Abstract
Human papillomavirus type 16 (HPV16) E6 and E7 oncoproteins are required for cellular transformation and represent candidate targets for HPV-specific and major histocompatibility complex class I-restricted CD8+-T-cell responses in patients with cervical cancer. Recent evidence suggests that cross-reactivity represents the inherent nature of the T-cell repertoire. We identified HLA-A2 binding HPV16 E7 variant peptides from human, bacterial, or viral origin which are able to drive CD8+-T-cell responses directed against wild-type HPV16 E7 amino acid 11 to 19/20 (E711-19/20) epitope YMLDLQPET(T) in vitro. CD8+ T cells reacting to the HLA-A2-presented peptide from HPV16 E711-19(20) recognized also the HLA-A2 binding peptide TMLDIQPED (amino acids 52 to 60) from the human coronavirus OC43 NS2 gene product. Establishment of coronavirus NS2-specific, HLA-A2-restricted CD8+-T-cell clones and ex vivo analysis of HPV16 E7 specific T cells obtained by HLA-A2 tetramer-guided sorting from PBL or tumor-infiltrating lymphocytes obtained from patients with cervical cancer showed that cross-reactivity with HPV16 E711-19(20) and coronavirus NS252-60 represents a common feature of this antiviral immune response defined by cytokine production. Zero of 10 patients with carcinoma in situ neoplasia and 3 of 18 patients with cervical cancer showed ≥0.1% HPV16 E7-reactive T cells in CD8+ peripheral blood lymphocytes. In vivo priming with HPV16 was confirmed in patients with cervical cancer or preinvasive HPV16-positive lesions using HLA-A2 tetramer complexes loaded with the E6-derived epitope KLPQLCTEL. In contrast, we could not detect E6-reactive T cells in healthy individuals. These data imply that the measurement of the HPV16 E711-19(20) CD8+-T-cell response may reflect cross-reactivity with a common pathogen and that variant peptides may be employed to drive an effective cellular immune response against HPV.
Anti-human papillomavirus (HPV)-directed and major histocompatibility complex (MHC)-restricted cellular immune responses have been detected in patients with cervical cancer (1, 2, 6, 10, 11, 15, 16, 34-37, 49). The quality and quantity of T cells directed against MHC-presented HPV epitopes may determine the clinical outcome of patients with HPV-positive tumors. Induction of HPV-reactive T cells also represents one of the surrogate markers in vaccine strategies. However, HPV type 16 (HPV16) E711-19(20)-specific T cells are scant or absent in peripheral blood lymphocytes (PBL) from patients with cervical cancer (1, 15, 16, 35, 36, 49). Several immunological factors may have an impact on either eradication or containment of HPV infection mediated by T lymphocytes (1, 5). For instance, the T-cell repertoire available at the time of viral infection may limit the successful outcome of an effective antiviral cellular immune response (46). We hypothesize that ineffective T-cell responses in patients with cervical cancer may, in part, be associated with low-affinity cytotoxic T-lymphocyte (CTL) responses due to cross-reactive recognition of T-cell epitopes shared among HPV16 E7 with peptides of human, bacterial, or viral origin. We have evaluated (i) HPV16 E7 peptide variants for the capacity to drive CD8+-T-cell responses directed against the HPV16 E7 gene product and (ii) HPV16 E7-peptide specific CD8+ T cells in PBL or tumor-infiltrating lymphocytes (TIL) from patients with cervical cancer using soluble tetramer complexes. We implemented the HPV16 E711-19, as well as the HPV16 E711-20 epitope loaded onto tetramer complexes, since the latter T-cell epitope (E711-20) may be superior compared to the E711-19 epitope in regard to HPV16 E7-specific CD8+-T-cell responses (24, 34, 49).
(This work was performed in fulfillment of the doctoral thesis of K. Nilges.)
MATERIALS AND METHODS
Flow cytometry.
Blood was obtained after obtaining informed consent of the patients and approval by the local ethics committee (reference number 837.210.00 [2576] dated 23 August 2000). Peripheral blood mononuclear cells (PBMCs) were obtained by separation over a Ficoll gradient and stored in liquid nitrogen at 1 × 107 to 5 × 107 cells/vial in 90% fetal calf serum (FCS) and 10% dimethyl sulfoxide (DMSO). TIL or T cells obtained from tumor-draining lymph nodes were obtained from patients undergoing intentive curative surgery for cervical cancer. Frozen PBL or TIL were thawed and washed in RPMI 1640 supplemented with 10% FCS and incubated with 1 μg of phycoerythrin (PE)-labeled tetramer reagent for 1 h, followed by incubation with the directly PE-cyanin (PC5)-labeled anti-CD8 alpha monoclonal antibody (MAb) clone T8 and fluorescein isothiocyanate-coupled anti-CD3 clone UCHT1. Cells were analyzed using the Coulter Epics XL and XL system software (version 2.1) as previously described (27). Cells were gated on CD3+ CD8+ cells, and tetramer-binding was evaluated. HLA-A2 tetramer-reagents loaded with the HPV16 E711-19 epitope YMLDLQPET, or alternatively with the HPV16 E711-20 epitope YMLDLQPETT, were obtained from the Immunomics Corporation (Beckman/Coulter, San Diego, Calif.). The HPV16 E6 epitope KLPQLCTEL (4) has also been used to prepare HLA-A2 tetramer complexes in order to gauge the cellular immune response to the HPV E6 gene product. HLA-A2 tetramer complexes loaded with the CMVpp65 epitope NLVPMVATV served as controls. A total of 20,000 events per sample were collected.
T-cell cloning.
CD8+ T cells obtained from healthy (HPV-negative) individuals were isolated by positive selection using immunomagnetic beads and initially stimulated two times with autologous interleukin-4 (IL-4)/granulocyte-macrophage colony-stimulating factor (GM-CSF)-generated dendritic cells pulsed with HPV16 E7 variant peptides as described in detail elsewhere (44). T cells were cloned at 100, 10, 1, or 0.1 cell/well in 96-well plates using HLA-A2-matched irradiated PBMCs as feeder cells in medium containing 50% AIM-V (GIBCO, Eggenstein, Germany) and 50% Dulbecco's modified Eagle medium (high glucose) supplemented with IL-2 (50 IU/ml) and IL-7 (50 ng/ml). T cells were restimulated at weekly intervals with irradiated HLA-A2-matched allogeneic PBMCs loaded with the appropriate peptide. T-cell clones or oligoclonal lines could only be sufficiently expanded in cultures stimulated with the coronavirus-derived peptide. T cells were seeded in 48-well plates and tested for target cell recognition in functional assays. Purity of peptides (listed in Table 1) was >97%, and amino acid composition was confirmed by mass spectroscopy. T-cell receptor (TCR) variable alpha (VA) and beta (VB) chain TCR usage of individual T-cell lines was determined using a PCR-based approach covering the entire TCR VA/VB repertoire (19, 21, 33).
TABLE 1.
HPV16-E7 variant peptidesa
| Peptide no. | Accession no. (reference[s]) | Sequencee | Origin | location |
|---|---|---|---|---|
| 1 | U76404, P03129 (13, 14) | YMLDLQPET | HPV16-E7b | 11-19 |
| 2 | Q15150 | YILDIQPQG | Human platelet glycoprotein IIBd | 99-107 |
| 3 | AG010038 | YMLSLHPED | Human chromosome 21q regiond | 80-88 |
| 4 | AL008719 | YMLILHPET | Human DNA sequence clone 342 BIId | 30507-30515 |
| 5 | AC004060 | YMLGLKPEV | Human chromosome 4, BAC cloned | 25844-25852 |
| 6 | P44987 | AKLDLEPET | Haemophilus sp. biotin synthetasec | 319-327 |
| 7 | P94189 | YILDLQPEN | Alcaligenes sp. polymerasec | 230-238 |
| 8 | Q80872 (15) | TMLDIQPED | Coronavirus NS2 proteinb | 52-60 |
| 9 | Control (17) | VLTDGNPPEV | M. tuberculosis 19-kDa antigen | 88-97 |
| 10 | Control (18) | YLEPGPVTA | Melanoma-associated antigen gp 100 | 280-288 |
HPV16-E7 variant peptides were selected for similarity to the wildtype HPV peptide using a computer algorithm (SYFPEITHI). The HLA-A2 anchor position two was either preserved or showed an amino acid residue which would still allow HLA-A2 binding.
Peptide from viral sources.
Peptide from bacterial origin.
Peptide from human origin.
The amino acid residues which differ compared to the HPV16 wild-type peptide are shown in boldface type.
Immunomagnetic cell sorting and functional assays.
CD4+ T cells were separated from 3 × 107 to 5 × 107 PMBCs using anti-CD4 coated immunomagnetic beads (Miltenyi, Bergisch Gladbach, Germany). The CD4+ T-cell-depleted PBMC population was incubated with the respective PE-labeled tetramer reagents (1 μg of tetramer/2 × 107 cells) for 1 h, washed once, and positively selected using anti-PE-directed immunomagnetic beads (Miltenyi). T cells were rested for 24 h in 96-well plates containing 50% AIM-V-medium and 50% Dulbecco's modified Eagle medium (high glucose) obtained from Gibco (Eggenstein, Germany) supplemented with 10% FCS and human recombinant IL-7 (50 ng/ml) generously provided by Adrian Minty, Sanofi, Paris, France. Tetramer-sorted cells using the HPV16 E711-19 or alternatively the HPV16 E711-20 peptide loaded on HLA-A2 molecules were pulsed onto T2 cells loaded either with diluent alone (10% DMSO, 90% RPMI) or 1 μg of HPV analogue peptides as indicated, supplemented with human β2-microglobulin (20 μg/106 cells/ml) obtained from Sigma (Deisenhofen, Germany). Fifty microliters of this target cell suspension was used per well; the effector/target ratio was 1:1. Cells were incubated with target antigens for 48 h, and supernatants harvested and tested for gamma interferon (IFN-γ) or GM-CSF using the enzyme-linked immunosorbent assay system obtained from Diaclone, Besançon, France. Additional target cell lines included the HLA-A2+, HPV16+ cervical cancer cell line Caski (38, 42) or the HLA-A2+ B-cell line C1R-A2 (see below). C1R-A2 cells were either transfected with the entire coronavirus OC43 NS2 gene (25) to test for recognition of naturally processed and presented peptides or with the melanoma-associated target antigen NY-ESO1 (22) as a control in the CMV-driven TA expression vector (Invitrogen, Groningen, The Netherlands). Peptides (listed in Table 1) were pulsed onto T2 cells plus human β2-microglobulin to test for peptide-specific T-cell recognition. MHC class I (MHC-I)-restricted T-cell responses were blocked with the anti-MHC-I specific MAb W6/32 (10 μg/well); the anti-DR specific MAb L243 served as the control. The peptide VLTDGNPPEV from the Mycobacterium tuberculosis 19-kDa antigen (31), as well as the peptide YLEPGPVTA from the melanoma-associated antigen gp100 (3), was used as non-HPV-related control peptides.
TCR-CDR3-spectratyping of in vitro-stimulated T-cell cultures.
CD8+ T cells from PBL obtained from patients with cervical cancer or with carcinoma in situ neoplasia (CIN) were positively sorted using anti-CD8 coated immunomagnetic beads. CD8+ T cells were stimulated in the presence of IL-2 (50 IU/ml) and IL-7 (50 ng/ml) at weekly intervals using autologous irradiated dendritic cells (generated with 1,000 IU of IL-4 and 1,000 IU GM-CSF for 7 days) pulsed with individual HLA-A2 peptides as indicated for 1 h at room temperature plus human β2-microglobulin. After four rounds of restimulation, T-cell cultures were tested for target cell recognition in a cytokine release assay. A different aliquot from these cultures was subjected for quantitative and qualitative TCR analysis in order to detect shared expansion of VB families. Briefly, RNA was extracted and reverse transcribed into cDNA, extracted RNA was amplified by individual TCR VA and VB-specific primer pairs, and a runoff reaction using a fluorophore-labeled TCR-CA-, or -CB-specific primer was performed as described in detail previously (19, 21, 33). Quantitative analysis and differences in the TCR CDR3 regions were assessed as described recently in detail elsewhere (32).
In order to identify monoclonal and oligoclonal TCR transcripts, amplicons were subcloned into the TA sequencing vector (Invitrogen) followed by DNA sequence analysis. Each CDR3 peak represents 3 bp coding for 1 amino acid (aa). The area under the curve represents the frequency of a distinct CDR3 length in an individual TCR VB family. In order to condense the information from a single sample analysis, the individual TCR VB families were grouped into a single figure, with VB1 to VB24 along with the CDR3 length expressed as the number of amino acids. This TCR-CDR3 landscape provides the structural anatomy as defined by the TCR-CDR3 length for each TCR family in a T-cell subpopulation.
HLA-A2 binding assay.
Endogenous peptides on HLA-A2 molecules expressed as a transgene on C1R-A2 cells generously provided from Russell Salter, Department of Pathology, University of Pittsburgh Medical School, Pittsburgh, Pa., were removed by mild acid treatment, and individual peptides were tested for the capacity to reconstitute the HLA-A2 molecule on the C1R-A2+ indicator cell line (23, 50). MHC-I/peptide/β2-microglobulin complexes were dissociated by incubating cells for 2 min in acid buffer, pH 3.3 (total volume, 5 ml). After neutralizing the pH by adding RPMI 1640 without serum, cells were pelleted and resuspended at 106 cells/ml in RPMI 1640 with 1% FCS, supplemented with β2-microglobulin (5 μg/ml; Sigma). One milliliter of this single-cell suspension was immediately added to individual assay tubes containing peptides at different concentrations. Peptides had been dissolved in DMSO and adjusted to the appropriate concentration as indicated, added in a total volume of 10 μl to each individual test tube, and incubated for 16 h at room temperature, and this was followed by the addition of 100 μl of MAb W6/32 (which recognizes a conformational determinant on the MHC-I product) for 30 min and a polyclonal goat fluorescein isothiocyanate-conjugated F(ab)′2 fragment anti-mouse immunoglobulin G as the secondary reagent. Control test tubes contained non-acid-treated HLA-A2-transfected C1R cells and acid-treated cells without peptides. Flow cytometric analysis was performed, and results are reported as a percentage of HLA-A2 reconstitution based on the following calculation: (mean fluorescence channel of non-acid-treated cells − mean fluorescence channel of acid-treated cells plus peptide)/(mean fluorescence channel of non-acid-treated cells − mean fluorescence channel of acid-treated cells without peptide) × 100.
The HLA-A1 binding peptide from MAGE-1 (EADPTGHSY) (45) was used as a negative control.
RESULTS
Identification of HPV16 E7 variant peptides.
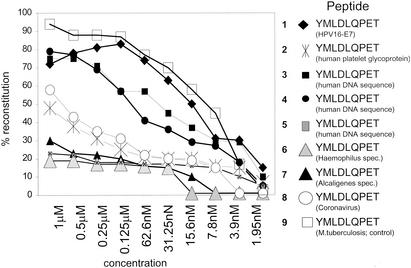
Peptides can be grouped according to their effect on CD8+ T cells (including proliferation, cytokine secretion, and cytotoxicity) in agonists, antagonists, or superagonists (43). Since mimicry or cross-reactive T-cell epitopes appear to exist in nature (26), we examined the accessible gene bank data for similarity with the HLA-A2 binding peptide YMLDLQPET provided from the HPV16 E7 gene product. If four permutations were allowed, eight different peptides (designated as analogue peptides [Table 1]) of either viral, bacterial, or human origin were identified which retained the ability to bind to HLA-A2 (Fig. 1).
FIG. 1.
HPV16 E7 variant peptides bind to HLA-A2. Individual peptides (numbering is identical to that in Table 1) were serially diluted and tested for the capacity to reconstitute empty HLA-A2 complexes on C1R-A2 cells in the presence of human β2-microglobulin as described in detail previously (23, 50). The HPV16 E7 wild-type peptide shows high affinity to HLA-A2 compared to the coronavirus derived peptide TMLDIQPED. The high-affinity HLA-A2 binding peptide VLTDGNPPEV from an M. tuberculosis antigen (19-kDa antigen) served as the positive control. Results are expressed as percent HLA-A2 reconstitution on C1R-A2 cells. An HLA-A1 binding peptide (EADPTGHSY) derived from the melanoma-associated MAGE-1 protein did not yield detectable HLA-A2 refolding (data not shown).
Detection of HPV and coronavirus peptide cross-reactive T cells.
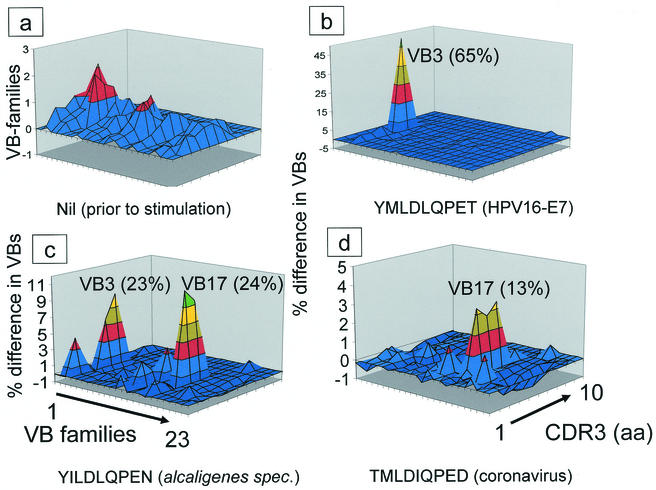
It is, however, unclear whether these peptides are naturally processed and presented and if they are capable of driving a peptide-specific CD8+-T-cell response. Next, we evaluated whether the HPV16 E7 peptide YMLDLQPET, or the panel of closely related peptides, would allow the establishment of HLA-A2+ restricted and peptide-specific T-cell lines. CD8+ T cells obtained from HPV-healthy individuals were stimulated using 7-day IL-4/GM-CSF-activated autologous dendritic cells with each of the peptides listed in Table 1. Briefly, T cells stimulated with the HPV16 E7 wild-type peptide were able to recognize the epitope used for stimulation but also peptides derived from viral (coronavirus) or bacterial (Alcaligenes sp.) origin (Table 2). A similar observation was found to be true for a T-cell line generated with a peptide (YMLSLHPED) deduced from the human chromosome 21q region. This T-cell line recognized the stimulating peptide but also HLA-A2 binding target peptides from the coronavirus NS2 gene product or from Alcaligenes sp. Similarly, the coronavirus peptide TMLDIQPED stimulated T cells which reacted against the coronavirus NS2 peptide, as well as the HPV16 E711-19 wild-type peptide, the “self peptide” YMLSLHPED, and peptides derived from Alcaligenes sp. TCR-repertoire analysis (Fig. 2) of these T-cell lines showed that each individual peptide expanded distinct as well as common T-cell populations; e.g., the HPV16 E711-19 peptide resulted in expansion of the TCR VB3 family (up to 65% after in vitro expansion), and the coronavirus NS2 peptide yielded preferential expansion of the TCR VB17 subset. In contrast, the peptide from Alcaligenes sp. resulted in TCR VB3 (similar to the HPV16 E7 peptide) and TCR VB17 (similar to the coronavirus peptide) expansion. Thus, T-cell cross-reactivity observed in polyclonal T-cell lines could in part be derived from preferential expansion of shared TCR VB families. T-cell recognition of HPV peptides could be observed not only for synthetic peptide epitopes but also for recognition of naturally processed and presented peptides provided from HPV either from the HPV16+, HLA-A2+ cervical cancer cell lines Caski or the human (HLA-A-, -B-negative) B-cell line C1R, which expressed HLA-A2 and the coronavirus NS2 gene product as transgenes (Table 3).
TABLE 2.
Definition of peptide-specific cross-reactive CD8+ T cellsa
| Peptide no. | Stimulating peptide | IFN-γ secretion (pg/ml/48 h) with peptide:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| nil | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| 1 | YMLDLQPET | 0 | 1,131 | 0 | 0 | 0 | 0 | 0 | 714 | 176 | 0 |
| 2 | YILDIQPQG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | YMLSLHPED | 0 | 0 | 0 | 152 | 0 | 0 | 0 | 517 | 739 | 0 |
| 4 | YMLILHPET | 0 | 371 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | YMLGLKPEV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | AKLDLEPET | 0 | 0 | 0 | 0 | 0 | 0 | 152 | 0 | 0 | 0 |
| 7 | YILDLQPEN | 0 | 298 | 0 | 298 | 0 | 0 | 0 | 1,055 | 0 | 0 |
| 8 | TMLDIQPED | 0 | 517 | 0 | 125 | 0 | 0 | 0 | 982 | 1,030 | 0 |
| 9 | VLTDGNPPEV | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | 1,179 |
Each individual T-cell line generated with either the HPV16 E7 wild-type or variant peptide listed in Table 1 was tested against the stimulating peptide as well as for recognition of HPV16 E7 analogue (target) peptides in an IFN-γ cytokine release assay (numbers refer to peptides listed in Table 1). Most of the peptides were able to drive a T-cell response directed against the stimulating peptide. T cells stimulated with the HPV16 E7 wild-type peptide were able to recognize the stimulating epitope but also the peptide derived from viral (coronavirus) or bacterial (Alcaligenes) origin. Similarly, the coronavirus peptide TMLDIQPED stimulated T cells which reacted against the HPV16 E7 peptide, the self-peptide YMLSLHPED and the Alcaligenes- and coronavirus-derived peptides. The M. tuberculosis-derived peptide VLTDGNPPEV served as the control. T-cell lines were generated from HPV-negative individuals.
FIG. 2.
In vitro expansion of similar TCR VB families by HPV16 E7 variant peptides. (a) Qualitative and quantitative assessment of the TCR VB composition in freshly harvested CD3+ CD8+ T cells. Note that not the raw data derived from the TCR VB-repertoire analysis in each individual T-cell culture, but the difference (in percent) in regard to quality (CDR3 analysis) and quantity of each TCR VB family compared to the freshly isolated T-population is provided. The perturbations of CD8+ T cells induced by repetitive stimulation with HPV16 E7 variant peptides (indicated underneath each figure) depicts the difference of the CDR3 profiles in respect to over- or underrepresentation of individual peaks in the TCR VB family. Each color represents 10% difference of the area under the curve in individual CDR3 peaks. An unperturbed repertoire (depicted here in blue) represents a smooth landscape surface. This implies that no difference compared to the qualitative or quantitative TCR landscape from the freshly harvested sample occurred. Individual TCR VB families are indicated (in boldface type and underlined), and the percentage of the respective VB family within the T-cell culture is provided in parentheses. The numbers in parentheses are absolute numbers of VB-positive staining CD8+ T cells. (a) Freshly harvested CD8+ T cells prior to peptide stimulation. (b) Note the prominent monoclonal TCR VB3 expansion in T cells stimulated with the HPV16 E7 wild-type peptide as well as in the T-cell culture stimulated with the peptide derived from Alcaligenes sp. (c) along with a prominent expansion of the TCR VB17 family. (d) Similar oligoclonal TCR VB17 expansion in T-cell cultures stimulated the peptide derived from coronavirus. The T-cell culture stimulated with a nonrelated peptide from a M. tuberculosis 19-kDa antigen showed an entirely different perturbation of the TCR VB repertoire (data not shown), indicating that expansion of TCR VB families is related not to culture conditions but to the stimulating peptide. PBL used to expand peptide-reactive T-cell lines were obtained from a patient with CIN. Monoclonality of the TCR VB3 expansion has been determined by DNA-sequence analysis. We could not generate T-cell lines from healthy blood donors or from patients with invasive cervical cancer.
TABLE 3.
Peptide-specific CD8+ T-cell lines recognize naturally processed and presented peptidesa
| Stimulating peptide no. | Target | Amt of cytokine produced (pg/ml/48 h)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cervical cancer cell line (A2+ HPV16+)
|
C1R-A2 NS2
|
C1R-A2 NY-ESO1
|
|||||||||
| Caski
|
Caski+ W6/32
|
Caski+ L243
|
|||||||||
| GM-CSF | IFN-γ | GM-CSF | IFN-γ | GM-CSF | IFN-γ | GM-CSF | IFN-γ | GM-CSF | IFN-γ | ||
| 1 | YMLDLQPET | 7,058 | 274 | 152 | 0 | 3,386 | 152 | 0 | 0 | 200 | 0 |
| 2 | YILDIQPQG | 201 | 0 | 249 | 0 | 450 | 0 | 0 | 0 | 200 | 0 |
| 3 | YMLSLHPED | 2,553 | 0 | 201 | 0 | 2,158 | 0 | 0 | 0 | 0 | 0 |
| 4 | YMLILHPET | 0 | 0 | 200 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | YMLGLKPEV | 200 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | AKLDLEPET | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | YILDLQPEN | 5,149 | 763 | 0 | 0 | 2,504 | 0 | 2,894 | 0 | 200 | 0 |
| 8 | TMLDIQPED | 5,976 | 517 | 249 | 0 | 4,754 | 0 | 8,036 | 1495 | 250 | 0 |
Peptide-reactive T-cell lines (Table 2) were tested for recognition of naturally processed and presented epitopes by secretion defined by GM-CSF and IFN-γ production. Stimulating peptide numbers refer to the listing of epitopes in Table 1. The HLA-A2+ and HPV16+ cervical cancer cell line Caski and the (HLA-A-, B-negative) B-cell line C1R, which expressed HLA-A2 and the coronavirus protein NS2 as a transgene, were used as target cells. The irrelevant melanoma-associated antigen NY-ESO1 expressed in HLA-A2+ C1R cells served as a control. Anti-MHC-I-specific T-cell recognition was blocked with the anti-MHC-I-directed MAb W6/32. The MAb L243, directed against MHC-II DR alleles, was used as a control reagent. The T-cell line generated with the HPV16 E7 wild-type peptide and T cells stimulated with the peptide from the coronavirus NS2 gene product recognized the HLA-A2+. HPV16+ cell line Caski in an MHC--I-restricted fashion. The HLA-A2+ NS2+ target C1R cell line was exclusively recognized by T cells stimulated with either the coronavirus peptide epitope or the peptide derived from Alcaligenes sp. In contrast, HLA-A2+ C1R cells transgenic for a different target antigen (the melanoma-associated antigen NY-ESO1) were not effectively recognized. Reduction of cytokine production by the anti-MHC-II-MAb reflects suppression of the allo-recognition of the cell line which has not been completely HLA-A matched.
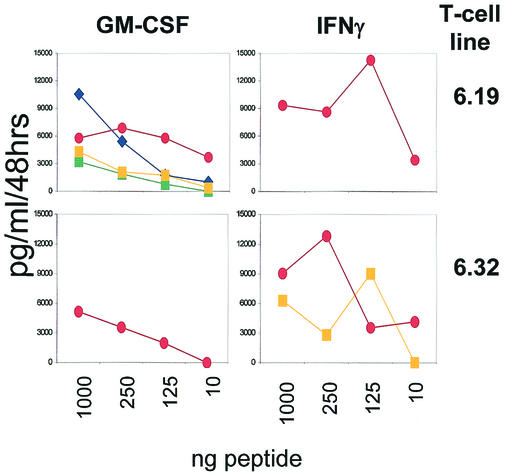
The notion of cross-reactive CD8+ T cells recognizing HPV16 E7 could be consolidated by molecularly defined clonal and oligoclonal T-cell lines reacting to the coronavirus peptide TMLDIQPED: this peptide is able to expand T-cell populations which recognize the stimulatory peptide but also closely related peptides, including an HLA-A2 binding peptide from the HPV16 E7 gene product. Each T-cell line was tested with the peptide listed in Table 1. Representative results are depicted in Fig. 3 as defined by either GM-CSF or IFN-γ release in response to peptides titrated onto T2 cells. Similar to polyclonal T-cell lines (Tables 2 and 3), the T-cell clone 6.19 reacted to the stimulating coronavirus peptide measured by IFN-γ release but secreted GM-CSF in a dose-dependent manner in response to the closely related HPV16 E7 peptide and recognized naturally processed and presented peptides provided from HPV16 on the cervical cancer cell line Caski and peptides presented on C1R cells transgenic for the NS2 gene and the restricting HLA-A2 molecule (data not shown). Cross-reactivity with HPV16 E7 could not be demonstrated in every individual TMLDIQPED (coronavirus)-specific T-cell line: the T-cell line 6.32 (Table 4) recognized the stimulating peptide and the self peptide YMLILHPET, but not the HPV16 E711-19 epitope.
FIG. 3.
CD8+-T-cell lines recognize the HPV16 E7 and the coronavirus-derived peptide. T-cell lines (Table 4) were obtained by repetitive stimulation with the coronavirus-derived peptide TMLDIQPED as described and tested for recognition of each of the peptides listed in Table 1. Each peptide was pulsed in different concentrations on surrogate antigen-presenting T2 cells and tested for the capacity to induce GM-CSF or IFN-γ in responder T cells. Exclusively positive results are depicted. Note that also self peptides were recognized. T-cell clones or lines were derived from an apparently healthy HLA-A2+ individual. We could not establish T-cell clones from patients with CIN or cervical cancer. These T-cell lines recognized also naturally processed and HLA-A2 presented peptides on HPV16+ A2+ Caski cells or C1R A2+ cells expressing the coronavirus NS2 gene product, respectively (data not shown). Other HPV16 E7 variant peptides (listed in Table 1) did not yield detectable cytokine production. Symbols: red ovals and blue diamonds, viral antigens (coronavirus NS2 and HPV16 E7, respectively; squares, self peptides (YMLILHPET [yellow] and YMLSLHPED [green]).
TABLE 4.
TCR composition of anti-coronavirus-directed T-cell clonesa
| T-cell clone | TCR composition of:
|
||||
|---|---|---|---|---|---|
| v family | Variable | CDR3 | Joining | j family | |
| 6.19 | VB 5S8 | LCASS | PRW | DTQYF | BJ2.3 |
| 6.32 | VB 1 | FCASS | LTQG | YEQYF | BJ2.7 |
| 6.32 | VB 2 | FYICS | AELAGN | EQYFG | BJ2.7 |
T-cell clones were established using the coronavirus NS2-derived epitope TMLDIQPED as a stimulus and tested for TCR VB composition. The TCR complementarity determining region 3 (CDR3) of individual T-cell lines designated 6.19 and 6.32 is depicted. Each individual T-cell line was tested for recognition of HPV16 E7 variant peptides (Fig. 3). The T-cell line 6.19 used the TCR VB (5S8) chain; analysis of the T-cell line 6.32 revealed usage of the TCR VB 1 and VB 2 chains. Both T-cell lines recognized naturally processed and presented epitopes on Caski cells and on C1R-A2+ cells, which expressed the coronavirus NS2 gene product as a transgene (data not shown).
Isolation of HPV16 E7 cross-reactive T cells from PBL using HLA-A2 tetramer complexes.
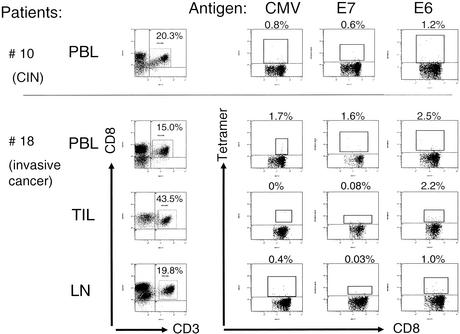
In order to test whether the phenomenon of cross-reactive T cells would also be observed in freshly harvested PBL, we took advantage of soluble HLA-A2 complexes loaded with the HPV16 E7 (aa 11 to 19)-derived peptide YMLDLQPET. Since reports suggested that HPV16 E7 (aa 11 to 20) YMLDLQPETT would be superior to the peptide YMLDLQPET, we implemented tetramer complexes with the HPV16 E7 (aa 11 to 20) epitope as a control. We could not utilize HLA-A2 tetramer complexes loaded with the coronavirus-derived peptide TMLDIQPED, since the binding affinity of this peptide to HLA-A2 (Fig. 1) is lower than that to the HPV16 peptide: this did not allow the formation of a stable tetramer complex. We have also considered altering the coronavirus peptide at the HLA-A2 anchor positions in order to enhance HLA-A2 binding. However, since the experiments in this report suggest that subtle changes in the peptide composition are sufficient to induce alternate T-cell effector functions, we resorted to the HPV16 E7 wild-type peptide to detect HPV16 E7-binding T cells, or to alternatively sort HLA-A2-restricted and YMLDLQPET(T)-specific CD8+ T cells from three individuals either suffering from CIN or from frank invasive cervical cancer. Noteworthy, 5 of 10 apparently healthy blood donors showed ≥0.1% tetramer-staining T cells in CD3+ CD8+ PBL (Table 5). This magnitude could not be observed in 10 patients with CIN (Table 5). In contrast to the lower numbers of tetramer-positive T cells in PBL obtained from patients with CIN, 3 of 18 patients with invasive cervical cancer showed ≥0.1% of CD3+ CD8+ T cells (Fig. 4a and Table 6) staining with the HPV16 E7 tetramer reagent. The CMV pp56 epitope loaded onto HLA-A2 tetramer complexes served as a control to a non-HPV-related CD8+-T-cell response directed against a viral epitope in PBL. Of note, the frequency of HPV tetramer-binding T cells was not elevated in freshly isolated TIL, nor was it elevated in T cells obtained from tumor-draining lymph nodes (Table 6) compared to PBL. PBL obtained from one patient with CIN (CIN patient 7) and from two patients with cervical cancer (patients 13 and 14) were subjected to tetramer-guided sorting using the HPV16 E711-19 epitope. Sorted T cells were tested for recognition of the nominal HPV16 E7 ligand, the coronavirus-derived NS2 peptide, as well as for recognition of the self peptide YMLILHPET, which has been shown to be recognized by clonal/oligoclonal coronavirus NS2-specific T-cell lines (Fig. 3). Tetramer-sorted T cells from all of these three patients showed GM-CSF, but not IFN-γ-secretion, in response to the nominal ligand, as well as to the coronavirus-derived peptide (Table 7). The self peptide or target cells without peptide were not recognized. Of note, these data have been obtained using the HPV16 E711-19 epitope, ex vivo-isolated T cells using the HPV16 E711-20 epitope (Table 8), and analysis of T cells in PBL (see Fig. 4b and Table 9). Thus, CD8+ T-cell cross-reactivity with the coronavirus NS2 gene product could be detected in HPV16 E711-19 and in HPV16 E711-20 ex vivo-sorted T cells. In order to show that HPV16 E711-19/20-reactive T cells in patients with HPV+ lesions reflect in vivo priming, we tested the presence of CD8+ T cells recognizing an HLA-A2 presented epitope from HPV16 E6. All HPV16+ patients showed an E6-directed T-cell response defined by tetramer analysis (Fig. 5). In contrast, we could not detect HPV16 E6-reactive immune responses in HPV-healthy individuals (data not shown), suggesting that the ex vivo detection of E7-reactive T cells in patients with HPV+ lesions reflects in vivo priming, since E6-specific responses are absent in HPV-negative individuals.
TABLE 5.
Detection of HPV16 E7-reactive CD8+ T cells in healthy individuals and patients with carcinoma in situa
| Group and donor no. | CD3+ CD8+ (%) | HPV (%) | CMV (%) |
|---|---|---|---|
| Healthy individuals | |||
| 1 | 18 | 0 | 0.03 |
| 2 | 24 | 0 | 0 |
| 3 | 16 | 0 | 0.1 |
| 4 | 15 | 0 | 0.12 |
| 5 | 14.6 | 0.02 | NDb |
| 6 | 8.37 | 0.41 | ND |
| 7 | 19.1 | 0.11 | ND |
| 8 | 11.5 | 0.13 | ND |
| 9 | 18.2 | 0.18 | ND |
| 10 | 38 | 0.1 | 0 |
| Patients with carcinoma | |||
| 1 | 21.8 | 0.06 | 0.10 |
| 2 | 15.4 | 0.03 | 0.03 |
| 3 | 21.2 | 0.03 | 0.01 |
| 4 | 8.89 | 0.04 | 0.03 |
| 5 | 15.9 | 0.01 | 0.00 |
| 6 | 27.4 | 0.04 | 0.00 |
| 7 | 11.9 | 0.08 | 0.32 |
| 8 | 9.73 | 0.02 | 0.02 |
| 9 | 16.1 | 0.06 | 0.00 |
| 10 | 20.3 | 0.6 | 0.8 |
PBL, TIL, or T cells from tumor-draining lymph nodes were isolated, gated on CD3+ CD8+ T cells as indicated, and tested for the frequency of HPV16-E711-19 or CMVpp65 binding T cells using HLA-A2 tetramer complexes. Note the high frequency of HPV16 E7 staining T cells in healthy individuals and the low numbers of HPV-reactive T-cells in the CD8+ T-cell population in patients with preinvasive cervical cancer or in PBL from patients with invasive cervical cancer (see Table 6). Data for tetramer-reactive T cells showing ≥ 0.1 frequency in CD3+ CD8+ are shown in bold face type. In contrast to HPV-specific T cells, 7 of 18 patients with cervical cancer showed CD8+ T cells (≥ 0.1%) binding to the CMVpp65 T-cell epitope. Low numbers of HPV-reactive T cells could be detected in TIL or in tumor draining lymph nodes (see Table 6). All individuals are HLA-A2+, and HPV16 DNA was present in the tumor lesions of all patients with CIN, as detected by PCR analysis. PBL from healthy individuals (donors 5 to 7), patients with CIN (donors 4 and 7), or patients with cervical cancer (donors 6, 8, 13, and 16 in Table 6) were restimulated in vitro either with the HPV16 E7 wild-type epitope YMLDLQPET or with the coronavirus-derived peptide TMLDIQPED using HLA-A2-matched dendritic cells as antigen-presenting cells in order to test if the number of HPV tetramer-reactive T cells could be increased. No significant increase of tetramer-binding T cells could be observed in any of the in vitro culture assays (data not shown).
ND, not determined.
FIG. 4.
Detection of HPV16 E711-19/20YMLDLQPET(T)-reactive T cells in PBL or TIL from patients with cervical cancer. (a) PBL or TIL were tested for binding to HLA-A2 tetramer complexes loaded with the HPV16 E711-19 wild-type epitope. Tetramer-positive staining T cells are shown in red. Samples correspond to those from patients listed in Table 6. Note the difference of tetramer-reactive T cells in freshly harvested TIL compared to PBL. Detection of CMVpp65-binding T cells served as a positive control. TIL or T cells obtained from tumor-draining lymph nodes were obtained from patients who underwent surgery for cervical cancer. Similar results were obtained using the HPV16 E711-20 epitope (YMLDLQPETT) loaded onto HLA-A2 complexes compared to the HPV16 E711-19 peptide. (b) The data correspond to tetramer-positive T cells in PBL, TIL, or T cells obtained from tumor draining lymph nodes from patient A listed in Table 9.
TABLE 6.
Detection of HPV16 E7-reactive CD8+ T cells in patients with cervical cancera
| Donor no. | PBL
|
TIL
|
Lymph node T cells
|
HPV16 status of tumor | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CD3+ CD8+ (%) | HPV (%) | CMV (%) | CD3+ CD8+ (%) | HPV (%) | CMV (%) | CD3+ CD8+ (%) | HPV (%) | CMV (%) | ||
| 1 | 3.45 | 0.02 | ND | 14.7 | 0.00 | ND | 6.45 | 0.00 | ND | + |
| 2 | 14.2 | 0.02 | 0.06 | 52.4 | 0.01 | 0.02 | ND | ND | ND | + |
| 3 | ND | ND | ND | 13.7 | 0.02 | 0.01 | ND | ND | ND | + |
| 4 | 16.7 | 0.04 | 1.00 | ND | ND | ND | ND | ND | ND | + |
| 5 | 6.74 | 0.02 | ND | ND | ND | ND | ND | ND | ND | + |
| 6 | 23.2 | 0.00 | 0.10 | 46.4 | 0.00 | 0.03 | 4.28 | 0.00 | 0.01 | + |
| 7 | 15.1 | 0.06 | 0.72 | ND | ND | ND | ND | ND | ND | + |
| 8 | 17.6 | 0.02 | 0.03 | 14.8 | 0.02 | ND | ND | ND | ND | + |
| 9 | ND | ND | ND | 8.57 | 0.00 | 0.02 | ND | ND | ND | + |
| 10 | ND | ND | ND | 14.4 | 0.00 | ND | ND | ND | ND | − |
| 11 | 18.6 | 0.00 | 0.00 | ND | ND | ND | ND | ND | ND | + |
| 12 | 22.2 | 0.03 | ND | ND | ND | ND | ND | ND | ND | + |
| 13 | 9.57 | 0.47 | 0.30 | 26.6 | 0.02 | 0.01 | 24.7 | 0.04 | 0.01 | + |
| 14 | 38.1 | 0.18 | 0.69 | ND | ND | ND | ND | ND | ND | + |
| 15 | 15.1 | 0.01 | 0.26 | 20.0 | 0.04 | 0.37 | ND | ND | ND | + |
| 16 | 14.2 | 0.00 | 0.00 | 15.7 | 0.03 | ND | ND | ND | ND | + |
| 17 | 17.1 | 0.00 | 0.02 | 9.02 | 0.01 | 0.00 | 8.91 | 0.00 | 0.01 | − |
| 18 | 15 | 1.6 | 1.7 | 43.5 | 0.08 | 0.00 | 19.81 | 0.03 | 0.4 | + |
See footnotes to Table 5 for details.
TABLE 7.
YMLDLQPET tetramer-sorted CD8+ T cells recognize a peptide provided from the coronavirus NS2 gene producta
| Target | Peptide | Amt of GM-CSF produced (pg/ml/48 h) by T cells from patient in indicated expt
|
|||||
|---|---|---|---|---|---|---|---|
| 7
|
13
|
14
|
|||||
| A | B | A | B | A | B | ||
| T2 cells pulsed with: | |||||||
| Medium alone | 0 | 0 | 0 | 0 | 0 | 0 | |
| HPV16 E7 | YMLDLQPET | 37 | 43 | 51 | 12 | 27 | 30 |
| Human DNA clone 342 | YMLILHPET | 10 | 7 | 0 | 0 | 13 | 0 |
| Coronavirus NS2 | TMLDIOPED | 139 | 50 | 11 | 5 | 123 | NDb |
| gp100 (control) | YLEPGPVTA | 0 | 0 | 0 | 0 | 0 | 0 |
YMLDLQPET tetramer-reactive T cells from PBL from patient 7 with CIN (Table 5) or patients 13 and 14 with invasive cervical cancer (Table 6) were isolated as described in detail in Materials and Methods. The patient designation corresponds to the numbering in Tables 5 and 6. Two independent experiments (this table and Table 8) have been performed. HPV16 E7-isolated T cells recognize the nominal T-cell epitope as well as the peptide derived from the coronavirus NS2 gene product. T2 cells alone, or those pulsed with a self peptide, or a non-HPV-related (melanoma-associated peptide, i.e., gp 100) peptide, were not recognized. T cells responded with GM-CSF production but not with IFN-γ production. Due to the low number of tetramer-sorted T cells, blocking experiments using the anti-MHC-I-specific MAb W6/32 could not be performed. A and B indicate two independent experiments.
ND, not determined.
TABLE 8.
YMLDLQPETT tetramer-sorted T cells recognize a peptide provided from the coronavirus NS2 gene producta
| Target | T-cell epitope | Blocking MAb | Amt of GM-CSF produced (pg/ml/48 h) by T cells from patient:
|
|
|---|---|---|---|---|
| A | C | |||
| Medium alone | Nil | 0 | 0 | |
| HPV16 E711-20 | YMLDLQPETT | Nil | 4,000 | 2,500 |
| Anti-MHC-I | 0 | 0 | ||
| Anti-MHC-II | 4,000 | 1,800 | ||
| Human DNA clone 342 | YMLILHPET | Nil | 0 | 0 |
| Coronavirus NS2 | TMLDIQPED | Nil | 4,000 | 3,025 |
| Anti-MHC-I | 0 | 0 | ||
| Anti-MHC-II | 4,000 | 3,000 | ||
| HPV16 E711-19 | YMLDLQPET | Nil | 1,000 | 1,940 |
| gp100 (control) | YLEPGPVTA | 0 | 0 | 0 |
Experimental conditions were similar to those described in footnote a of Table 7 except that the HPV16-E711-20 epitope YMLDLQPETT was utilized to sort antigen-specific T cells. Note that HPV16 E711-20 tetramer-sorted T cells also recognize the HPV16 E711-19 epitope. Patients designations were respond to patients A to C listed in Table 9. MHC restriction could be demonstrated by blocking with the anti-MHC-I-directed MAb w6/32, but not with the anti-MHC-II (HLA-DR)-directed MAb L243. YMLDLQPETT-reactive T cells also recognize the related coronavirus peptide in an MHC-I-restricted fashion.
TABLE 9.
Similar HPV16 E7-specific CD8+ T-cell numbers in PBL directed against the HPV16 E711-19 or HPV16 E711-20 peptide epitopesa
| Patient | Compartment | % Tetramer+ T cells within CD3+ CD8+ T cells
|
|
|---|---|---|---|
| HPV16 E711-19 | HPV16 E711-20 | ||
| A | PBL | 0.6 | 0.5 |
| TIL | 0.1 | 0 | |
| LN | 0.2 | 0.2 | |
| B | PBL | 0.2 | 0.3 |
| TIL | 0.1 | 0 | |
| LN | 0.2 | 0.2 | |
| C | PBL | 0.3 | 0.3 |
| TIL | 0.1 | 0 | |
| LN | 0.1 | 0.1 | |
PBL, TIL, or T cells from tumor-draining lymph nodes obtained from three additional patients suffering from cervical cancer were tested for binding either to the HLA-A2 binding peptide HPV16 E711-19 YMLDLQPET or, alternatively, to the HPV16 E711-20 peptide YMLDLQPETT. T cells were first gated on CD3+ CD8+ cells and tested for tetramer binding. No substantial difference of E711-19 compared to E711-20 peptide-binding T cells was observed Note that the HPV16 E711-19 peptide, but not the HPV16 E711-20 peptide loaded onto tetramer complexes, yielded a low but detectable number (0.1%) of tetramer-positive. T cells in TIL. Patients A to C do not match the patients listed in Tables 5 and 6. Flow cytometry data from patient A are depicted in Fig. 4b. The analysis of YMLDLQPETT-sorted T cells indicated that HPV16 E711-20-reactive T cells also recognize the HPV16 E711-19 T-cell epitope (Table 8).
FIG. 5.
Evidence for in vivo priming with HPV. PBL, TIL, or T cells from tumor-draining lymph nodes (Tables 5 and 6) were analyzed for CD8+ T cells binding to an HPV16 E6 epitope. Each HPV+ patient exhibited detectable levels of E6-reactive T cells. Two representative samples are shown: E7- and E6-reactive T cells from patient 10 with CIN (top panel) (Table 5) and E7- and E6-reactive T cells obtained from patient 18 with invasive cervical cancer (bottom panel) (Table 6). None of the HPV-healthy individuals showed E6-tetramer binding T cells (data not shown).
DISCUSSION
Ex vivo-sorted HPV16 E711-19/20-reactive T-cell populations as well as in vitro-generated T-cell clones recognize not only the stimulating index peptide but also variant peptides derived from different (human, viral, or bacterial) origins. It is unclear if these peptides are presented in vivo, but T-cell recognition of HPV16+, HLA-A2+ Caski cells, as well as B cells transgenic for HLA-A2 and the NS2 gene product suggest that T-cell cross-reactivity affects not only synthetic peptides but also naturally processed and presented T-cell epitopes (Tables 2 and 3). Is this cross-reactivity a single fortuitous finding, or does it reflect a more common pattern of T-cell specificity?
Recent studies revealed that each individual experiences a series of bacterial or viral infections which shape the quality and quantity of the memory T-cell pool (39, 48). This preexisting T-cell (memory) pool may be activated and expanded by subsequent viral infections: T-cell cross-reactivity may represent a common event (28) and reflect the inherent nature of the T-cell recognition of humans. Indeed, a detailed analysis of anti-Epstein-Barr virus-directed and HLA-B8-restricted CD8+-T-cell responses demonstrated CTL clones which are able to react not only to the wild-type (Epstein-Barr virus)-peptide but also to a human self peptide provided from a serine/threonine kinase and a peptide from the Staphylococcus aureus replication initiation protein (30). Similarly, antigenic peptides provided from self proteins—including CD9, glutamyl transferase, a G-protein-coupled receptor, or from bacterial or viral (e.g., herpes simplex virus) sources (26)—may explain why antimelanoma (i.e., anti-Melan-A/MART-1)-directed CD8+ T cells are present in apparently healthy HLA-A2 individuals. Not mutually exclusive, the high degree of cross-reactivity in Melan-A/MART-1-reactive T cells defining several specificities (i.e., self, peptides from viral or bacterial origin) may be associated with thymic selection of a broad T-cell repertoire in naïve CD8+ T cells in humans (13). A similarsituation may, in part, be true for anti-HPV-directed immune responses. This list could be expanded; e.g., CD8+ T cells specific for a hemagglutinin epitope (aa 210 to 219) recognize a peptide provided by a human immunoglobulin VH gene product (7, 8). It is likely that such cross-reactive T-cell populations may also have an impact on viral clearance and/or disease progression: a more recent study showed that cross-reactive T cells have an impact on the clinical course of disease if the cellular immune response is inflicted with inherent cross-recognition of unrelated pathogens in mice (40).
The observation that cross-reactive T cells directed against the HPV16 E711-19/20 or the coronavirus peptide exist may explain why HPV16 E711-19/20-directed CD8+-T-cell responses are present in apparently healthy individuals as illustrated in the present as well as in a previous report (49), showing low frequencies of HPV16 E711-20 tetramer-binding T cells in the range of 1 of 1,855 to 1 of 42,004 PBMCs in the HPV-negative control population. The HPV16 E711-20-directed immune response has been described to be present in the (CD45RO+) memory T-cell population in patients with cancer (36). Until now, the CD8 differentiation status of T cells recognizing HPV16 E7 variant peptides has not yet been determined.
Rhinovirus and coronavirus are most frequently associated with the common cold and contribute up to 5.1% of all hospital admission related to acute respiratory illnesses (14). Thus, anti-HPV16 E7-directed CD8+ T cells in healthy individuals may have originated from an anticoronavirus-directed T-cell response. This notion is supported by our observation that healthy donors testing positive for E7-directed immune responses do not exhibit evidence for in vivo priming with HPV: we could not detect anti-HPV16 E6-directed CD8+ T cells in this population, but we could detect them in patients with HPV+ lesions (Fig. 5). Of interest, HPV16-E2 reactive CD4+-T-cell responses have also recently been reported to be frequent in healthy individuals (12). The high frequency of E2-reactive memory T cells has been attributed to cross-reactivity with peptides from HPV types other than HPV16. Not mutually exclusive, closely related peptides of different origin may also contribute to sustain strong HPVE2 CD4+ memory T-cell responses.
Is this apparent T-cell cross-reactivity beneficial or deleterious for the host? In theory, the HPV peptide may behave as a low-affinity ligand which leads to a weak (re)activation of antigen-specific clonotypic T cells due to (i) peripheral tolerance of T cells either to self peptides or (ii) tolerance to ligands which had been encountered in previous infections provided by viruses or bacteria. The ligation of the T-cell receptor by such altered peptide ligands may result in diminished or altered TCR signaling events (29). This type of T-cell recognition is also associated with different downstream events, i.e., different cytokine release as detected in our assay system (GM-CSF and/or IFN-γ production; see Fig. 3). Thus, cross-reactive peptides may perform a role in contributing to maintenance and survival of CD8+ T cells which react to such mimicry ligands: T-cell cross-reactivity impact the quality of memory T-cell responses directed against unrelated pathogens (41). In contrast, previous encounters with closely related peptide epitopes may result in activation-induced cell death upon reexposure to the nominal or cross-reactive T-cell epitope (47). This would bear deleterious consequences for the host, a situation which has been termed exhaustion of the T-cell repertoire (18). Differences of the TCR repertoire reacting to variant HPV16 E7 peptides may represent the basis for an intrinsic inefficient CTL response in concert with other factors which are not favorable to mount an effective anti-HPV response, i.e., low density of peptide presentation on HPV-infected epithelial cells due to low E7 protein expression (20) or the absence of an inflammatory danger response (reviewed in reference 17).
Alternatively, cross-reactive T cells may also contribute to immune protection. For instance, memory CD8+ T cells specific for a lymphocytic choriomeningitis virus epitope are functionally activated and secrete IFN-γ in response to acute infection with (the unrelated) vaccinia virus associated with decreased mortality (9).
The data presented in the present study imply that T-cell responses directed against the HPV16 E711-19/20 epitope may not be particularly useful for gauging an HPV16 E7-specific CD8+-T-cell response. However, the existence of cross-reactive T-cell populations demonstrated in this report indicates the opportunity to implement variant peptides capable of driving HPV reactive CD8+ T cells which have not yet been activated by naturally presented peptides but are capable of successfully recognizing the wild type HPV-epitope displayed on tumor cells. This strategy may aid in the activation of strong and long-lived anti-HPV-directed cellular immune responses.
Acknowledgments
This work was in part supported by the Deutsche Forschungsgemeinschaft, grant SFB 432, A9 (to M.J.M.), and a core grant from the Deutsche Krebshilfe.
REFERENCES
- 1.Adams, M., L. Borysiewicz, A. Fiander, S. Man, B. Jasani, H. Navabi, C. Lipetz, A. S. Evans, and M. Mason. 2001. Clinical studies of human papilloma vaccines in pre-invasive and invasive cancer. Vaccine 19:2549-2556. [DOI] [PubMed] [Google Scholar]
- 2.al-Saleh, W., S. L. Giannini, N. Jacobs, M. Moutschen, J. Doyen, J. Boniver, and P. Delvenne. 1998. Correlation of T-helper secretory differentiation and types of antigen-presenting cells in squamous intraepithelial lesions of the uterine cervix. J. Pathol. 184:283-290. [DOI] [PubMed] [Google Scholar]
- 3.Bakker, A. B., M. W. Schreurs, G. Tafazzul, A. J. de Boer, Y. Kawakami, G. J. Adema, and C. G. Figdor. 1995. Identification of a novel peptide derived from the melanocyte-specific gp100 antigen as the dominant epitope recognized by an HLA-A2.1-restricted anti-melanoma CTL line. Int. J. Cancer 62:97-102. [DOI] [PubMed] [Google Scholar]
- 4.Bartholomew, J. S., S. N. Stacey, B. Coles, D. J. Burt, J. R. Arrand, and P. L. Stern. 1994. Identification of a naturally processed HLA A0201-restricted viral peptide from cells expressing human papillomavirus type 16 E6 oncoprotein. Eur. J. Immunol. 24:3175-3179. [DOI] [PubMed] [Google Scholar]
- 5.Borysiewicz, L. K., A. Fiander, M. Nimako, S. Man, G. W. Wilkinson, D. Westmoreland, A. S. Evans, M. Adams, S. N. Stacey, M. E. Boursnell, E. Rutherford, J. K. Hickling, and S. C. Inglis. 1996. A recombinant vaccinia virus encoding human papillomavirus types 16 and 18, E6 and E7 proteins as immunotherapy for cervical cancer. Lancet 347:1523-1527. [DOI] [PubMed] [Google Scholar]
- 6.Breitburd, F., N. Ramoz, J. Salmon, and G. Orth. 1996. HLA control in the progression of human papillomavirus infections. Semin. Cancer Biol. 7:359-371. [DOI] [PubMed] [Google Scholar]
- 7.Cao, W., B. A. Myers-Powell, and T. J. Braciale. 1994. Recognition of an immunoglobulin VH epitope by influenza virus-specific class I major histocompatibility complex-restricted cytolytic T lymphocytes. J. Exp. Med. 179:195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, W., B. A. Myers-Powell, and T. J. Braciale. 1996. The weak CD8+ CTL response to an influenza hemagglutinin epitope reflects limited T cell availability. J. Immunol. 157:505-511. [PubMed] [Google Scholar]
- 9.Chen, H. D., A. E. Fraire, I. Joris, M. A. Brehm, R. M. Welsh, and L. K. Selin. 2001. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat. Immunol. 2:1067-1076. [DOI] [PubMed] [Google Scholar]
- 10.De Bruijn, M. L., D. H. Schuurhuis, M. P. Vierboom, H. Vermeulen, K. A. de Cock, M. E. Ooms, M. E. Ressing, M. Toebes, K. L. Franken, J. W. Drijfhout, T. H. Ottenhoff, R. Offringa, and C. J. Melief. 1998. Immunization with human papillomavirus type 16 (HPV16) oncoprotein-loaded dendritic cells as well as protein in adjuvant induces MHC class I-restricted protection to HPV16-induced tumor cells. Cancer Res. 58:724-731. [PubMed] [Google Scholar]
- 11.de Gruijl, T. D., H. J. Bontkes, J. M. Walboomers, M. J. Stukart, F. S. Doekhie, A. J. Remmink, T. J. Helmerhorst, R. H. Verheijen, M. F. Duggan-Keen, P. L. Stern, C. J. Meijer, and R. J. Scheper. 1998. Differential T helper cell responses to human papillomavirus type 16 E7 related to viral clearance or persistence in patients with cervical neoplasia: a longitudinal study. Cancer Res. 58:1700-1706. [PubMed] [Google Scholar]
- 12.de Jong, A., S. H. van der Burg, K. M. Kwappenberg, J. M. van der Hulst, K. L. Franken, A. Geluk, K. E. van Meijgaarden, J. W. Drijfhout, G. Kenter, P. Vermeij, C. J. Melief, and R. Offringa. 2002. Frequent detection of human papillomavirus16. E2-specific T-helper immunity in healthy subjects. Cancer Res. 62:472-479. [PubMed] [Google Scholar]
- 13.Dutoit, V., V. Rubio-Godoy, M. J. Pittet, A. Zippelius, P. Y. Dietrich, F. A. Legal, P. Guillaume, P. Romero, J. C. Cerottini, R. A. Houghten, C. Pinilla, and D. Valmori. 2002. Degeneracy of antigen recognition as the molecular basis for the high frequency of naive A2/Melan-a peptide multimer+ CD8+ T cells in humans. J. Exp. Med. 196:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Sahly, H. M., R. L. Atmar, W. P. Glezen, and S. B. Greenberg. 2000. Spectrum of clinical illness in hospitalized patients with “common cold”virus infections. Clin. Infect. Dis. 31:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, C., S. Bauer, T. Grubert, C. Brucker, S. Baur, K. Heeg, H. Wagner, and G. B. Lipford. 1996. HLA-A2-restricted peripheral blood cytolytic T lymphocyte response to HPV type 16 proteins E6 and E7 from patients with neoplastic cervical lesions. Cancer Immunol. Immunother. 42:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, E. M., S. Man, A. S. Evans, and L. K. Borysiewicz. 1997. Infiltration of cervical cancer tissue with human papillomavirus-specific cytotoxic T-lymphocytes. Cancer Res. 57:2943-2950. [PubMed] [Google Scholar]
- 17.Frazer, I. H., R. Thomas, J. Zhou, G. R. Leggatt, L. Dunn, N. McMillan, R. W. Tindle, L. Filgueira, P. Manders, P. Barnard, and M. Sharkey. 1999. Potential strategies utilised by papillomavirus to evade host immunity. Immunol. Rev. 168:131-142. [DOI] [PubMed] [Google Scholar]
- 18.Gallimore, A., A. Glithero, A. Godkin, A. C. Tissot, A. Pluckthun, T. Elliott, H. Hengartner, and R. Zinkernagel. 1998. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J. Exp. Med. 187:1383-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genevee, C., A. Diu, J. Nierat, A. Caignard, P. Y. Dietrich, L. Ferradini, S. Roman-Roman, F. Triebel, and T. Hercend. 1992. An experimentally validated panel of subfamily-specific oligonucleotide primers (V alpha 1-w29/V beta 1-w24) for the study of human T cell receptor variable V gene segment usage by polymerase chain reaction. Eur. J. Immunol. 22:1261-1269. [DOI] [PubMed] [Google Scholar]
- 20.Greenfield, I., J. Nickerson, S. Penman, and M. Stanley. 1991. Human papillomavirus 16 E7 protein is associated with the nuclear matrix. Proc. Natl. Acad. Sci. USA 88:11217-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohn, H., T. Reichert, C. Neukirch, H. Pilch, and M. J. Maeurer. 1999. Monoclonal TCR mRNA transcripts are preferentially detected in the TCR variable alpha chain in CD8+ T-lymphocytes: implications for immunomonitoring. Int. J. Mol. Med. 3:139-144. [DOI] [PubMed] [Google Scholar]
- 22.Jager, E., S. Gnjatic, Y. Nagata, E. Stockert, D. Jager, J. Karbach, A. Neumann, J. Rieckenberg, Y. T. Chen, G. Ritter, E. Hoffman, M. Arand, L. J. Old, and A. Knuth. 2000. Induction of primary NY-ESO-1 immunity: CD8+ T lymphocyte and antibody responses in peptide-vaccinated patients with NY-ESO-1+ cancers. Proc. Natl. Acad. Sci. USA 97:12198-12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jager, E., H. Hohn, J. Karbach, F. Momburg, C. Castelli, A. Knuth, B. Seliger, and M. J. Maeurer. 1999. Cytotoxic T lymphocytes define multiple peptide isoforms derived from the melanoma-associated antigen MART-1/Melan-A. Int. J. Cancer 81:979-984. [DOI] [PubMed] [Google Scholar]
- 24.Kast, W. M., R. M. Brandt, J. Sidney, J. W. Drijfhout, R. T. Kubo, H. M. Grey, C. J. Melief, and A. Sette. 1994. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J. Immunol. 152:3904-3912. [PubMed] [Google Scholar]
- 25.Labonte, P., S. Mounir, and P. J. Talbot. 1995. Sequence and expression of the ns2 protein gene of human coronavirus OC43. J. Gen. Virol. 76:431-435. [DOI] [PubMed] [Google Scholar]
- 26.Loftus, D. J., C. Castelli, T. M. Clay, P. Squarcina, F. M. Marincola, M. I. Nishimura, G. Parmiani, E. Appella, and L. Rivoltini. 1996. Identification of epitope mimics recognized by CTL reactive to the melanoma/melanocyte-derived peptide MART-1(27-35). J. Exp. Med. 184:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeurer, M. J., A. Necker, R. D. Salter, C. Castelli, H. Hohn, J. Karbach, K. Freitag, C. Neukirch, A. Knuth, and E. Jager. 2002. Improved detection of melanoma antigen-specific T cells expressing low or high levels of CD8 by HLA-A2 tetramers presenting a Melan-A/Mart-1 peptide analogue. Int. J. Cancer 97:64-71. [DOI] [PubMed] [Google Scholar]
- 28.Mason, D. 1998. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today 19:395-404. [DOI] [PubMed] [Google Scholar]
- 29.McKeithan, T. W. 1995. Kinetic proofreading in T-cell receptor signal transduction. Proc. Natl. Acad. Sci. USA 92:5042-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misko, I. S., S. M. Cross, R. Khanna, S. L. Elliott, C. Schmidt, S. J. Pye, and S. L. Silins. 1999. Crossreactive recognition of viral, self, and bacterial peptide ligands by human class I-restricted cytotoxic T lymphocyte clonotypes: implications for molecular mimicry in autoimmune disease. Proc. Natl. Acad. Sci. USA 96:2279-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohagheghpour, N., D. Gammon, L. M. Kawamura, A. van Vollenhoven, C. J. Benike, and E. G. Engleman. 1998. CTL response to Mycobacterium tuberculosis: identification of an immunogenic epitope in the 19-kDa lipoprotein. J. Immunol. 161:2400-2406. [PubMed] [Google Scholar]
- 32.Pilch, H., H. Hohn, K. Freitag, C. Neukirch, A. Necker, P. Haddad, B. Tanner, P. G. Knapstein, and M. J. Maeurer. 2002. Improved assessment of T-cell receptor (TCR) VB repertoire in clinical specimens: combination of TCR-CDR3 spectratyping with flow cytometry-based TCR VB frequency analysis. Clin. Diagn. Lab. Immunol. 9:257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puisieux, I., J. Even, C. Pannetier, F. Jotereau, M. Favrot, and P. Kourilsky. 1994. Oligoclonality of tumor-infiltrating lymphocytes from human melanomas. J. Immunol. 153:2807-2818. [PubMed] [Google Scholar]
- 34.Ressing, M. E., A. Sette, R. M. Brandt, J. Ruppert, P. A. Wentworth, M. Hartman, C. Oseroff, H. M. Grey, C. J. Melief, and W. M. Kast. 1995. Human CTL epitopes encoded by human papillomavirus type 16 E6 and E7 identified through in vivo and in vitro immunogenicity studies of HLA-A*0201-binding peptides. J. Immunol. 154:5934-5943. [PubMed] [Google Scholar]
- 35.Ressing, M. E., W. J. van Driel, R. M. Brandt, G. G. Kenter, J. H. de Jong, T. Bauknecht, G. J. Fleuren, P. Hoogerhout, R. Offringa, A. Sette, E. Celis, H. Grey, B. J. Trimbos, W. M. Kast, and C. J. Melief. 2000. Detection of T helper responses, but not of human papillomavirus-specific cytotoxic T lymphocyte responses, after peptide vaccination of patients with cervical carcinoma. J. Immunother. 23:255-266. [DOI] [PubMed] [Google Scholar]
- 36.Ressing, M. E., W. J. van Driel, E. Celis, A. Sette, M. P. Brandt, M. Hartman, J. D. Anholts, G. M. Schreuder, W. B. ter Harmsel, G. J. Fleuren, B. J. Trimbos, W. M. Kast, and C. J. Melief. 1996. Occasional memory cytotoxic T-cell responses of patients with human papillomavirus type 16-positive cervical lesions against a human leukocyte antigen-A *0201-restricted E7-encoded epitope. Cancer Res. 56:582-588. [PubMed] [Google Scholar]
- 37.Rudolf, M. P., S. Man, C. J. Melief, A. Sette, and W. M. Kast. 2001. Human T-cell responses to HLA-A-restricted high binding affinity peptides of human papillomavirus type 18 proteins E6 and E7. Clin. Cancer Res. 7:788s-795s. [PubMed] [Google Scholar]
- 38.Seedorf, K., G. Krammer, M. Durst, S. Suhai, and W. G. Rowekamp. 1985. Human papillomavirus type 16 DNA sequence. Virology 145:181-185. [DOI] [PubMed] [Google Scholar]
- 39.Selin, L. K., S. R. Nahill, and R. M. Welsh. 1994. Cross-reactivities in memory cytotoxic T lymphocyte recognition of heterologous viruses. J. Exp. Med. 179:1933-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selin, L. K., S. M. Varga, I. C. Wong, and R. M. Welsh. 1998. Protective heterologous antiviral immunity and enhanced immunopathogenesis mediated by memory T cell populations. J. Exp. Med. 188:1705-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, D. K., R. Dudani, J. A. Pedras-Vasconcelos, Y. Chapdelaine, H. Van Faassen, and S. Sad. 2002. Cross-reactive antigen is required to prevent erosion of established T cell memory and tumor immunity: a heterologous bacterial model of attrition. J. Immunol. 169:1197-1206. [DOI] [PubMed] [Google Scholar]
- 42.Song, Y. S., S. H. Kee, J. W. Kim, N. H. Park, S. B. Kang, W. H. Chang, and H. P. Lee. 1997. Major sequence variants in E7 gene of human papillomavirus type 16 from cervical cancerous and noncancerous lesions of Korean women. Gynecol. Oncol. 66:275-281. [DOI] [PubMed] [Google Scholar]
- 43.Thomson, C. T., A. M. Kalergis, J. C. Sacchettini, and S. G. Nathenson. 2001. A structural difference limited to one residue of the antigenic peptide can profoundly alter the biological outcome of the TCR-peptide/MHC class I interaction. J. Immunol. 166:3994-3997. [DOI] [PubMed] [Google Scholar]
- 44.Tjandrawan, T., D. M. Martin, M. J. Maeurer, C. Castelli, M. T. Lotze, and W. J. Storkus. 1998. Autologous human dendriphages pulsed with synthetic or natural tumor peptides elicit tumor-specific CTLs in vitro. J. Immunother. 21:149-157. [DOI] [PubMed] [Google Scholar]
- 45.Traversari, C., P. van der Bruggen, I. F. Luescher, C. Lurquin, P. Chomez, A. Van Pel, E. De Plaen, A. Amar-Costesec, and T. Boon. 1992. A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J. Exp. Med. 176:1453-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varga, S. M., X. Wang, R. M. Welsh, and T. J. Braciale. 2001. Immunopathology in rsv infection is mediated by a discrete oligoclonal subset of antigen-specific cd4(+) t cells. Immunity 15:637-646. [DOI] [PubMed] [Google Scholar]
- 47.Wei, C. H., H. Yagita, M. G. Masucci, and V. Levitsky. 2001. Different programs of activation-induced cell death are triggered in mature activated CTL by immunogenic and partially agonistic peptide ligands. J. Immunol. 166:989-995. [DOI] [PubMed] [Google Scholar]
- 48.Yang, H. Y., P. L. Dundon, S. R. Nahill, and R. M. Welsh. 1989. Virus-induced polyclonal cytotoxic T lymphocyte stimulation. J. Immunol. 142:1710-1718. [PubMed] [Google Scholar]
- 49.Youde, S. J., P. R. Dunbar, E. M. Evans, A. N. Fiander, L. K. Borysiewicz, V. Cerundolo, and S. Man. 2000. Use of fluorogenic histocompatibility leukocyte antigen-A*0201/HPV 16 E7 peptide complexes to isolate rare human cytotoxic T-lymphocyte-recognizing endogenous human papillomavirus antigens. Cancer Res. 60:365-371. [PubMed] [Google Scholar]
- 50.Zeh, H. J., 3rd, G. H. Leder, M. T. Lotze, R. D. Salter, M. Tector, G. Stuber, S. Modrow, and W. J. Storkus. 1994. Flow-cytometric determination of peptide-class I complex formation. Identification of p53 peptides that bind to HLA-A2. Hum. Immunol. 39:79-86. [DOI] [PubMed] [Google Scholar]