Abstract
The human papillomavirus (HPV) life cycle is tightly linked to differentiation of the squamous epithelia that it infects. Capsid proteins, and hence mature virions, are produced in the outermost layer of differentiated cells. As late gene transcripts are produced in the lower layers, posttranscriptional mechanisms likely prevent capsid protein production in less differentiated cells. For HPV type 16 (HPV-16), a 79-nucleotide (nt) negative regulatory element (NRE) inhibits gene expression in basal epithelial cells. To identify key NRE sequences, we carried out transient transfection in basal epithelial cells with reporter constructs containing the HPV-16 late 3′ untranslated region with deletions and mutations of the NRE. Reporter gene expression was increased over 40-fold by deletion of the entire element, 10-fold by deletion of the 5′ portion of the NRE that contains four weak consensus 5′ splice sites, and only 3-fold by deletion of the 3′ GU-rich region. Both portions of the element appear to be necessary for full repression. Inactivating mutations in the 5′ splice sites in the 5′ NRE partially alleviated repression in the context of the 79-nt NRE but caused full derepression when assayed in a construct with the 3′ NRE deleted. All four contribute to the inhibitory effect, though the second splice site is most inhibitory. Sm proteins, U1A and U1 snRNA, but not U1 70K, could be affinity purified with the wild-type NRE but not with the NRE containing mutations in the 5′ splice sites, indicating that a U1 snRNP-like complex forms upon the element.
Human papillomaviruses (HPVs) are small DNA viruses that specifically infect squamous epithelial cells, giving rise to warts or papillomas (16). They may be divided into mucosal or cutaneous types and also into high- or low-risk types, depending on the probability of the lesions that they cause becoming malignant. High-risk mucosal types, which include HPV type 16 (HPV-16), HPV-18, HPV-31, HPV-33, and HPV-45, may give rise to cervical intraepithelial neoplasias and cervical carcinomas. HPV-16 is the most clinically significant of these, being found in over 50% of such cervical lesions (36).
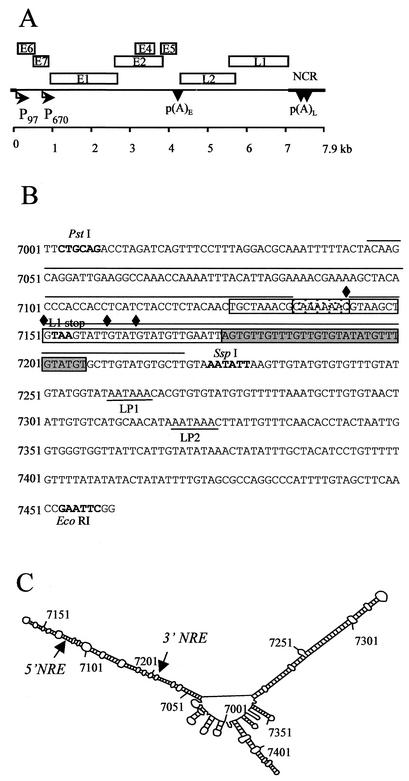
The early genes of the 8-kb HPV-16 genome encode proteins involved in the regulation of DNA replication, episomal maintenance of the genome, and control of host cell division. The late region encodes the major and minor capsid proteins, L1 and L2. The noncoding region contains the late gene 3′ untranslated region (late 3′ UTR) at its 5′ end and regulatory sequences controlling activity of the viral promoter toward its 3′ end (Fig. 1A). The major promoter in HPV-16, P97, is constitutively active from early in the viral life cycle (25). A second promoter, P670, also becomes active in differentiated cells (12). The genome consists of a single transcription unit, and alternative splicing generates individual mRNAs (7, 17, 23). Transcripts may be polyadenylated at the early polyadenylation [poly(A)] site; alternatively one of the tandem late poly(A) sites downstream of the L1 coding region may be used.
FIG. 1.
Diagram of the HPV-16 genome and sequence of the region containing the NRE and its putative secondary structure plot. (A) The linearized genomic structure of HPV-16, showing early (E) and late (L) gene coding regions (boxed). P97, constitutively active promoter; P670, promoter activated in differentiated cells; arrowheads [p(A)E and p(A)L], early and late poly(A) signals, respectively; heavy line, noncoding region (NCR). (B) The sequence of the 445-nt PstI-EcoRI fragment in the L1-late 3′ UTR of the viral genome. Boldface, L1 stop codon; boxed region, NRE; stippled box, loop of the predicted stem-loop structure; gray box, GU-rich region; diamonds, exon-intron boundaries of weak consensus 5′ splice sites; underlined sequences, late polyadenylation signals, LP1 and LP2; overlined sequences, extent of 5′ and 3′ stems of the stem-loop structure. (C) High-energy Zuker fold model of the 445-nt PstI-EcoRI fragment in the L1-late 3′ UTR of the viral genome. Every 50 bases are numbered. The 5′ and 3′ ends of the NRE are indicated with arrows.
Completion of the virus life cycle depends strictly on host cell differentiation. Capsid proteins are produced only in the outermost, differentiated cell layers; however, late mRNAs encoding the capsid proteins are detectable in partially differentiated cells in the lower layers of the epithelium (20, 29). This suggests that late gene expression in undifferentiated cells is regulated posttranscriptionally. Elements that inhibit gene expression in undifferentiated cells have been identified in the open reading frames of HPV-16 L1 (31) and L2 (4, 27) and of HPV-31 L1 (32). Inhibitory RNA elements also occur in the late 3′ UTRs of HPV-16 (18), bovine papillomavirus type 1 (BPV-1) (9), HPV-1 (26, 30), and HPV-31 (5). Most information is available for the 51-nucleotide (nt) BPV-1 inhibitory element. It contains a consensus 5′ splice site, which binds U1 snRNP (9, 10), the U1 70K component of which interacts with poly(A) polymerase and inhibits polyadenylation at the late site (15).
The HPV-16 late gene 3′ UTR contains a 79-nt negative regulatory element (NRE) overlapping the 3′ end of the L1 coding region and extending into the late gene 3′ UTR (18). In basal epithelial cells transfected with a reporter construct containing the HPV-16 late 3′ UTR, gene expression is very low, and deletion of the NRE allows up to a 100-fold increase in gene expression (18, 19). The 5′ portion of the NRE contains four weak consensus 5′ splice sites and a predicted stem-loop structure (Fig. 1B). The 3′ portion of the element is very GU rich. There is evidence that the NRE is a complex inhibitory element, acting via more than one mechanism. It acts as an RNA instability element in vitro (19), causes nuclear retention of transcripts (20, 31), and regulates polyadenylation (K. McGuire and S. V. Graham, unpublished data). UV cross-linking experiments and electrophoretic mobility shift assays (EMSAs) show that the NRE RNA binds several proteins present in cervical epithelial cell extracts (6, 20).
Two groups have previously analyzed the inhibitory effect of this element but have each reported different key sequences within the NRE responsible for the inhibitory effect (6, 10). In this study we attempted to resolve these differences and identify functionally important NRE sequences and the proteins which they bind. Deletion and mutation analysis showed that the whole 79-nt NRE is required for maximum repression of gene expression in undifferentiated epithelial cells but that the 5′ and 3′ NREs can independently inhibit gene expression, albeit to a lesser extent. We found that the weak consensus 5′ splice sites are the only functionally important sequences in the 5′ NRE and that all four contribute to NRE function. Splice site 2, which most closely resembles the consensus sequence, has the greatest effect. The NRE bound U1 snRNA, Sm proteins, and U1A. Inactivating mutations in all four 5′ splice sites reduced binding of these proteins, while converting splice site 2 to a perfect consensus sequence increased binding of the U1 snRNP proteins. This suggests that a U1 snRNP-like complex binds the 5′ NRE via the 5′ splice sites. We suggest that this complex, perhaps stabilized by interaction of other NRE-binding proteins such as U2AF65 with the 3′ NRE, could interact with the polyadenylation machinery, reducing the overall efficiency of 3′-end processing in undifferentiated epithelial cells.
MATERIALS AND METHODS
Plasmids.
Plasmid pLW1 is a chloramphenicol acetyltransferase (CAT) expression vector (11), which lacks 3′-end processing signals. Plasmid pCATPE445 contains a 445-nt PstI-EcoRI (PE) fragment of the 3′ end of the HPV-16 L1 coding region extending into the late 3′ UTR (nt 7008 to 7453) cloned downstream of the CAT reporter gene in pLW1. Plasmid pCATSE227 contains a 227-nt SspI-EcoRI fragment of the HPV-16 late 3′ UTR (nt 7226 to 7453) cloned downstream of the CAT reporter in pLW1 (18). The plasmid pCATPE445 was used as a template for PCR to generate mutant NRE constructs. A PCR protocol was used to make point mutations or deletions (21). Specific forward or reverse primers were used with T3 or T7 primers to generate two overlapping PCR products each incorporating the mutation at one end. PCRs contained 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 10 pmol of each primer, and 1 U of Taq polymerase, in 1× PCR buffer supplied by the manufacturer (ABgene). PCR conditions were 95°C for 3 min; 30 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 30 s; 55°C for 5 min; and 72°C for 10 min. The two products were annealed at 65°C for 10 min and then used as a template for a second PCR with T3 and T7 primers. Reaction mixtures contained 10 pmol of each primer, 3 mM MgCl2, 0.5 mM dNTPs, and 1 U of Taq polymerase, in 1× PCR buffer as supplied by the manufacturer (ABgene). PCR conditions were 95°C for 3 min; 20 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 4 min; 55°C for 5 min; and 72°C for 10 min. The sequences of the primers used to make NRE mutant plasmids are available on request. Mutant PCR products were checked by sequencing and then cloned downstream of the CAT reporter gene in the plasmid pLW1.
Cell culture.
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 2 mM l-glutamine (Invitrogen), at 37°C and 5% CO2.
Nuclear extracts.
Nuclear extracts from HeLa cells were prepared as described previously (35) or were purchased from 4C Biotech, Seneffe, Belgium.
Transient transfections and CAT assays.
HeLa cells (2 × 105/35-mm-diameter well) were transfected with 2 μg of cesium chloride-purified plasmid DNA, by using Lipofectamine reagent (Invitrogen) in accordance with the manufacturer's instructions. Cells were harvested after 48 h by scraping and then lysed by freeze-thaw. CAT assays were performed for 2 h at 37°C in the presence of [3H]chloramphenicol (NEN). CAT activity was determined as described previously (24).
UV cross-linking and EMSA.
Primer sequences used to generate riboprobe templates are shown in Table 1. Probe templates were made by PCR amplification of pCATPE445, or its mutant derivatives as required, by using forward primers with a 5′ extension encoding a T3 promoter sequence. PCR products were purified by fractionation through a 6% polyacrylamide gel. Riboprobes were synthesized by in vitro transcription with the Stratagene RNA transcription kit in the presence of 25 μCi of [α-32P]UTP or [α-32P]ATP (800 mCi/mmol; NEN), according to the manufacturer's protocol. Full-length transcripts were purified from a 5% denaturing polyacrylamide gel for use in EMSA or purified with Mini Quick Spin Sephadex columns (Roche) for UV cross-linking experiments. UV cross-linking experiments were carried out as described previously (22); a Stratalinker (Stratagene) at a setting of 250 mJ was used to cross-link the samples. Where required, binding buffer composition was modified by increasing KCl or NaCl concentration. EMSAs were carried out as described previously (2). For EMSA competition experiments, specific competitor RNA was in vitro transcribed by using 0.5 μg of NRE DNA template as described above, with the exception that unlabeled UTP was substituted for [32P]UTP. Nonspecific competitor RNA was Escherichia coli tRNA or a 65-nt RNA homologous to the pBluescript KS(+) (Stratagene) polylinker transcribed from a plasmid template linearized with EcoRI. For supershift EMSA, nuclear extracts were preincubated with antibodies for 30 min on ice. Complexes were resolved on native 5% polyacrylamide gels.
TABLE 1.
Sequences, and annealing positions within the HPV-16 genome, of the PCR primers used to make riboprobe templatesa
| Primer name | Promoter | Genomic location (nt) | Sequence |
|---|---|---|---|
| NRE probe forward | T3 | 7128-7144 | 5′ CAGAGATGCAATTAACCCTCACTAAAGGGGCTAAACGCAAAAAACG 3′ |
| NRE probe reverse | None | 7186-7207 | 5′ CTGAGCTCACATACAAACATATACACAAC 3′ |
| 5′ NRE probe reverse | None | 7160-7176 | 5′ TAATTCAACATACATAC 3′ |
5′ NRE probe reverse NRE probe reverse |
None | 7171-7185 | 5′ TAGAGCTCACAACAAACAACACTAATTC 3′ |
| 3′ NRE probe forward | T3 | 7177-7192 | 5′ CAGAGATGCAATTAACCCTCACTAAAGGGAGAATTCGTGTTGTTTGTTGTGT 3′ |
3′ NRE probe forward NRE probe forward |
T3 | 7161-7180 | 5′ CAGAGATGCAATTAACCCTCACTAAAGGGAGAATTCTATGTATGTTGAATTAGTGT 3′ |
| Mutant 5′ NRE probe reverse | None | 7159-7176 | 5′ TAATTCAACATTCATCCAA 3′ |
Bacteriophage RNA polymerase promoter sequences are underlined.
Affinity purification of RNA-binding proteins.
Proteins binding to wild-type or mutant NRE RNAs were purified as described previously (3). Briefly, 500 pmol of RNA was prepared by in vitro transcription, treated with 5 mM sodium m-periodate and 100 mM sodium acetate (pH 5.0), and then precipitated and incubated with 400 μl of adipic acid dihydrazide agarose beads (Sigma) for 12 h at 4°C to cross-link the RNA to the beads. The beads were washed three times with 2 M NaCl and then equilibrated with buffer D (20 mM HEPES-KOH [pH 7.6], 5% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol). Two hundred fifty microliters of HeLa cell nuclear extract (1.25 mg) in buffer D was applied, and the mixture was incubated at 30°C for 20 min. The beads were washed four times in buffer D containing 4 mM MgCl2. The bound proteins were eluted in protein loading buffer by being heated to 90°C for 5 min. The affinity-purified proteins were electrophoresed on a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel. For isolation of NRE-bound U1 snRNA, following incubation of the beads with nuclear extract and washing, the beads were digested with proteinase K (1 mg/ml) exactly as described previously (8). Beads were pelleted, the supernatant was extracted with phenol-chloroform and ethanol precipitated in the presence of glycogen as a carrier molecule, and finally recovered nucleic acids were digested with RQ1 DNase I (Promega). Following a further round of phenol-chloroform extraction and ethanol precipitation, first-strand cDNA was synthesized by incubation at 42°C for 40 min in the presence of Superscript II (Invitrogen), with a U1 snRNA-specific reverse primer (5′ AAAGCGCGAACGCAGTCCCC 3′). RNA was removed by RNase H digestion at 37°C for 30 min. Phenol-chloroform-extracted, ethanol-precipitated cDNA was PCR amplified in a reaction mixture containing 3 mM MgCl2, 20 pmol of each primer, 0.2 mM dNTPs, and 1 U of Taq polymerase, in 1× buffer as supplied by the manufacturer (ABgene). PCR conditions were 94°C for 2 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and 72°C for 5 min. The U1 snRNA-specific primers were (forward) 5′ CCACAAATTATGCAGTCGAG 3′ and (nested reverse) 5′ CTTACCTGGCAGGGGAGATA 3′. PCR products were blunt ended with T4 DNA polymerase and then cloned into pBluescript KS(+) (Stratagene) linearized with EcoRV, and inserts were checked by sequencing.
Coimmunoprecipitation.
Beads for immunoprecipitation were prepared by mixing 50 μl of protein A-Sepharose beads with anti-Sm (Y12) antibody in binding buffer (5 mM Tris-HCl [pH 7.4], 250 mM NaCl, 1 mM EDTA, and 0.05% Nonidet P-40) at 4°C for 1 h (1 μl of antibody/10 μl of beads). Beads were then mixed with 50 μg of nuclear extract for 2 h in binding buffer at 4°C. Beads were pelleted and washed three times in binding buffer. Finally, 30 μl of protein loading buffer was added and the beads were boiled to release bound protein.
Western blotting.
SDS-polyacrylamide gels were electroblotted onto nitrocellulose (Hybond ECL) or polyvinylidene difluoride (Hybond P) membrane (Amersham Pharmacia Biotech). The blots were blocked overnight at 4°C in 5% (wt/vol) dried milk powder in phosphate-buffered saline (PBS)-0.05% Tween. Primary antibodies were diluted in PBS-0.05% Tween-5% dried milk powder. The anti-Sm antibody Y12 (gift of Iain Mattaj, European Molecular Biology Laboratory) was used at a dilution of 1/500, the U1 70K polyclonal antibody 35 sc-9569 (Santa Cruz Biotechnology) was used at a dilution of 1/250, and the anti-U1A polyclonal antibody (gift of Iain Mattaj) was used at a dilution of 1/500. Blots were incubated in the primary antibody for 1 h at room temperature with shaking. After being washed in PBS-0.05% Tween, the membrane was incubated in the secondary antibody for 1 h at room temperature with shaking. The secondary antibody, either anti-mouse antibody-horseradish peroxidase or protein A-horseradish peroxidase (Sigma), was diluted 1/1,000 in PBS-0.05% Tween plus 5% (wt/vol) dried milk powder. After being washed, the membranes were visualized with ECL reagents (Amersham Pharmacia Biotech) in accordance with the manufacturer's protocol.
RESULTS
Site-directed and deletion mutant analysis of HPV-16 NRE function.
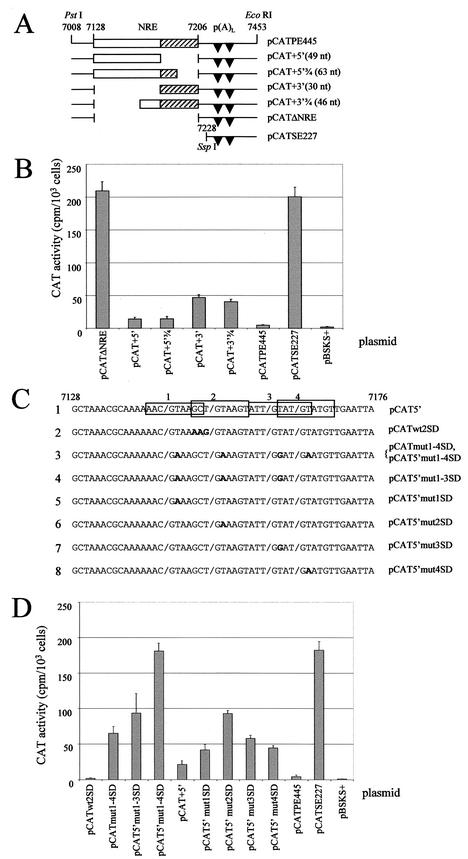
Using CAT reporter gene constructs in transient transfection of HeLa cells, Furth et al. (10) showed that the 5′-most 51 nt of the HPV-16 NRE in concert with a simian virus 40 polyadenylation signal was sufficient to inhibit gene expression. In contrast, using a very similar approach, Dietrich-Goetz et al. (6) found that, in the context of the HPV-16 late 3′ UTR, the whole 79-nt NRE was required for maximal repression. In order to clarify and delineate which regions of the NRE are the most important in control of gene expression, we carried out site-directed and deletion mutagenesis of the NRE. First we introduced groups of five random substitution mutations sequentially along the 79-nt NRE in the expression plasmid pCATPE445. This plasmid has as its 3′ UTR the 445-bp region of the HPV-16 genome (Fig. 1B) that contains the 3′ end of the L1 coding region, the NRE, and the tandem HPV-16 late polyadenylation signals. Fifteen such pCATPE445-derived NRE mutated plasmids were transiently transfected into HeLa cells, and CAT activity was assayed after 48 h. The positive control, pCATSE227, which has the NRE region deleted (18), gave high levels of CAT activity, and pCATPE445, which retains the NRE intact, gave very low levels of CAT activity as expected. However, all of the plasmids containing site-directed mutations of the NRE also gave very low CAT activities that did not differ significantly from those obtained with pCATPE445 (data not shown).
As this result could indicate that there are multiply redundant elements within the NRE necessary for its negative effect, we tested different portions of the NRE separately, by making a series of deletion mutant constructs. Firstly, we checked that the 79-nt region predicted to comprise the whole NRE had a significant repressive effect on gene expression. The NRE was precisely deleted from pCATPE445 to make plasmid pCATΔNRE (Fig. 2A) and assayed by transient transfection as before. Figure 2B shows that this deletion plasmid gives a similar alleviation of the repressive effect as pCATSE227 (which lacks all sequences upstream of the SspI site [Fig. 1B]), demonstrating that the 79-nt fragment contains repressive sequences. Next, we made plasmid pCAT+5′ (Fig. 2A), which contains the 5′ portion of the NRE (49 nt) that includes the four weak consensus 5′ splice sites and the predicted loop of the stem-loop structure (Fig. 1C). This region is very similar to the 51-nt fragment used by Furth et al., which they concluded gave a strong repressive effect (10). The plasmid pCAT+3′ (Fig. 2A) contains only the 3′ GU-rich portion (30 nt). In addition, we made the constructs pCAT+5′[3/4] (Fig. 2A), which contains the first 63 nt of the NRE, and pCAT+3′[3/4] (Fig. 2A), which contains the last 46 nt of the NRE. Transient transfection of HeLa cells followed by CAT assays showed that each of the NRE deletion mutants gave elevated reporter activity compared to that of the wild-type construct pCATPE445 (Fig. 2B; Table 2); however, each was still significantly inhibitory. The plasmids pCAT+5′ and pCAT+5′[3/4] each gave a threefold increase over pCATPE445, but CAT expression with pCAT+5′ was still approximately 15-fold lower than that obtained with the positive control pCATSE227. In contrast, the plasmids pCAT+3′ and pCAT+3′[3/4] gave increases of 10- and 8.5-fold, respectively. These results show that, while the whole 79-nt NRE is required for maximum repression of reporter gene expression in undifferentiated epithelial cells, the 5′ NRE in particular must contain significant inhibitory sequences.
FIG.2.
Functional analysis of mutant NRE constructs. (A) NRE deletion mutants. Open box, 5′ NRE; hatched box, 3′ GU-rich portion of NRE; arrowheads, late polyadenylation sites [p(A)L]. (B) Bar chart of CAT activity per 103 HeLa cells of deletion mutant constructs assayed in the presence of [3H]chloramphenicol, as the means plus standard deviations of duplicate transfections from three separate experiments. (C) Splice site mutant constructs. Line 1, wild-type sequence of the 5′ NRE in the expression plasmid pCAT5′. Boxed sequences, weak consensus 5′ splice sites (1 to 4). The shill marks the position of the exon-intron boundary. Line 2, pCATwt2SD contains a group of mutations (boldface) that convert 5′ splice site 2 to a perfect consensus 5′ splice site sequence. Line 3, pCATmut1-4SD contains inactivating point mutations (boldface) in all four 5′ splice sites; pCAT5′mut1-4SD also lacks the GU-rich 3′ NRE. Line 4, pCAT5′mut1-3SD contains inactivating point mutations in three 5′ splice sites (boldface) but retains wild-type sequence at splice site 4. Lines 5 to 8, pCAT5′mut1SD, pCAT5′mut2SD, pCAT5′mut3SD, and pCAT5′mut4SD contain inactivating point mutations (boldface) in splice sites 1 to 4, respectively. (D) Bar chart of CAT activity per 103 HeLa cells of 5′ splice site mutant constructs assayed in the presence of [3H]chloramphenicol, as the means plus standard deviations of duplicate transfections from three separate experiments.
TABLE 2.
Results of CAT assay of transiently transfected HeLa cells with NRE deletion mutant constructs
| Construct | Position (size of deletion in genome) (both in nt) | CAT activity (cpm/103 cells; mean ± SD) | Fold increase over wild-type construct (pCATPE445) |
|---|---|---|---|
| pCATPE445 | 4.71 ± 0.56 | ||
| pCAT5′ | 7177-7206 (30) | 13.90 ± 2.48 | 3.0 |
pCAT5′
|
7191-7206 (16) | 14.48 ± 3.55 | 3.1 |
| pCAT3′ | 7129-7176 (49) | 46.54 ± 3.95 | 9.8 |
pCAT3′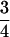
|
7129-7160 (33) | 40.38 ± 3.48 | 8.6 |
| pCATSE227 | 7008-7227 (219) | 200.46 ± 14.53 | 42.5 |
| pBluescript KS(+) | 1.70 ± 0.9 |
All four weak consensus 5′ splice sites, but not the predicted stem-loop structure, contribute to the inhibitory effect of the 5′ NRE.
In the BPV-1 late inhibitory element, a single strong consensus 5′ splice site binds U1 snRNP, whose U1 70K moiety acts to inhibit poly(A) polymerase in the downstream polyadenylation reaction (15). Barksdale and Baker demonstrated that the multiple adjacent 5′ splice sites could be additive in their inhibitory activity (1). Thus, we tested whether the four weak 5′ splice sites in the HPV-16 NRE cooperate to recapitulate the role of the single strong 5′ splice site in the BPV-1 NRE. We systematically tested each 5′ splice site in isolation and in combination for its ability to repress gene expression. First, we introduced inactivating substitution mutations at the +2 position in all four 5′ splice sites simultaneously, to make plasmid pCATmut1-4SD (Fig. 2C). As a positive control, splice site 2, the closest match to the consensus sequence (9), was converted to a perfect consensus 5′ splice site (AAG/GTAAGT) in plasmid pCATwt2SD (Fig. 2C). The CAT activity of pCATwt2SD was significantly lower than that obtained with the control plasmid pCATPE445 (Table 3; Fig. 2D), confirming that a good consensus 5′ splice site is inhibitory of gene expression in this assay. The CAT activity of pCATmut1-4SD, however, was approximately 15-fold higher than that of pCATPE445 (Table 3; Fig. 2D), though still significantly (2.8-fold) lower than that obtained with pCATSE227. These results show that the weak consensus 5′ splice sites are indeed important determinants of the inhibition of gene expression by the NRE but do not constitute the entire inhibitory activity, the remainder residing in the separable inhibitory activity of the 3′ NRE.
TABLE 3.
Results of CAT assay of transiently transfected HeLa cells with splice site mutant constructs
| Construct | CAT activity (cpm/103 cells; mean ± SD) | Fold increase over wild-type construct (pCATPE445) |
|---|---|---|
| pCATPE445 | 4.45 ± 2.19 | |
| pCATwt2SD | 1.74 ± 0.75 | 0.4 |
| pCATmut1-4SD | 65.15 ± 9.70 | 14.6 |
| pCAT5′mut1-3SD | 93.84 ± 26.27 | 21 |
| pCAT5′mut1-4SD | 180.54 ± 11.58 | 40.5 |
| pCAT5′ | 21.56 ± 4.97 | 4.8 |
| pCAT5′mut1SD | 41.62 ± 8.03 | 9.3 |
| pCAT5′mut2SD | 93.13 ± 3.53 | 20.9 |
| pCAT5′mut3SD | 57.94 ± 4.49 | 13.0 |
| pCAT5′mut4SD | 44.98 ± 3.07 | 10.1 |
| pCATSE227 | 182.37 ± 11.67 | 40.9 |
| pBluescript KS(+) | 1.22 ± 0.10 |
We next asked whether the 5′ splice sites are the sole determinant of the inhibitory activity of the 5′ NRE. To test this, we deleted the 3′ NRE from pCATmut1-4SD to make pCAT5′mut1-4SD (Fig. 2C). By a fortuitous PCR error, another construct, pCAT5′mut1-3SD (Fig. 2C), retained wild-type sequence at splice site 4. These two plasmids, when transiently transfected into HeLa cells, gave CAT activities 40- and 21-fold, respectively, higher than that of pCATPE445 (Table 3; Fig. 2D). The CAT activity of pCAT5′mut1-4SD was very similar to that of the control construct pCATSE227, which entirely lacks the NRE. These results suggest that the 5′ splice sites are the only sequences in the 5′ NRE that are required to inhibit gene expression. They further suggest that splice site 2 is not the sole determinant of repression by the 5′ NRE (10), since pCAT5′mut1-3SD, which has a functionally inactive splice site 2 but retains wild-type sequence at splice site 4, still significantly inhibits gene expression.
To investigate the role of the individual 5′ splice sites further, we introduced an inactivating point mutation into each 5′ splice site separately, to make plasmids pCAT5′mut1SD, pCAT5′mut2SD, pCAT5′mut3SD, and pCAT5′mut4SD (Fig. 2C).
We found that inactivation of each site separately caused a significant increase in gene expression over that of the control plasmid pCAT+5′ (Table 3; Fig. 2D). Plasmid pCAT5′mut1SD gave expression 1.9-fold higher, pCAT5′mut2SD gave expression 4.3-fold higher, pCAT5′mut3SD gave expression 2.7-fold higher, and pCAT5′mut4SD gave expression 2.1-fold higher than that of pCAT5′. Thus, splice site 2 is, as previously shown (10), the most important of the four sites, but all four sites contribute to the inhibitory effect of the NRE in an additive manner.
Finally, although our site-directed mutagenesis studies and the subsequent analysis of the weak 5′ splice sites in the 5′ portion of the NRE appeared to rule out any contributory effect of the predicted stem-loop structure (Fig. 1C), we also tested this directly. Deletion or mutation of the 8 nt in the loop of the stem-loop would be predicted to destroy its structure, but neither mutation introduced into pCATPE445 gave any alleviation of report gene expression (data not shown). This could indicate that the stem-loop structure is not a functionally significant element; however, we have not tested whether the secondary structure induced by the stem-loop has some further repressive effect on the 5′ splice sites.
Protein binding to the NRE is specific, and several proteins bind with high affinity.
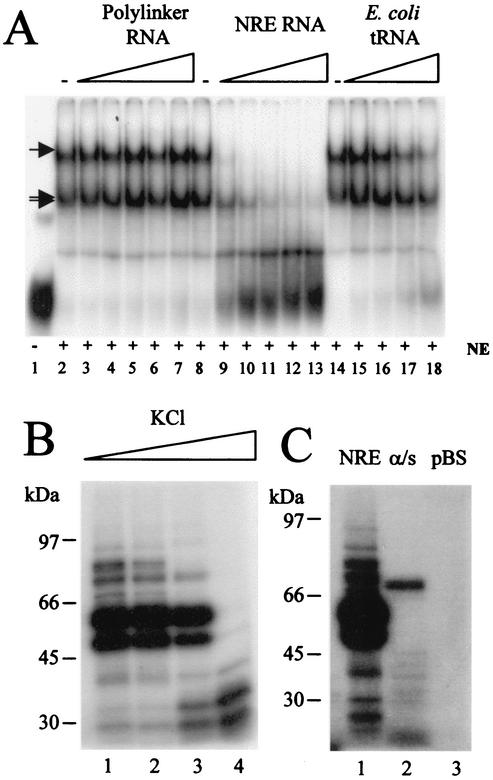
A number of HeLa and W12 cell proteins bind the NRE, forming two complexes of similar size and a third, more retarded EMSA complex. Previous studies indicated that three RNA-processing proteins, U2AF65, CstF-64, and HuR, contribute to formation of these complexes (20). Binding of these proteins is specific, as RNA-protein complexes were disrupted when a twofold, or greater, molar excess of unlabeled NRE RNA was added to an EMSA mixture with HeLa cell nuclear extracts and an NRE probe (Fig. 3A, lanes 9 to 13). Addition of unrelated RNA [2- to 16-fold molar excess of in vitro-transcribed RNA homologous to the pBluescript KS(+) polylinker or E. coli tRNA did not disrupt the complexes (Fig. 3A, lanes 3 to 7 and 15 to 18). These protein complexes bind with reasonably high affinity, as they are still present in 400 mM salt, indicating that they represent specific binding to the NRE (data not shown). By using UV cross-linking, the affinity of proteins that bind the NRE directly was determined. Figure 3B shows that by using similar probes and HeLa cell nuclear extracts as used in the EMSAs the major 65-kDa and 50-kDa bands that are products of the UV cross-linking experiment were still bound at 250 mM KCl (Fig. 3B, lane 3) or NaCl (data not shown). However, these no longer bound in 500 mM KCl (Fig. 3B, lane 4). At this high salt concentration, binding of two proteins around 30 to 35 kDa was greatly increased, suggesting that individual proteins may compete for binding sites on the NRE. In case any of the proteins binding directly to the NRE even in high-salt conditions were nonspecific, we tested binding in low-salt conditions (60 mM KCl) of nuclear extract to the sense NRE, the antisense NRE, and a pBluescript polylinker-transcribed RNA of similar size. While strong binding to the NRE was detected as before (Fig. 3C, lane 1), only one major protein bound to the antisense strand and this protein was not of a size that bound the sense RNA strand (Fig. 3C, lane 2). No proteins bound the pBluescript polylinker RNA (Fig. 3C, lane 3).
FIG. 3.
Binding of protein to the NRE is specific and occurs with high binding affinity. (A) Specific competition EMSA. A 32P-labeled NRE probe (1.5 pmol) was incubated with HeLa cell nuclear extracts in the presence of specific or nonspecific competitor RNA. Lane 1, no extract; lane 2, no competitor; lanes 3 to 7, 1- to 16-fold molar excess of nonspecific competitor, i.e., 1.5 to 24 pmol of in vitro-transcribed, unlabeled pBluescript KS(+) polylinker RNA; lane 8, no competitor; lanes 9 to 13, 1- to 16-fold molar excess of specific competitor, i.e., 1.5 to 24 pmol of in vitro-transcribed NRE RNA; lane 14, no competitor RNA; lanes 15 to 18, nonspecific competitor, i.e., 500 ng to 2 μg of E. coli tRNA. Arrows, RNA-protein complexes; NE, HeLa cell nuclear extracts. (B) A 32P-labeled NRE probe was UV cross-linked to HeLa cell nuclear extracts in the presence of various concentrations of KCl (60, 120, 250, and 500 mM [lanes 1 to 4, respectively]). (C) 32P-labeled sense NRE (lane 1), antisense NRE (lane 2), and pBluescript polylinker probes (lane 3) were UV cross-linked to HeLa cell nuclear extracts in 60 mM KCl. In panels B and C cross-linked protein-RNA samples were RNase digested and fractionated on SDS-polyacrylamide gels, dried, and subjected to autoradiography.
NRE deletion mutants show differences in UV cross-linking of HeLa cell nuclear extract proteins.
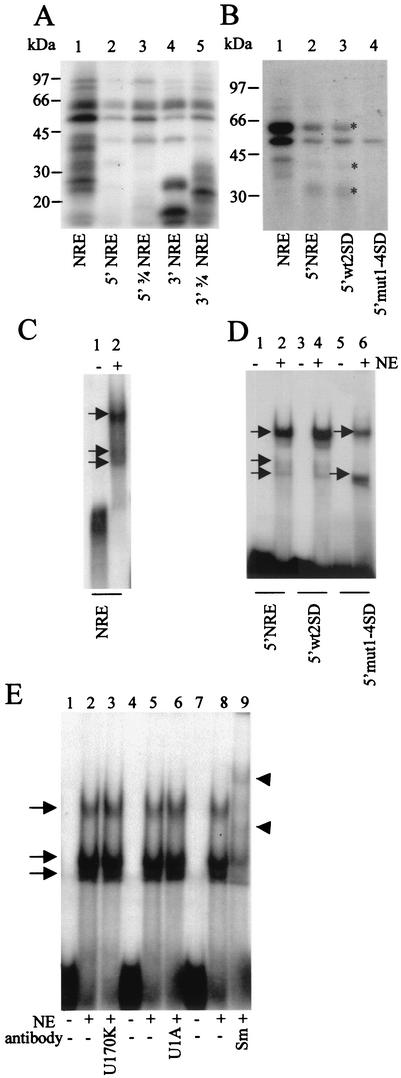
We wished to determine whether binding of any of these proteins was functionally important and whether the splice site mutant NREs that showed altered ability to inhibit reporter gene expression showed changes in protein binding. The NRE deletion mutant probes were 32P labeled and UV cross-linked to HeLa cell nuclear extracts. All the deletion constructs differed from the wild-type NRE in the number and relative intensity of bands produced, although several proteins between 40 and 70 kDa appeared to bind to every deletion mutant tested. The wild-type NRE UV cross-linked to at least four proteins between 25 and 40 kDa that did not bind any of the deletion mutants (Fig. 4A, lane 1). The 5′ NRE (49 nt), although longer than the 3′ NRE (30 nt), bound less protein overall (lanes 2 and 4). The 3′ NRE also bound three small (<30-kDa) proteins that bound none of the other probes (lane 4). The 3′[3/4] NRE generated a strong band of around 25 kDa and several other unique bands of around 30 kDa (lane 5). These results suggest that individual proteins compete for binding to the NRE and that, although the two halves of the NRE RNA element can bind similar proteins, they may cooperate to provide a stabilized binding site for these proteins.
FIG.4.
UV cross-linking and EMSA with mutant NRE probes and HeLa cell nuclear extracts. (A) UV cross-linking of 32P-labeled wild-type and deletion mutant NRE probes to HeLa cell nuclear extracts. Lane 1, NRE; lane 2, 5′ NRE; lane 3, 5′[3/4] NRE; lane 4, 3′ NRE; lane 5, 3′[3/4] NRE. (B) UV cross-linking of [32P]rATP-labeled whole-NRE, 5′ NRE, and 5′ splice site mutant probes to HeLa cell nuclear extracts. Lane 1, whole-NRE probe; lane 2, 5′ NRE probe; lane 3, 5′wt2SD probe; lane 4, 5′mut1-4SD probe. Asterisks, proteins binding to 5′wt2SD and 5′ NRE but not to 5′mut1-4SD. (C) EMSA with 32P-labeled whole-NRE probe and HeLa cell nuclear extracts. Lane 1, no nuclear extract; lane 2, with HeLa cell nuclear extract. Arrows, RNA-protein complexes. (D) EMSA with 32P-labeled short (49-nt) 5′ splice site mutant NRE probes and HeLa cell nuclear extracts. Lanes 1 and 2, 5′ NRE probe; lanes 3 and 4, 5′wt2SD probe; lanes 5 and 6, 5′mut1-4SD probe. Arrows, RNA-protein complexes; NE, HeLa cell nuclear extracts. (E) EMSA with 32P-labeled whole NRE probe and HeLa cell nuclear extracts preincubated with various antibodies. Lanes 1, 4, and 7, no nuclear extract and no antibody; lanes 2, 5, and 8, nuclear extract (NE) and no antibody; lane 3, nuclear extract with anti-U1 70K antibody; lane 6, nuclear extract with anti-U1A antibody; lane 9, nuclear extract with anti-Sm antibody. Arrows, RNA-protein complexes. Arrowheads mark the positions of shifted complexes.
The splice site mutants show altered patterns of protein binding in UV cross-linking experiments.
We hypothesized that, if the four weak 5′ splice sites in the 5′ portion of the NRE were functionally important, protein binding patterns to wild-type and mutant NRE probes might be different. UV cross-linking analysis with wt79ntNRE, wt2SD, and mut1-4SD probes gave a broadly similar pattern of bands for each probe, possibly due to 3′ NRE-mediated stabilization of partial complexes (data not shown). In contrast, UV cross-linking of 32P-labeled short (49-nt) 5′wtNRE and 5′mut2SD probes to HeLa cell nuclear extracts generated clear differences (Fig. 4B). As before, overall protein binding to the shorter RNA probes was weaker than that to the full NRE (Fig. 4B, compare lanes 1 and 2), but a very similar protein binding pattern was observed for the 5′ NRE and 5′wt2SD probes, indicating that the wild-type NRE with the four weak 5′ splice sites binds proteins that also bind a single strong consensus 5′ splice site (Fig. 4B, lanes 2 and 3). However, protein binding to the 5′mut1-4SD probe was markedly different, with most of these proteins showing much-reduced binding (Fig. 4B, lane 4). The mutations introduced to inactivate the four weak 5′ splice sites changed T residues to A residues; thus, this experiment was carried out with both [32P]rUTP and [32P]rATP. Very similar results were obtained in each case, and Fig. 4B shows the data with [32P]rATP. Taken together, these UV cross-linking experiments show distinct differences in protein binding between wild-type NRE and the NRE splice site mutant probes, indicating that protein binding to the weak 5′ splice sites is likely to be functionally important.
NRE deletion mutants each generate three retarded EMSA complexes.
In an EMSA with HeLa cell nuclear extracts and NRE deletion mutant probes, three shifted complexes were formed on each probe (Fig. 4C). Titration experiments with increasing amounts of HeLa cell nuclear extracts with an NRE probe show a progressive loss of the smaller complexes and an increase in the intensity of the largest complex, suggesting that the lower bands may represent partially formed intermediates (S. A. Cumming and S. V. Graham, unpublished data). In an EMSA with 32P-labeled splice site mutant 5′ NRE probes and a threefold-higher ratio of nuclear extract to probe, the upper complex clearly predominated in each case. In Fig. 4D the intensity of the upper complex with the 5′mut1-4SD probe was significantly reduced, and one of the lower complexes was absent (lane 6). This is consistent with the UV cross-linking data with similar probes, again indicating that the protein complexes that form on the 5′ NRE interact via the four weak 5′ splice sites. As the strong consensus 5′ splice site in the BPV-1 late inhibitory element binds U1 snRNP, the protein complex that most likely interacted with the NRE was also U1 snRNP. We carried out supershift EMSAs to determine if we could detect the presence of any key U1 snRNP proteins in these complexes. Figure 4E shows that preincubation of nuclear extracts with antibodies against U1 70K and U1A proteins did not alter the pattern of complexes formed upon the NRE. However, preincubation with an anti-Sm antibody dramatically altered the mobility of the larger and smaller complexes. This confirms that an Sm protein-containing complex binds the NRE and indicates that at least the lowest complex is a precursor complex for the most retarded complex. Failure to detect supershifted complexes or any reduction in intensity of complexes formed with the anti-U1 70K and U1A antibodies could mean that these proteins were not present in the NRE-bound complexes or that these proteins when in the complexes were inaccessible to the antibodies.
Splice site mutants show altered binding of certain splicing factors.
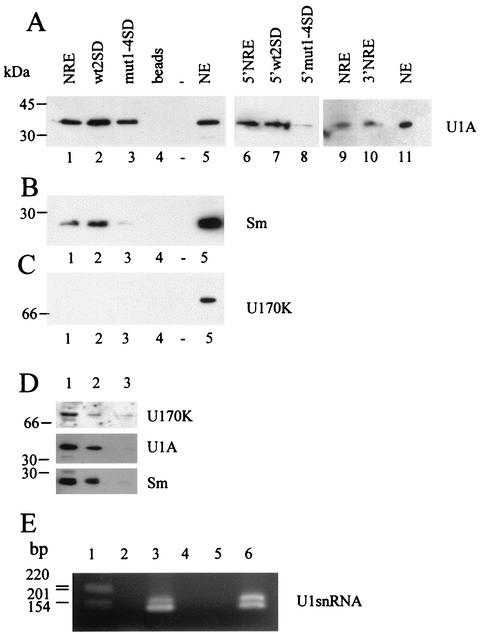
Next we used affinity chromatography to ask if the four weak 5′ splice sites in HPV-16 bound components of U1 snRNP. As a positive control, we used the NRE mutant with the second weak 5′ splice site mutated to a strong consensus sequence, and as a negative control we used mut1-4SD, where all four splice sites were inactivated. The NRE and splice site mutant RNAs were chemically cross-linked to agarose beads to purify RNA-binding proteins from HeLa cell nuclear extracts (3). These proteins were eluted and Western blotted with antibodies to RNA-processing factors that bind the NRE. Figure 5 shows that the components of U1 snRNP, U1A (Fig. 5A), Sm proteins (with anti-Sm monoclonal antibody Y12, which detects the B/B′, D1, and D3 Sm proteins) (Fig. 5B), and U1 snRNA (Fig. 5E), but not U1 70K (Fig. 5C), bind the NRE. Although U1A bound to all three RNAs (Fig. 5A, lanes 1 to 3), slightly more U1A was detected in the wt2SD sample (Fig. 5A, lane 2). When the experiment was repeated with only the 5′ NRE probes to purify proteins, very little U1A could be detected in the 5′mut1-4SD reaction (Fig. 5A, lane 8), indicating that U1A does bind the four weak 5′ splice sites but may also bind the 3′ portion of the NRE. We tested this directly by comparing binding of U1A in nuclear extracts to the whole NRE and to its 3′ portion. We detected strong binding both to the whole NRE and to its 3′ end (Fig. 5A, lanes 9 and 10). With the anti-Sm antibody, we obtained a major band at approximately 30 kDa (Fig. 5B), which from its size is most likely B/B′ (33). The wt2SD RNA binds slightly more B/B′ Sm protein (lane 2) and in addition binds a protein of approximately 15 kDa (data not shown), most likely D1 (33). The eluate from mut1-4SD contains almost no Sm protein; only a very faint band is visible in lane 3. We were able to detect U1 70K protein in HeLa cell nuclear extracts, though not in any of our eluted protein samples (Fig. 5C). To check that U1 70K was present in complexes with Sm protein and U1A in the nuclear extracts that we used, we carried out coimmunoprecipitation experiments with the anti-Sm antibody Y12. Figure 5D shows that Y12-linked beads immunoprecipitated Sm proteins as expected and also U1A protein. Coimmunoprecipitation of U1 70K was less efficient, but more protein was pulled out with antibody-linked beads than with beads alone. The detection of a U1 70K-specific band in the lane containing beads alone was most likely due to cross-reactivity of this polyclonal antibody (Fig. 5D, lane 3). As expected U1 snRNA could be reverse transcription PCR amplified from NRE affinity-purified RNA, though not from RNA purified with beads alone (Fig. 5E). The two PCR products were cloned, sequenced, and confirmed to be U1 snRNA specific. The shorter product (130 bp) is exactly the size and sequence predicted by using the nested U1 snRNA-specific reverse primer, while the longer product (161 bp) extends to the 3′ end of the U1 snRNA. These data suggest that a U1 snRNA-like complex binds the NRE via the weak consensus 5′ splice sites at the 5′ end.
FIG. 5.
Affinity purification and identification of proteins and RNA that interact with NRE and splice site mutant RNAs. (A) Western blotting with a polyclonal U1A antiserum. Lanes 1 and 9, NRE-binding proteins isolated from HeLa cell nuclear extracts; lane 2, wt2SD-binding proteins; lane 3, mut1-4SD-binding proteins; lane 4, proteins purified with beads alone; lane 5, 20 μg of HeLa cell nuclear extracts; lane 6, 5′ NRE-binding proteins; lane 7, 5′wt2SD-binding proteins; lane 8, 5′mut1-4SD-binding proteins (the exposure of lanes 6 to 8 is 10 times longer than that of lanes 1 to 5); lane 10, 3′ NRE-binding proteins; lane 11, 2 μg of HeLa nuclear extracts. (B) Western blotting with the Y12 Sm protein antibody. Lane 1, NRE-binding proteins; lane 2, wt2SD-binding proteins; lane 3, mut1-4SD-binding proteins; lane 4, proteins purified with beads alone; lane 5, 20 μg of HeLa nuclear extracts. (C) Western blotting with the U1 70K antibody. Lanes are as in panel B. (D) Western blotting of proteins coimmunoprecipitated with anti-Sm antibody from HeLa cell nuclear extracts. Lane 1, nuclear extract alone; lane 2, nuclear extract incubated with Sm antibody Y12-bound protein A-Sepharose beads; lane 3, nuclear extract incubated with protein A-Sepharose beads without anti-Sm antibody. The top panel was probed with anti-U1 70K antibody, the middle panel was probed with anti-U1A antibody, and the bottom panel was probed with anti-Sm antibody. (E) Ethidium bromide-stained agarose gel of reverse transcription-PCR-amplified U1 snRNA affinity purified on NRE RNA. Lane 1, 1-kb DNA ladder (Invitrogen); lane 2, no cDNA added; lane 3, NRE affinity-purified U1 snRNA cDNA; lane 4, NRE affinity-purified RNA, no reverse transcriptase; lane 5, affinity-purified cDNA from beads alone; lane 6, affinity-purified cDNA from HeLa cell nuclear extract.
DISCUSSION
The HPV-16 late 3′ UTR NRE is proposed to regulate viral late gene expression posttranscriptionally. We have shown here that the entire 79-nt NRE is required for maximum repression of gene expression in undifferentiated epithelial cells. All deletion and site-directed mutants of the NRE that we synthesized still significantly inhibited gene expression, demonstrating that no individual short sequence(s) is responsible for the repressive effect. Further, comparison of the relative repressive effects of the 5′ portion, which contains the four weak 5′ splice sites, with the GU-rich 3′ portion revealed that the former sequence on its own has a stronger repressive effect than does the 3′ GU-rich region, and yet both portions are required for full repressive effect. Within the 5′ portion, we have demonstrated that the four weak consensus 5′ splice sites are the only functionally important sequences. Although all four contribute to NRE function, splice site 2, which most closely resembles the consensus sequence, has the largest single effect. Taken together, these data suggest that the HPV-16 NRE is a surprisingly complex regulatory element. This contrasts with the BPV-1 NRE, which comprises only a single good consensus 5′ splice site (10), and the HPV-1 NRE, which consists of an AU-rich element that appears to act like other eukaryotic 3′ UTR AU-rich elements by binding the RNA stability regulator protein HuR and hnRNPC1/C2 (26, 28).
Previous studies of the HPV-16 NRE yielded differing results. Furth et al. (10) concluded that a 51-nt region (very similar to that defined by us as the 5′ portion of the NRE), and in particular the second weak 5′ splice site that it contained, was the primary determinant of NRE function. Dietrich-Goetz et al. (6) showed that constructs containing deletions of splice sites 1 and 2 or 1 to 4 yielded increases in gene expression of 14 and 50%, respectively. Point mutations affecting splice site 1 alone, or sites 3 and 4 together, showed little effect, but these were assayed in constructs retaining the 3′ NRE, as well as the wild-type sequence at splice site 2. In the present study, we have analyzed the effect of the splice sites in isolation, by combining 5′ splice site mutations with deletions of the 3′ NRE. By using this approach, we have shown that the 5′ splice sites are the only functionally important sequences in the 5′ NRE. Although Furth et al. (10) demonstrated that only the strong splice site 2 controls gene expression, this was in the context of a heterologous poly(A) signal. As we have evidence that the NRE cross talks to the late poly(A) sites (McGuire and Graham, unpublished), our analysis of the inhibitory effects of the NRE within its natural context in the HPV-16 late 3′ UTR may have uncovered subsidiary roles of the weaker first, third, and fourth 5′ splice sites.
A number of RNA-processing factors, including U1 snRNP proteins, U2AF65, CstF-64, and HuR, bind NRE RNA specifically, and with high affinity (20; this work). The pattern of binding in EMSAs and UV cross-linking experiments is quite similar for our deletion and site-directed mutant RNAs. This could indicate that binding of at least some of these proteins was nonspecific. However, our EMSA competition experiments and UV cross-linking salt titrations demonstrate a reasonable degree of specificity. We suggest that our data indicate that an NRE-binding complex may have several points of contact with the sequence. Overall, protein binding to the 5′ NRE was less than that to the 3′ NRE, suggesting that the 3′ portion may stabilize complex formation over the NRE, although the 5′ splice sites clearly bind a U1 snRNP-like complex. In EMSA, the full-length and 5′ NRE each generated three retarded complexes of similar mobilities but of varying intensities between experiments. Titration experiments with increasing amounts of HeLa cell nuclear extracts with an NRE probe show a progressive loss of the smaller complexes and an increase in the intensity of the largest complex, suggesting that the lower bands may represent partially formed intermediates (Cumming and Graham, unpublished). This can be observed in Fig. 4C, where a threefold-higher ratio of nuclear extract to probe was used than for Fig. 4E. This may indicate that at high protein concentrations a single multiprotein complex covers the whole NRE, and although a similar complex may still form on a shorter RNA, it will be less stable, resulting in less complex formation overall and in relative underrepresentation of the most retarded EMSA band. If similar complexes also form in vivo, reduced complex stability on deletion mutants might explain how truncated NREs can still inhibit gene expression, albeit less efficiently.
Our in vitro protein binding experiments with splice site mutant RNAs show a reduction in binding of cellular proteins in UV cross-linking experiments and EMSAs. Using affinity chromatography, we showed that, although U1A binds the full-length mut1-4SD RNA, it does not bind 5′mut1-4SD. This suggests that, at least in vitro, U1A can bind the 3′ GU-rich portion of the NRE but that binding to the 5′ NRE requires 5′ splice site homology. Sm proteins, in contrast, do not bind mut1-4SD RNA, though they do bind the full-length NRE RNA. This suggests that Sm proteins do not normally bind the GU-rich 3′ NRE but do bind the 5′ splice sites in the 5′ NRE. As we have affinity purified U1 snRNA with NRE RNA, these results suggest that a U1 snRNP-like protein complex binds via the 5′ splice sites to the 5′ NRE.
In the case of the BPV-1 inhibitory element, the U1 70K component of U1 snRNP interacts with poly(A) polymerase, reducing polyadenylation efficiency (15). For the NRE, binding of U1 70K cannot be detected despite its presence in a complex with Sm and U1A proteins in our nuclear extracts. Levels of U1 70K proteins are low in our extracts, and we cannot rule out the possibility that it is present at very low levels in an NRE-binding complex. A second U1 snRNP component, U1A, however, does bind the NRE. U1A regulates its own production by binding simultaneously to poly(A) polymerase and to the U1A RNA 3′ UTR, reducing polyadenylation efficiency (13, 14). U1A could therefore substitute for U1 70K in this system. U2AF65 has also been shown elsewhere to bind poly(A) polymerase (34) and could also inhibit polyadenylation under certain conditions. Since U2AF65 also binds the NRE (6, 20), it could potentially act to reduce the polyadenylation efficiency of NRE-containing transcripts.
Taken together, our results suggest that, although the NRE is more complex than the BPV-1 inhibitory element, it may work via a broadly similar mechanism. We have shown that the weak consensus 5′ splice sites in the 5′ NRE are very important but do not wholly account for NRE function. In vitro protein binding to truncated or full-length NRE RNAs is remarkably consistent, which suggests that a single multiprotein complex might form over the whole NRE. We have shown that binding of U1A and Sm to the 5′ NRE is stabilized by the introduction of a perfect consensus 5′ splice site sequence but abolished by inactivating mutations. These changes correlate with changes in gene expression in a functional assay. This suggests that the 5′ NRE binds a U1 snRNP-like complex, most likely via base pairing between U1 snRNA and the 5′ splice sites. Binding could be stabilized by binding of U2AF65 to the 3′ NRE (6, 20), as well as by bridging interactions with other NRE-binding proteins. We propose that, in undifferentiated cells, the U1A or U2AF65 components of the complex reduce the efficiency of polyadenylation at the downstream sites, thus inhibiting late gene expression.
Acknowledgments
This work was supported by the Wellcome Trust and the Biotechnology and Biological Sciences Research Council.
We thank Iain Mattaj for the anti-Sm antibody Y12 and for the U1A antiserum. We also thank Barklie Clements for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Barksdale, S. K., and C. C. Baker. 1995. Differentiation-specific alternative splicing of bovine papillomavirus late mRNAs. J. Virol. 69:6553-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, D. J., R. Chan, H. Min, J. Wang, and L. Bell. 1998. The electrophoretic mobility shift assay for RNA binding proteins, p. 109-136. In C. W. J. Smith (ed.), RNA-protein interactions: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 3.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier, B., L. Goobar-Larsson, M. Sokolowski, and S. Schwartz. 1998. Translational inhibition of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogeneous ribonucleoprotein K and poly(rC) binding proteins 1 and 2. J. Biol. Chem. 273:22648-22656. [DOI] [PubMed] [Google Scholar]
- 5.Cumming, S. A., C. E. Repellin, M. McPhillips, J. C. Radford, J. B. Clements, and S. V. Graham. 2002. The human papillomavirus type 31 late 3′ untranslated region contains a complex bipartite negative regulatory element. J. Virol. 76:5993-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietrich-Goetz, W., I. M. Kennedy, B. Levins, M. A. Stanley, and J. B. Clements. 1997. A cellular 65-kDa protein recognizes the negative regulatory element of human papillomavirus late mRNA. Proc. Natl. Acad. Sci. USA 94:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doorbar, J., A. Parton, K. Hartley, L. Banks, T. Crook, M. Stanley, and L. Crawford. 1990. Detection of novel splicing patterns in a HPV-16-containing keratinocyte cell line. Virology 178:254-262. [DOI] [PubMed] [Google Scholar]
- 8.Evans, D., I. Perez, M. MacMorris, D. Leake, C. J. Wilusz, and T. Blumenthal. 2001. A complex containing CstF-64 and the SL2 snRNP connects mRNA 3′ end formation and trans-splicing in C. elegans operons. Genes Dev. 15:2562-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furth, P. A., and C. C. Baker. 1991. An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J. Virol. 65:5806-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furth, P. A., W.-T. Choe, J. H. Rex, J. C. Byrne, and C. C. Baker. 1994. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol. Cell. Biol. 14:5278-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaffney, D., J. L. Whitton, M. Lynch, J. McLauchlan, and J. B. Clements. 1985. A modular system for the assay of transcriptional regulatory signals: the sequence TAATGARAT is required for herpes simplex virus immediate early gene activation. Nucleic Acids Res. 13:7847-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grassmann, K., B. Rapp, H. Maschek, K. U. Petry, and T. Iftner. 1996. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol. 70:2339-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunderson, S. I., K. Beyer, G. Martin, W. Keller, W. C. Boelens, and I. W. Mattaj. 1994. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 76:531-541. [DOI] [PubMed] [Google Scholar]
- 14.Gunderson, S. I., S. Vagner, M. Polycarpou-Schwartz, and I. W. Mattaj. 1997. Involvement of the carboxy terminus of vertebrate poly(A) polymerase in U1A autoregulation and in coupling of splicing and polyadenylation. Genes Dev. 11:761-773. [DOI] [PubMed] [Google Scholar]
- 15.Gunderson, S. I., M. Polycarpou-Schwartz, and I. W. Mattaj. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell 1:255-264. [DOI] [PubMed] [Google Scholar]
- 16.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive expression of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy, I. M., J. K. Haddow, and J. B. Clements. 1990. Analysis of human papillomavirus type 16 late mRNA 3′ processing signals in vitro and in vivo. J. Virol. 64:1825-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy, I. M., J. K. Haddow, and J. B. Clements. 1991. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J. Virol. 65:2093-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koffa, M. D., S. V. Graham, Y. Takagaki, J. L. Manley, and J. B. Clements. 2000. The human papillomavirus type 16 negative regulatory element interacts with three proteins that act at different posttranscriptional levels. Proc. Natl. Acad. Sci. USA 97:4677-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews, K. R., C. Tschudi, and E. Ullu. 1994. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8:491-501. [DOI] [PubMed] [Google Scholar]
- 22.Moore, M. J., and C. C. Query. 1998. Use of site-specifically modified RNAs constructed by RNA ligation, p. 75-108. In C. W. J. Smith (ed.), RNA-protein interactions: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 23.Ozbun, M. A., and C. Meyers. 1997. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 71:5161-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seed, B., and S.-Y. Sheen. 1988. A simple phase-extraction assay for chloramphenicol acetyltransferase activity. Gene 67:271-277. [DOI] [PubMed] [Google Scholar]
- 25.Smotkin, D., and F. O. Wettstein. 1986. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. USA 83:4680-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokolowski, M., C. Zhao, W. Tan, and S. Schwartz. 1997. AU-rich mRNA instability elements on human papillomavirus type 1 late mRNAs and c-fos mRNAs interact with the same cellular factors. Oncogene 15:2303-2319. [DOI] [PubMed] [Google Scholar]
- 27.Sokolowski, M., W. Tan, M. Jellne, and S. Schwartz. 1998. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J. Virol. 72:1504-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokolowski, M., H. Furneaux, and S. Schwartz. 1999. The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3′ untranslated region correlates with its affinity for the elav-like HuR protein. J. Virol. 73:1080-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoler, M. H., S. M. Wolinsky, A. Whitbeck, T. R. Broker, and L. T. Chow. 1989. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridisation with message-specific RNA probes. Virology 172:331-340. [DOI] [PubMed] [Google Scholar]
- 30.Tan, W., and S. Schwartz. 1995. The Rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3′ untranslated region. J. Virol. 69:2932-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan, W., B. K. Felber, A. S. Zolotukhin, G. N. Pavlakis, and S. Schwartz. 1995. Efficient expression of the human papillomavirus type 16 L1 protein in epithelial cells by using Rev and the Rev-responsive element of human immunodeficiency virus or the cis-acting transactivation element of simian retrovirus type 1. J. Virol 69:5607-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terhune, S. S., W. G. Hubert, J. T. Thomas, and L. A. Laimins. 2001. Early polyadenylation signals of human papillomavirus type 31 negatively regulate capsid gene expression. J. Virol. 75:8147-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urlaub, H., V. A. Raker, S. Kostka, and R. Lührmann. 2001. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 20:187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vagner, S., C. Vagner, and I. W. Mattaj. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 14:403-413. [PMC free article] [PubMed] [Google Scholar]
- 35.Wahle, E., and W. Keller. 1994. 3′ end-processing of mRNA, p. 1-34. In S. J. Higgins and B. D. Hames (ed.), RNA processing—a practical approach. Oxford University Press, New York, N.Y.
- 36.zur Hausen, H. 1989. Papillomaviruses in anogenital cancer as a model to understand the role of viruses in human cancers. Cancer Res. 49:4677-4681. [PubMed] [Google Scholar]