I. Sundquist, submitted for publication). We therefore conclude that altered Gag expression or stability does not account for any of the phenotypes we observed.
Gag processing, particle release, and viral protein packaging were monitored by Western blotting of sucrose cushion-pelleted virions from culture supernatants (Fig. 2). Fully processed MA and CA proteins were detectable in all virions, indicating that none of the mutations blocked Gag processing (although K70A and E128A/R132A did increase the levels of Gag processing intermediates; see Fig. 2, upper panel). All of the mutant viruses also packaged processed Pol (RT p51) and Env (TM gp41) proteins (Fig. 2, bottom), and the levels of these proteins generally paralleled Gag protein levels, indicating that the virion stoichiometries of RT and Env were not grossly altered by any of the mutations. Levels of pelletable virus were significantly reduced or eliminated by mutations at nine different sites, namely, E75A/E76A, R100A/S102A, T107A/T108A, T110A/Q112A, K158A(D,Q), W184A (in some experiments), M185A, D197A(N), and P224A. These mutations presumably reduced immature particle production or stability by adversely affecting proper Gag assembly.
Virion morphology.
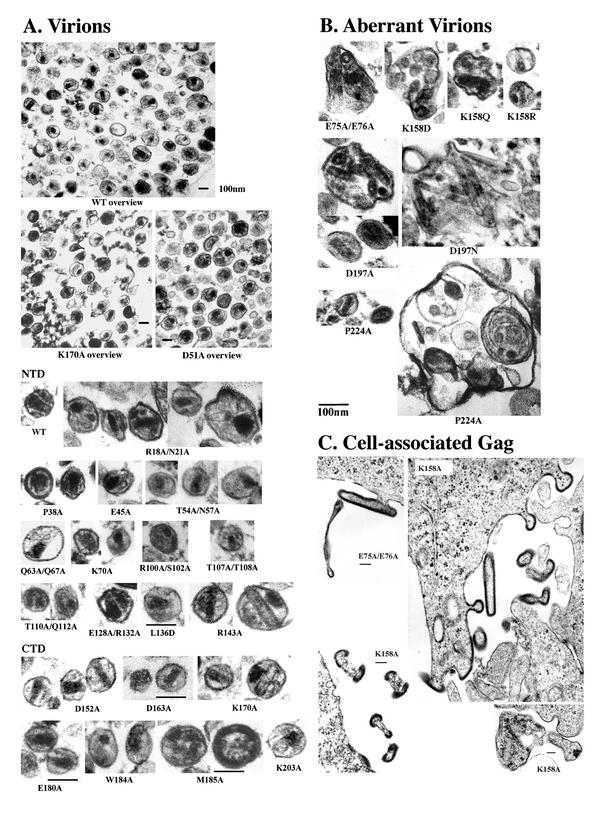
Viral particle morphology was analyzed in EM images of pelleted, thin-sectioned virions (Fig. 3). Mutations at four of nine sites that reduced overall particle release also grossly altered particle morphology (E75A/E76A, K158A, K158D, K158Q, D197A, D197N, and P224A). These aberrant particles were enlarged (up to fivefold in diameter), irregularly shaped, and frequently contained multiple capsids that could be tubular and kinked (Fig. 3B). Normal-sized virions with conical capsids were very rarely observed in this mutant class.
FIG. 3.
Electron micrographs of mutant virions with reduced infectivity. (A) Shown are overviews over fields of virions photographed at ×50,000 to demonstrate variability of capsid shapes (top panels) and particle morphologies resulting from CA mutations in the NTD (middle panels) and CTD (bottom panels). Mutants with phenotypes are ordered from the N to C terminus except for those that produced gross defects (shown in Fig. 3B and C). (B) Mutant virions with gross particle assembly defects. (C) Cell-associated Gag assemblies with gross defects in particle formation. Shown are CA E75A/E76A and K158A mutants expressed in 293T cells (upper panels and lower right) and CA K158A mutant expressed in HeLa cells (lower left). Note the aberrant curvature produced by Gag molecules assembling beneath the plasma membrane. Scale bars in all figures are 100 nm.
The unusual sizes and shapes of these mutant viruses again suggested defective Gag lattice interactions, and EM images of HeLa and 293T cells expressing the E75A/E76A, K158A, D197A, and P224A mutants further supported this idea (Fig. 3C and data not shown). In all mutants and cell types, electron-dense patches of associated Gag molecules accumulated beneath the plasma membrane but lacked the proper curvature necessary to create closed particles. Moreover, when particles were released, they frequently exhibited discontinuities in the Gag layer (Fig. 3C). We therefore conclude that accumulations of Gag patches beneath the plasma membrane, reductions in particle release, and enlarged virions likely represent different manifestations of the same basic defect, i.e., the inability of the assembling Gag lattice to assemble, curve, and close properly.
Viral maturation.
Nine mutations in the N-terminal domain of CA were classified as defective in viral maturation because they supported normal levels of particle release but either eliminated (A22D, E28A/E29A, M39D, A42D, and D51A) or reduced (R18A/N21A, T54A/N57A, K70A, and L136D) the efficiency of capsid formation (Fig. 3A and 4B and Table 2). All of these mutations also reduced infectivity more than 20-fold.
Although conical capsids predominate in WT HIV-1, capsids can adopt a range of different shapes, including cylinders (2, 37, 53, 65, 93), and it is therefore difficult to define a typical HIV-1 capsid morphology (see Fig. 3A, overviews). Nevertheless, the noninfectious R18A/N21A mutation clearly produced a range of abnormally shaped and multiple capsids in addition to reducing the overall efficiency of capsid assembly (Fig. 3A).
Other defects.
Five CA mutations supported normal or almost normal capsid assembly (see footnote d to Table 2) but were nevertheless noninfectious (P38A, E45A, Q63A/Q67A, K170A, and K203A, colored blue in Fig. 4B and C).
DISCUSSION
The importance of the CA protein in HIV-1 replication is underscored by our finding that over half (27 of 48) of our CA surface point mutations reduced viral infectivity at least 20-fold, and nearly all (43 of 48) reduced infectivity at least twofold. Thus, most of the CA surface cannot tolerate substitution without a reduction in viral fitness. This presumably reflects the fact that CA must engage in a series of different interactions as the virion assembles, matures, and disassembles. The effects of various CA mutations at different stages of the viral life cycle are discussed below.
Immature particle assembly.
Nine mutations significantly diminished or altered immature particle production, presumably by disrupting important interactions during Gag assembly. These mutations mapped to three different CA surfaces, namely, (i) the base of the CA CTD surrounding the loop preceding helix 8, (ii) the crystallographic C-terminal dimer interface (helix 9), and (iii) a surface surrounding helix 5 in the CA NTD. These observations suggest that CA makes at least three different kinds of interactions during HIV particle assembly (discussed in turn below).
A Gag assembly element at the base of the CA CTD.
The base of CA contains determinant(s) that are critical for retroviral Gag assembly (21, 41, 82), and our most detrimental Gag assembly mutations map to residues in this region (K158A/D/Q, D197A/E/N, and P224A) (see Fig. 4B). All three mutations reduce particle production severely and also alter the shape of particles that are produced. These mutations do not block Gag association altogether, as sheets of Gag molecules can be seen assembling beneath the plasma membrane (Fig. 3C); rather, they appear to disrupt important Gag-Gag interactions that are essential for creating a tight, closed immature particle lattice.
The K158 side chain is critical for Gag assembly, and Ala, Asp, and Asn mutations, but not Arg mutations, at this position all impair virion release. The exposed K158 side chain extends away from the type I β-turn in the center of the MHR in the mature HIV-1 CA structure, and mutations at this position do not reduce mature CA protein assembly in vitro (B. K. Ganser, et al., submitted). We therefore presume that this residue either makes an intramolecular contact in a distinct, immature CA conformation or, alternatively, contacts another protein. Although the lysine side chain is poorly conserved in other retroviruses, mutation of the equivalent glutamate residue in MPMV CA (to Tyr or Asn) also blocks infectivity, albeit with only modestly impaired virion assembly (82).
HIV-1 CA residue Asp197 also makes critical Gag assembly contact(s) and cannot be mutated to Ala, Asn, or even Glu without impairing virion release. Asp197 is located near the N terminus of helix 10, which is outside the continuous MHR motif, but the Asp197 and Lys158 side chains project out in the same direction from the bottom of the CA CTD and are only 5 Å apart. Moreover, the two residues seem to make functionally analogous contacts as judged by their similar phenotypes. P224A also impairs Gag assembly; it is located in the disordered C-terminal tail in structures of the mature HIV-1 CA CTD.
We and others have observed similar Gag assembly defects for additional mutations in retroviral CA CTD residues (11, 59, 63; O.W. Pornillos and K. M. Stray, personal communication; D. Muriaux and A. Rein, personal communication), as well as for mutations in the SP1 spacer peptide (1, 43, 55, 58) and in NC (5, 18). We therefore speculate that these downstream elements may interact with the base of the CA CTD domain to create a continuous structure that is critical for immature Gag lattice assembly (but which may be unstable following Gag proteolysis or in the absence of lattice contacts). Indeed, there is evidence that the CA-SP1 junction forms a helix that is lost upon Gag proteolysis (1, 58, 59). Formation of a continuous Gag structure spanning the CA CTD-SP1-NC junctions and involving the MHR fold (11) would explain the common phenotypes of mutations in this region, and it might also provide a mechanism for coupling RNA packaging with viral assembly and/or for ensuring incorporation of intact Gag molecules.
The CA dimer interface in Gag assembly.
The CA CTD dimerizes in solution (35, 78) and crystallizes as a symmetrical dimer created by parallel packing of CA helix 9 against its symmetry-related mate (98). One goal of our study was to test whether or not the crystallographically characterized CA CTD dimer interface functions during particle assembly and maturation. Residues W184 and M185 are buried in the core of the dimer interface, and alanine mutations at either position block CA dimerization in vitro (35). In addition, several pairs of residues on the periphery of the dimer interface make favorable hydrophilic contacts across the dyad (e.g., D152-E180 and K203-Q192). However, their energetic contributions to CA dimerization are not known.
Mutations in peripheral dimer interface residues (D152A, E180A, and Q192A) reduced infectivity (3- to 13-fold) but did not obviously alter the morphology of either immature or mature particles. These modest phenotypes may reflect the fact that surface side chain salt bridges often contribute very little to overall protein stability (83, 84). However, mutations in the two core dimer interface residues (W184A and M185A) measurably diminished (but did not eliminate) immature particle production. These same mutations also reduced intermolecular Gag-Gag interactions in vitro (10). Taken together, these data suggest that the CA dimer interface may form and contribute to the stability of the immature Gag lattice but is not absolutely required for particle assembly. An important role for Gag dimerization in HIV-1 assembly is also suggested by the fact that defects in particle release caused by deletion of the NC protein can be rescued by dimeric leucine zipper motifs (2, 48, 103).
The partial defect of the CA dimer interface mutants in immature particle assembly complicates evaluation of possible downstream role(s) of the dimer in mature capsid assembly. Nevertheless, both mutations in the core of the dimer interface inhibited CA assembly in vitro (B. K. Ganser, et al., submitted) and completely blocked mature capsid assembly and viral infectivity in cultured virus, supporting the idea that this interface is also essential at later stage(s) of the assembly-maturation pathway.
A Gag assembly element in the CA NTD.
Studies from several laboratories indicate that the NTD of CA may not play an essential role in immature particle assembly (reviewed in reference 41). Indeed, Gag proteins lacking this entire domain can assemble and bud from cells (2, 7, 67, 77). Nevertheless, we found that a series of mutations in CA helices 4 to 6 (E75A/E76A, R100A/S102A, T107A/T108A, and T110A/Q112A) reduced viral particle production, abolished capsid formation, and blocked viral infectivity. A similar phenotype was reported for an Ala mutation of the Pro99 residue that precedes helix 5 (28, 52). Despite strong viral phenotypes, however, none of our mutations in this region affected CA cylinder or cone formation in vitro (B. K. Ganser et al., submitted). Taken together, these data indicate that Gag assembly is sensitive to the composition of this CA NTD surface (at least when the CA NTD is present), and they define a previously unrecognized contiguous CA surface located beneath the cyclophilin A binding loop that participates in Gag assembly. We speculate that this region of CA may form a weak Gag-Gag interface or, alternatively, bind a cellular factor necessary for efficient assembly in vivo.
Capsid assembly elements.
Strikingly, all of the mutant virions that retained infectivity formed conical capsids, whereas all mutants lacking cones were severely reduced in infectivity (>20-fold). This correlation strongly supports the long-standing, but still unproven, assumption that the proper assembly of a mature conical capsid is essential for viral infectivity. Both the NTD and CTD of HIV-1 CA perform essential structural roles in the mature viral capsid (41), and we have proposed that this is because the NTD forms hexameric rings while the CTD links the hexamers into a continuous lattice (57). Consistent with this model, two mutations in the CA CTD dimer interface blocked conical capsid formation (W184A and M185A, discussed above), as did a series of mutations in the CA NTD. The latter mutants clustered about N-terminal helices 1 to 3 (mutants R18A/N21A, A22D, E28A/E29A, M39D, A42D, D51A, and N57A; two exceptions were K70A [helix 4] and L136D [helix 7]) (Table 2 and Fig. 4B). These mutants likely affected capsid assembly (and not prior steps) because many had no effect on particle release and because most diminished CA tube and cone assembly in vitro (B. K. Ganser et al., submitted).
Our mutational analysis agrees well with the structural model for the CA hexamer in which helices 1 and 2 form the primary intermolecular contacts at the center of the CA hexamer (Fig. 4C). Helix 1/2 residues R18, A22, E28, E29, M39, and A42 are positioned so they could make intermolecular contacts in the CA hexamer, and mutations in each block capsid assembly. An exception to this correlation is residue Pro38 (helix 2), which also appears positioned to make intermolecular contacts but can be mutated to Ala without blocking capsid assembly (albeit with a 27-fold reduction in infectivity). However, a P38L mutation does block capsid assembly (28), indicating that the CA hexamer likely cannot accommodate larger side chains at this position. Similarly, an F40A mutation is also defective in CA assembly (87).
Mutation R18A/N21A (at the top of helix 1) was unique in that it frequently caused assembly of multiple aberrant capsids within the same enlarged virions, including tubes, cones, and possibly spheres (which are difficult to discern unambiguously in thin section). The assembly phenotype of this CA mutant in vitro was similarly aberrant (B. K. Ganser et al., submitted). We suggest that this mutation alters the proper distribution of CA pentamers and hexamers required to form a cone (B. K. Ganser et al., submitted).
Helix 3 serves to buttress the first two CA helices and also anchors the β-hairpin, and this may explain the diminished capsid assembly of the D51A and N57A mutants. In contrast, mutations in residues within the N-terminal β-hairpin that precedes helices 1 and 2 (Q4A, Q7A/Q9A, and Q13A) had no effect on cone formation and only modest effects on infectivity (≤10-fold), consistent with other experiments indicating that the hairpin itself does not make essential contacts in the assembled capsids (44). Several alternative models for the CA hexamer that have been proposed recently (56, 62) are discussed in detail elsewhere (B. K. Ganser et al., submitted).
Other defects.
Five mutant viruses (P38A, E45A, Q63A/Q67A, K170A, and K203A) were not obviously defective in assembly, maturation, or TM and RT packaging yet were significantly reduced in infectivity (>20-fold). These mutants are therefore of interest for understanding how CA performs its essential nonstructural function(s).
Further studies have revealed that these mutations all package normal levels of viral RNA and exhibit normal levels endogenous reverse transcription activity (29). With the exception of E45A, however, all of these mutations appeared to reduce viral capsid stability as judged by the fact that intact cores from these mutants could not be isolated (29). Intriguingly, the E45A mutation actually appeared to stabilize the viral capsid because this mutation increased yields of isolated cores and reduced the rate of core dissociation in vitro. The reduced infectivity of all of these mutants suggests that proper disassembly of a capsid of normal stability is an essential early event in viral replication. Thus, our studies are consistent with the idea that the principal role of the capsid is to organize the viral replication complex for ordered uncoating and replication upon viral entry (8, 11, 15, 29, 87). Further support for this idea comes from the phenotype of the Q63A/Q67A mutant, which is actually reverse transcribed more rapidly than the WT virus upon entry yet is very poorly infectious-perhaps because it does not follow the proper viral uncoating pathway.
In summary, our mutational analyses have identified a series of residues in both domains of CA that help to establish the immature Gag lattice and to build the mature viral capsid. Our studies also support the idea that an essential function of the viral capsid is to organize the viral RNA and its associated enzymes for ordered disassembly and reverse transcription upon entry of the new host cell.
Acknowledgments
We thank Victor Klishko for technical assistance with sample preparation and EM, Miguel Knochel and Ryan Brady for assistance with mutagenesis, Su (Sam) Li and Felix Vajdos for help with Fig. 1 and 4, Klaus Wiegers, Gabriel Rutter, and Hans-Georg Kräusslich for electron microscopy of HeLa cells transfected with mutants, and Chris Hill, Hans-Georg Kräusslich, and Volker Vogt for helpful discussions and suggestions.
This work was supported by an NIH grant (to W.I.S.).
REFERENCES
- 1.Accola, M. A., S. Hoglund, and H. G. Göttlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alin, K., and S. P. Goff. 1996. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology 222:339-351. [DOI] [PubMed] [Google Scholar]
- 4.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthoux, L., C. Pechoux, M. Ottmann, G. Morel, and J. L. Darlix. 1997. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J. Virol. 71:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bess, J. W., Jr., R. J. Gorelick, W. J. Bosche, L. E. Henderson, and L. O. Arthur. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230:134-144. [DOI] [PubMed] [Google Scholar]
- 7.Borsetti, A., A. Ohagen, and H. G. Göttlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowzard, J. B., J. W. Wills, and R. C. Craven. 2001. Second-site suppressors of Rous sarcoma virus Ca mutations: evidence for interdomain interactions. J. Virol. 75:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns, T. M., and R. C. Craven. 2001. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. J. Virol. 75:242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos-Olivas, R., J. L. Newman, and M. F. Summers. 2000. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol. 296:633-649. [DOI] [PubMed] [Google Scholar]
- 14.Carter, C. A. 2002. Tsg101: HIV-1's ticket to ride. Trends Microbiol. 10:203-205. [DOI] [PubMed] [Google Scholar]
- 15.Cartier, C., P. Sivard, C. Tranchat, D. Decimo, C. Desgranges, and V. Boyer. 1999. Identification of three major phosphorylation sites within HIV-1 capsid. Role of phosphorylation during the early steps of infection. J. Biol. Chem. 274:19434-19440. [DOI] [PubMed] [Google Scholar]
- 16.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 17.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Rescue of multiple viral functions by a second-site suppressor of a human immunodeficiency virus type 1 nucleocapsid mutation. J. Virol. 74:4273-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornilescu, C. C., F. Bouamr, X. Yao, C. Carter, and N. Tjandra. 2001. Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein. J. Mol. Biol. 306:783-797. [DOI] [PubMed] [Google Scholar]
- 21.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawson, L., and X. F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251:141-157. [DOI] [PubMed] [Google Scholar]
- 23.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Göttlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebbesen, T. 1998. Cones and tubes: geometry in the chemistry of carbon. Acc. Chem. Res. 31:558-566. [Google Scholar]
- 25.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Facke, M., A. Janetzko, R. L. Shoeman, and H. G. Krausslich. 1993. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J. Virol. 67:4972-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferris, A. L., A. Hizi, S. D. Showalter, S. Pichuantes, L. Babe, C. S. Craik, and S. H. Hughes. 1990. Immunologic and proteolytic analysis of HIV-1 reverse transcriptase structure. Virology 175:456-464. [DOI] [PubMed] [Google Scholar]
- 28.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schroder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294-307. [DOI] [PubMed] [Google Scholar]
- 29.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 31.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 32.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuller, S. D., T. Wilk, B. E. Gowen, H. G. Kräusslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 34.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 35.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 36.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 37.Gelderblom, H. R., E. H. Hausmann, M. Ozel, G. Pauli, and M. A. Koch. 1987. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology 156:171-176. [DOI] [PubMed] [Google Scholar]
- 38.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 39.Gluschankof, P., I. Mondor, H. R. Gelderblom, and Q. J. Sattentau. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology 230:125-133. [DOI] [PubMed] [Google Scholar]
- 40.Goff, S., P. Traktman, and D. Baltimore. 1981. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J. Virol. 38:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Göttlinger, H. G. 2001. The HIV-1 assembly machine. AIDS 15:S13-S20. [DOI] [PubMed] [Google Scholar]
- 42.Göttlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gross, I., H. Hohenberg, C. Huckhagel, and H. G. Kräusslich. 1998. N-Terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gross, I., H. Hohenberg, and H. G. Kräusslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 46.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin, Z., L. Jin, D. L. Peterson, and C. L. Lawson. 1999. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J. Mol. Biol. 286:83-93. [DOI] [PubMed] [Google Scholar]
- 48.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khorasanizadeh, S., R. Campos-Olivas, and M. F. Summers. 1999. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 291:491-505. [DOI] [PubMed] [Google Scholar]
- 50.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kingston, R. L., T. Fitzon-Ostendorp, E. Z. Eisenmesser, G. W. Schatz, V. M. Vogt, C. B. Post, and M. G. Rossmann. 2000. Structure and self-association of the rous sarcoma virus capsid protein. Structure Fold. Des. 8:617-628. [DOI] [PubMed] [Google Scholar]
- 52.Kong, L. B., D. An, B. Ackerson, J. Canon, O. Rey, I. S. Chen, P. Krogstad, and P. L. Stewart. 1998. Cryoelectron microscopic examination of human immunodeficiency virus type 1 virions with mutations in the cyclophilin A binding loop. J. Virol. 72:4403-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kotov, A., J. Zhou, P. Flicker, and C. Aiken. 1999. Association of Nef with the human immunodeficiency virus type 1 core. J. Virol. 73:8824-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kräusslich, H. G. (ed.). 1996. Morphogenesis and maturation of retroviruses. Springer-Verlag, Berlin, Germany.
- 55.Kräusslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lanman, J., T. T. Lam, S. Barnes, M. Sakalian, M. R. Emmett, A. G. Marshall, and P. E. Prevelige. 2003. Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J. Mol. Biol. 325:759-772. [DOI] [PubMed] [Google Scholar]
- 57.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 58.Liang, C., J. Hu, R. S. Russell, A. Roldan, L. Kleiman, and M. A. Wainberg. 2002. Characterization of a putative α-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J. Virol. 76:11729-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang, C., J. Hu, J. B. Whitney, L. Kleiman, and M. A. Wainberg. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the Gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luban, J. 2001. HIV-1 and Ebola virus: the getaway driver nabbed. Nat. Med. 7:1278-1280. [DOI] [PubMed] [Google Scholar]
- 61.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Göttlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mayo, K., D. Huseby, J. McDermott, B. Arvidson, L. Finlay, and E. Barklis. 2003. Retrovirus capsid protein assembly arrangements. J. Mol. Biol. 325:225-237. [DOI] [PubMed] [Google Scholar]
- 63.McDermott, J., L. Farrell, R. Ross, and E. Barklis. 1996. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J. Virol. 70:5106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 65.Nakai, M., and T. Goto. 1996. Ultrastructure and morphogenesis of human immunodeficiency virus. J. Electron Microsc. (Tokyo) 45:247-257. [DOI] [PubMed] [Google Scholar]
- 66.Nermut, M. V., P. Bron, D. Thomas, M. Rumlova, T. Ruml, and E. Hunter. 2002. Molecular organization of Mason-Pfizer monkey virus capsids assembled from Gag polyprotein in Escherichia coli. J. Virol. 76:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ono, A., D. Demirov, and E. O. Freed. 2000. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 74:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orlinsky, K. J., J. Gu, M. Hoyt, S. Sandmeyer, and T. M. Menees. 1996. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J. Virol. 70:3440-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paillart, J. C., and H. G. Göttlinger. 1999. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73:2604-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez, O. D., and G. P. Nolan. 2001. Resistance is futile. Assimilation of cellular machinery by HIV-1. Immunity 15:687-690. [DOI] [PubMed] [Google Scholar]
- 73.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 74.Rao, Z., A. S. Belyaev, E. Fry, P. Roy, I. M. Jones, and D. I. Stuart. 1995. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature 378:743-747. [DOI] [PubMed] [Google Scholar]
- 75.Reicin, A. S., A. Ohagen, L. Yin, S. Hoglund, and S. P. Goff. 1996. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 70:8645-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Göttlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rose, S., P. Hensley, D. J. O'Shannessy, J. Culp, C. Debouck, and I. Chaiken. 1992. Characterization of HIV-1 p24 self-association using analytical affinity chromatography. Proteins 13:112-119. [DOI] [PubMed] [Google Scholar]
- 79.Sakalian, M., and E. Hunter. 1998. Molecular events in the assembly of retrovirus particles. Adv. Exp. Med. Biol. 440:329-339. [DOI] [PubMed] [Google Scholar]
- 80.Spearman, P., R. Horton, L. Ratner, and I. Kuli-Zade. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Srinivasakumar, N., M. L. Hammarskjold, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strambio-de-Castillia, C., and E. Hunter. 1992. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J. Virol. 66:7021-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strop, P., and S. L. Mayo. 2000. Contribution of surface salt bridges to protein stability. Biochemistry 39:1251-1255. [DOI] [PubMed] [Google Scholar]
- 84.Sun, D. P., U. Sauer, H. Nicholson, and B. W. Matthews. 1991. Contributions of engineered surface salt bridges to the stability of T4 lysozyme determined by directed mutagenesis. Biochemistry 30:7142-7153. [DOI] [PubMed] [Google Scholar]
- 85.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Press, Plainview, N.Y. [PubMed]
- 86.Swingler, S., P. Gallay, D. Camaur, J. Song, A. Abo, and D. Trono. 1997. The Nef protein of human immunodeficiency virus type 1 enhances serine phosphorylation of the viral matrix. J. Virol. 71:4372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 89.Turner, B. G., and M. F. Summers. 1999. Structural biology of HIV. J. Mol. Biol. 285:1-32. [DOI] [PubMed] [Google Scholar]
- 90.Vogt, V. M. 1997. Retroviral virions and genomes, p. 27-70. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Press, Plainview, N.Y. [PubMed]
- 91.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Welker, R., H. Hohenberg, U. Tessmer, C. Huckhagel, and H. G. Kräusslich. 2000. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 74:1168-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Kräusslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wiegers, K., G. Rutter, U. Schubert, M. Grattinger, and H. G. Kräusslich. 1999. Cyclophilin A incorporation is not required for human immunodeficiency virus type 1 particle maturation and does not destabilize the mature capsid. Virology 257:261-274. [DOI] [PubMed] [Google Scholar]
- 96.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H. G. Kräusslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 98.Worthylake, D. K., H. Wang, S. Yoo, W. I. Sundquist, and C. P. Hill. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr. D Biol. Crystallogr. 55:85-92. [DOI] [PubMed] [Google Scholar]
- 99.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]
- 101.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zabransky, A., E. Hunter, and M. Sakalian. 2002. Identification of a minimal HIV-1 gag domain sufficient for self-association. Virology 294:141-150. [DOI] [PubMed] [Google Scholar]
- 103.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou, W., and M. D. Resh. 1996. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 70:8540-8548. [DOI] [PMC free article] [PubMed] [Google Scholar]