Abstract
The Tat protein of human immunodeficiency virus type 1 (HIV-1) plays a key role as inducer of viral gene expression. We report that Tat function can be potently inhibited in human microglial cells by the recently described nuclear receptor cofactor chicken ovalbumin upstream promoter transcription factor-interacting protein 2 (CTIP2). Overexpression of CTIP2 leads to repression of HIV-1 replication, as a result of inhibition of Tat-mediated transactivation. In contrast, the related CTIP1 was unable to affect Tat function and viral replication. Using confocal microscopy to visualize Tat subcellular distribution in the presence of the CTIPs, we found that overexpression of CTIP2, and not of CTIP1, leads to disruption of Tat nuclear localization and recruitment of Tat within CTIP2-induced nuclear ball-like structures. In addition, our studies demonstrate that CTIP2 colocalizes and associates with the heterochromatin-associated protein HP1α. The CTIP2 protein harbors two Tat and HP1 interaction interfaces, the 145-434 and the 717-813 domains. CTIP2 and HP1α associate with Tat to form a three-protein complex in which the 145-434 CTIP2 domain interacts with the N-terminal region of Tat, while the 717-813 domain binds to HP1. The importance of this Tat binding interface and of Tat subnuclear relocation was confirmed by analysis of CTIP2 deletion mutants. Our findings suggest that inhibition of HIV-1 expression by CTIP2 correlates with recruitment of Tat within CTIP2-induced structures and relocalization within inactive regions of the chromatin via formation of the Tat-CTIP2-HP1α complex. These data highlight a new mechanism of Tat inactivation through subnuclear relocalization that may ultimately lead to inhibition of viral pathogenesis.
Regulation of human immunodeficiency virus type 1 (HIV-1) gene transcription is governed by a complex interplay between chromatin-associated proviral DNA, host cell proteins, and the virus-encoded transactivator protein, Tat. In the immediate-early phase of HIV infection, cellular transcription factors activate transcription from the viral long terminal repeat (LTR) (for a review, see references 24 and 32). This leads to the accumulation of the viral protein Tat that leads to a potent increase in transcription and is required for viral replication and a high viral load (for a review, see reference 37). The ability of Tat to function as a transcriptional activator is mediated by multiple interactions with cellular proteins and requires the concerted action of Tat and upstream nuclear factors that bind to the Sp1 and κB region of the LTR (for reviews, see references 16 and 17). Tat forms a ternary complex with the coactivators P/CAF and p300 which helps Tat activate transcription of integrated viral DNA and derepress the HIV-1 chromatin structure in response to histone acetylation (3). Mechanisms that inhibit HIV-1 LTR expression are largely unexplored. Recent studies have shown that Tat activation can be inhibited by the overexpression of the host factors YY1 and LSF, which recruit histone deacetylase 1 to the LTR (20).
We have previously reported that Tat also interacts and cooperates with an orphan member of the nuclear receptor superfamily, the chicken ovalbumin upstream promoter transcription factor (COUP-TF) that together with Sp1 activates HIV-1 LTR-driven transcription (34, 35). Members of the COUP-TF family were recently shown to bind to novel and related zinc finger proteins and COUP TF-interacting protein 1 (CTIP1) and CTIP2. CTIP1 was found to induce transcriptional silencing by relocating COUP-TF to distinct nuclear structures, possibly associated with heterochromatic regions (1). These studies revealed a novel mechanism for transcriptional repression, by recruitment of a transcription factor to distinct nuclear loci, instead of acting through recruitment of trichostatin A-sensitive histone deacetylases. It was therefore intriguing to examine whether the CTIP proteins possess the ability to repress the action of transcription factors involved in HIV-1 LTR-driven transcription.
We have investigated the functional effect of the nuclear proteins CTIP1 and CTIP2 on HIV-1 gene expression in human microglial cells. These resident macrophages are the primary target of HIV-1 productive infection within the central nervous system (33), which results in a wide range of neurological complications (18, 26). Our findings reveal the ability of CTIP2 to specifically act as a potent inhibitor of Tat function, leading to repression of viral replication. A combination of functional, in vitro and confocal microscopy data demonstrate how interactions between Tat, CTIP2 and the heterochromatin-associated protein HP1α contribute to the relocalization of Tat within a nuclear transcriptionally nonpermissive environment.
MATERIALS AND METHODS
Plasmids.
HIV-1 LTR-chloramphenicol acetyltransferase (LTR-CAT) and pCMV-Tat vectors were described previously (35). Full-length hemagglutinin (HA)-CTIP1 and Flag-CTIP2 were described previously (1). The Flag-CTIP2 deletion plasmids (domain containing residues 1 to 354 [1-354], 145-434, 350-716, 350-813, 610-813, and 717-813) were constructed by producing the cDNA fragments by PCR and subcloning into pcDNA3 using the pcDNA3.1/V5-His TOPO TA expression kit from Invitrogen. pTat-green fluorescent protein (GFP) was a generous gift from G. Pavlakis (National Cancer Institute, National Institutes of Health [NIH], Frederick, Md.) (36). Glutathione S-transferase (GST)-Tat expression vectors were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH, from A. Rice (21). GST-HP1α and Flag-HP1α vectors were a generous gift from R. Losson (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France) (29).
Cell culture, transfections, and CAT assays.
The human microglial cell line (obtained from M. Tardieu, Faculté de Médecine Paris-Sud, Paris, France) (22) was grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum in the presence of penicillin-streptomycin (100 U/ml) and was transfected by the calcium phosphate coprecipitation method. Each transfection was done in duplicate and repeated a minimum of three separate times with two different plasmid preparations. CAT assays were done using standard techniques. Input extracts were normalized by equal amounts of protein as measured by Bradford assay (Bio-Rad). Radioactivity was detected and quantified using a phosphorimager (Fuji).
Infectivity analysis.
Microglial cells cultured in 12-well plates were transfected in duplicate with HIV-1 pNL4-3 (1.5 μg) and the indicated CTIP1 and CTIP2 expression vectors. Two days after transfection, HIV-1 replication was monitored by measuring p24 Gag levels in the culture supernatants using an enzyme-linked immunosorbent assay (Innogenetics).
Western blot analysis.
Nuclear proteins (20 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose paper. Membranes were preincubated with 3% bovine serum albumin in phosphate-buffered saline (PBS) overnight at 4°C and were probed with monoclonal anti-Tat antibodies 8D1.8 (1:1,000 dilution; obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from J. Karn), or anti-Flag antibodies (1:10,000 dilution) for 1 h in PBS-0.1% Tween 20. After three washes in PBS-0.1% Tween 20, membranes were incubated with peroxidase-labeled anti-mouse antibodies (1:7,500 and 1:20,000 dilution, respectively; Santa Cruz Biotechnology) for 40 min and extensively washed. The signal was visualized by enhanced chemiluminescence (ECL+ detection system; Amersham).
GST pull down assays.
Bacterially expressed GST fusion proteins were bound on glutathione-Sepharose 8A beads (40 μl; Pharmacia) at 4°C overnight in NETN buffer (20 mM Tris [pH 8], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, containing protease inhibitors). Proteins were visualized by Coomassie blue staining, and protein content was normalized. The coated beads were washed with NETN and further incubated in a final volume of 300 μl of NETN for 2 h at 4°C with 15 μl of 35S-labeled input proteins prepared by in vitro translation using the TNT T7 coupled wheat germ extract system (Promega). After extensive washing, the bound proteins were dissociated by boiling for 3 min in Laemmli sample buffer and subjected to SDS-PAGE.
Coimmunoprecipitations.
Cell lysates prepared as described (34) were resuspended in 400 μl of TNE (50 mM Tris [pH 8.0], 1% Nonidet, 2 mM EDTA, a cocktail of protease inhibitors), and mixed with protein A-agarose beads (20 μl). After gentle shaking for 1 h at 4°C, the suspension was briefly centrifuged and the supernatant was mixed with monoclonal anti-HIV-1 Tat antibody (NT3 2D1.1; obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from J. Karn [10], or anti-HP1α antibodies [gift from R. Losson, Institut de Génétique et de Biologie Moléculaire et Cellulaire]). After overnight incubation at 4°C, protein A-agarose (20 μl) was added for 2 h. After extensive washing with TNE, the beads were processed for SDS-PAGE and Western blotting analysis.
Indirect immunofluorescence and confocal microscopy.
Microglial cells growing on coverslips were transfected with full-length HA-CTIP1 and Flag-CTIP2, various Flag-CTIP2 deletion mutants, pTat-GFP, and pCMV-Tat. At 24 or 48 h following transfection, cells were fixed and permeabilized in 4% paraformaldehyde and 0.1% Triton X-100. To localize endogenous lamin B and HP1α proteins, cells were preincubated with, respectively, anti-lamin B polyclonal antibodies and anti-HP1α polyclonal antibodies (both from Santa Cruz Biotechnology, Inc.) The corresponding immune complexes were detected by Alexa Fluor 568-conjugated donkey anti-goat antibodies (Molecular Probes, Eugene, Oreg.). After extensive washes, overexpressed HA-CTIP1, Flag-CTIP2, or Tat proteins were detected by incubations with anti-HA mouse monoclonal antibody (Covance Babco, Richmond, Calif.), anti-Flag M2 mouse monoclonal antibody (Upstate Biotechnology), or monoclonal antibody to HIV-1 Tat (NT3 2D1.1 obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from J. Karn [10]). HA-CTIP1, Flag-CTIP2, and Tat immune complexes were detected using cyanine 2-, cyanine 3-, or cyanine 5-conjugated goat anti-mouse antibodies (Jackson ImmunoResearch, West Grove, Pa.). To acquire optical sections of labeled cells we have used a Zeiss laser scanning microscope (model 510 invert) equipped with a Planapo oil (63×) immersion lens (numerical aperture = 1.4). As the fluorescent molecules were excited independently, no cross talk between the different fluorescent signals was detected during the triple channels acquisitions. Images were processed using Photoshop 5.0 (Adobe Systems, Inc.).
RESULTS
While both CTIP1 and CTIP2 cofactors inhibit HIV-1 LTR-driven transcription, only CTIP2 inhibits viral replication.
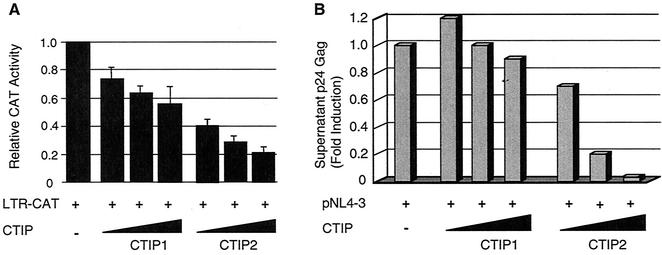
The functional effect of the transcriptional cofactors CTIP1 and CTIP2 on HIV-1 LTR-driven transcriptional activity was investigated by cotransfection of microglial cells with plasmids expressing HIV-1 LTR-CAT in the presence of increasing amounts of CTIP1 and CTIP2 (Fig. 1A). Both proteins were able to inhibit CAT activities in a dose-dependent manner, suggesting a repressive action through direct or indirect interactions with the LTR region.
FIG. 1.
Effect of CTIP1 and CTIP2 on HIV-1 LTR-driven transcription and on viral replication. (A) Microglial cells were cotransfected with vectors expressing HIV-1 LTR-CAT (1 μg) in the presence of increasing amounts (0.1, 0.5, and 1 μg) of CTIP1 and CTIP2, as indicated. After 2 days, CAT activities were measured and are expressed relative to the value obtained with LTR-CAT alone. Values correspond to an average of at least three independent experiments done in duplicate. Error bars, standard deviations. (B) Cells were cotransfected with HIV-1 pNL4-3 (1.5 μg) and with increasing amounts (0.1, 0.5, and 1 μg) of the indicated CTIP1 and CTIP2 expression vectors. Two days after transfection, samples of the culture supernatant were analyzed for p24 Gag content. Data represent an average of four independent experiments performed in duplicate. They are expressed relative to the value obtained with pNL4-3 alone taken as 1. Depending on the cell confluency, this value varied between 500 and 5,000 pg/ml.
To test whether this inhibitory effect on transcription was reflected at the level of viral replication, cells were cotransfected with HIV-1 pNL4-3 and increasing amounts of CTIP1 and CTIP2 expression vectors. Viral replication was monitored by measuring p24 Gag levels in the culture supernatants two days after transfection. While CTIP1 was unable to significantly affect replication, CTIP2 did inhibit viral production in a dose-dependent manner (Fig. 1B).
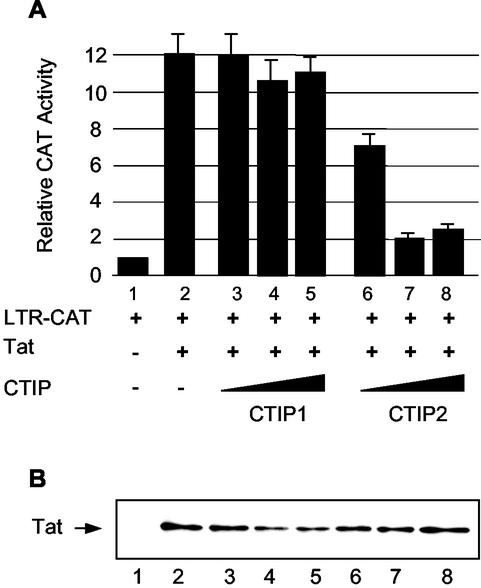
Since viral replication depends on Tat-mediated transcriptional stimulation, we examined whether CTIP1 and CTIP2 were able to affect the function of Tat. For this purpose, cells were cotransfected with HIV-1 LTR-CAT and a Tat expression vector in the presence of CTIP1 or CTIP2 (Fig. 2A). Results show that CTIP2, but not CTIP1, inhibits Tat-mediated HIV-1 transcriptional activity. As a control, Western blot analysis showed that the level of Tat expression is not affected by CTIP2 (Fig. 2B). Thus, CTIP2-mediated inhibition of Tat activity correlates with CTIP2 inhibition of viral replication.
FIG. 2.
CTIP2, but not CTIP1, inhibits Tat-mediated HIV-1 transcriptional activity. (A) Microglial cells were transfected with HIV-1 LTR-CAT (1 μg) in the presence of vectors expressing Tat (2 ng) and increasing amounts (0.1, 0.5, and 1 μg) of CTIP1 and CTIP2 as indicated. Histograms show CAT activities measured 2 days posttransfection and expressed relative to the value obtained with the LTR-CAT vector alone. Values correspond to an average of at least three independent experiments done in duplicate. Error bars, standard deviations. (B) Western blot analysis of nuclear proteins (20 μg) extracted from cells transfected with the plasmids corresponding to lanes 1 to 8 indicated in panel A. To detect Tat, 1 μg of expression vector was transfected. The blot was probed with monoclonal Tat antibodies.
CTIP2 and Tat interact in vitro and in cells.
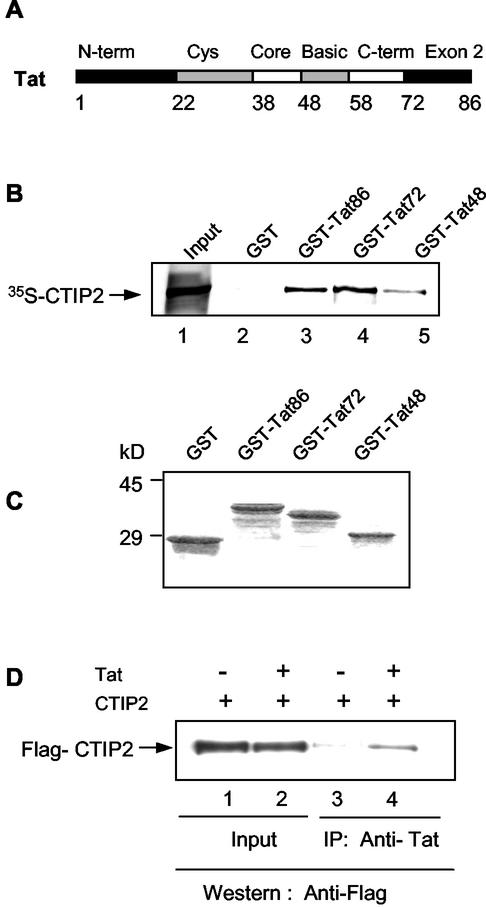
Since CTIP2, and not CTIP1, was able to affect both transcription and viral replication, we focused our studies on CTIP2. To decipher the mechanism whereby CTIP2 inhibits Tat-induced transcriptional stimulation, we first examined whether these two proteins were able to interact in vitro. GST pull down assays were performed with in vitro-translated 35S-labeled CTIP2 and full-length and truncated GST-Tat fusion proteins (Fig. 3B). A stained Coomassie gel shows that equivalent amounts of GST and GST fusion proteins were added to the reactions (Fig. 3C). Results showed that CTIP2 bound specifically to GST-Tat and not to the control GST protein. About 1 to 2% of CTIP2 bound to GST-Tat. Moreover the GST-Tat48 protein truncated after amino acid 48 was still able to mediate association with CTIP2, indicating that the N-terminal region of Tat is sufficient for interaction with CTIP2 in vitro.
FIG. 3.
CTIP2 and HIV-1 Tat interact in vitro and in cells. (A) Schematic representation of the domains present in the viral Tat protein. (B) The N-terminal region of Tat interacts with CTIP2. GST pull down assays were performed with 35S-CTIP2 and the indicated GST-Tat fusion proteins. 35S-CTIP2 was translated in vitro and incubated with GST (lane 2) or the indicated full-length and truncated GST-Tat proteins (lanes 3 to 5). After extensive washing, the bound proteins were eluted and analyzed by SDS-polyacrylamide gel electrophoresis. Lane 1, input 35S-CTIP2 (0.2 μl); lanes 2 to 5, GST and GST-Tat incubated with 35S-CTIP2 (10 μl). (C) SDS-PAGE of GST and GST-Tat fusion proteins used in panel B visualized by Coomassie brilliant blue staining. (D) Tat interacts with CTIP2 in vivo. Nuclear protein extracts from microglial cells, previously transfected with CTIP2 in the absence or presence of pCMV-Tat were immunoprecipitated with monoclonal anti-Tat antibodies (lanes 3 and 4). The presence of CTIP2 in the nuclear extracts (lanes 1 and 2) and in the immunoprecipitates (lanes 3 and 4) was detected with anti-Flag antibodies.
We next tested whether these two proteins were able to associate within cells. Microglial cells were transfected with CTIP2, in the absence or presence of Tat (Fig. 3D). Monoclonal anti-Tat antibodies did specifically coimmunoprecipitate CTIP2 in Tat-expressing extracts (lane 4) and not in nonexpressing Tat extracts (lane 3). These results confirm the existence of an association between Tat and CTIP2 in microglial cells.
CTIP2 harbors two Tat interaction interfaces.
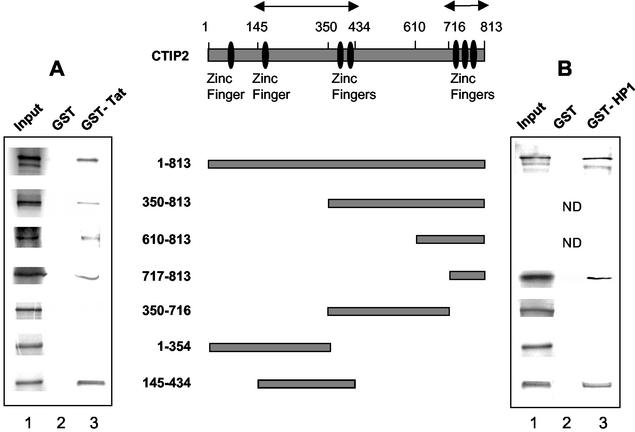
To precisely delineate the region of the CTIP2 protein which associates with Tat, GST pull down assays were performed with GST-Tat and a series of in vitro-translated 35S-labeled full-length and deletion mutants of CTIP2 (Fig. 4A). The full-length CTIP2 protein as well as the N-terminal deletion mutants starting at position 350, 610, and 717 did mediate interaction with GST-Tat and not with the control GST protein. This result shows that the C-terminal zinc finger region located between amino acids 717 and 813 is able to interact with Tat in vitro.
FIG. 4.
Definition of the in vitro interactions domains of CTIP2 with Tat (A) and HP1α (B). CTIP2 full-length and deletion mutants indicated in the schematic diagram were in vitro translated and used in GST pull down assays to examine their interaction with GST-Tat (A) (lane 3), GST-HP1α (B) (lane 3), and GST as a control (A and B) (lanes 2). Lanes 1, 35S-CTIP2 deletion mutants (0.2 μl); lanes 2 and 3, GST and GST-Tat incubated with the indicated 35S-CTIP2 proteins (10 μl). ND, not determined. Arrows delineate binding interfaces of Tat and HP1α.
We have previously shown that the related CTIP1 protein harbors two independent interaction domains with COUP-TF (1). This prompted us to examine whether an additional interaction interface was located in the N-terminal or central region of CTIP2. For this purpose we tested additional deletion constructs. The two CTIP2 deletion mutants 350-716 and 1-354 did abolish interaction with GST-Tat, confirming that the C-terminal region containing residues 717 to 813 mediates binding to Tat. Interestingly, when the central region localized between amino acids 145 and 434 was tested, it appeared able by itself to restore the interaction with GST-Tat.
Taken together, these findings clearly demonstrate that in vitro two interaction interfaces are implicated in CTIP2 binding to Tat, the central domain (residues 145 to 434) and the C-terminal domain (residues 717 to 813). The large 813-amino-acid CTIP2 may associate with one or two small 86-amino-acid Tat proteins, each Tat molecule binding to one of the two interfaces via its N-terminal region.
CTIP2 exhibits ball-like structures in the nucleus and recruits Tat to these structures.
To visualize the association of CTIP2 and Tat within the nucleus, cotransfected microglial cells were examined by immunofluorescence confocal microscopy (Fig. 5). The nuclear location was detected by staining of the endogenous lamin B protein, concentrated at the inner nuclear membrane (row 1) (19, 27). Tat linked to the GFP was expressed in the nucleoplasm and in nucleoli (row 1), in agreement with previous reports (7, 13, 36). Since it is believed that the functionally relevant and transcriptionally active Tat proteins are located in the nucleoplasm outside of the nucleoli (8), we have used conditions of minimal expression of Tat-GFP (5 ng of DNA) to minimize nucleolar localization. To correlate the results of confocal images with our transcription data, we have confirmed that both Tat and Tat-GFP stimulate LTR-driven transcription and that CTIP2 inhibits transactivation mediated by Tat and Tat-GFP to the same extent (results not shown).
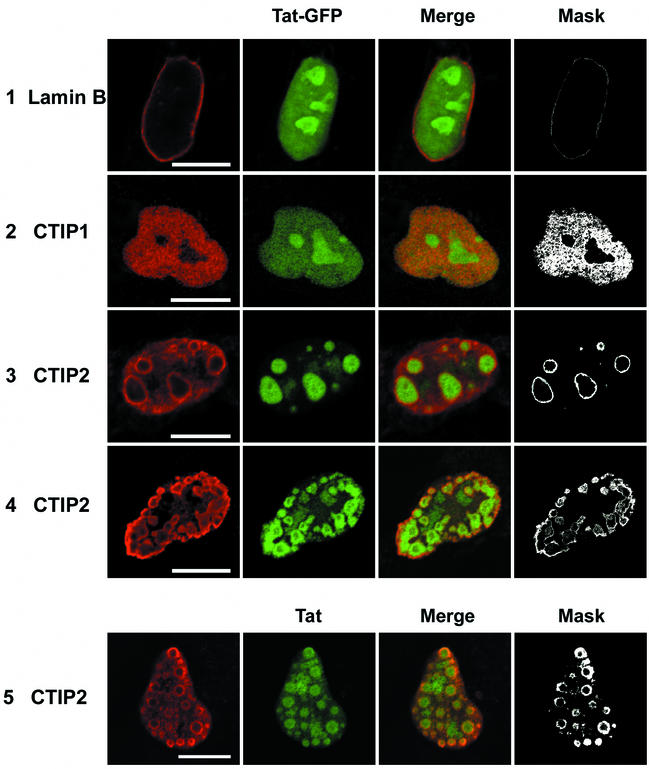
FIG. 5.
Localization of Tat-GFP and Tat in the presence of CTIP1 and CTIP2 in microglial cell nuclei. Cells were transfected with vectors expressing Tat-GFP (5 ng) in the absence (row 1) or the presence of HA-CTIP1 (row 2) and Flag-CTIP2 (rows 3 and 4). Cells were cotransfected with pCMV-Tat (500 ng) and Flag-CTIP2 (row 5). Cells were fixed 24 h after transfection, incubated with anti-lamin B, and stained with Alexa Fluor 568-conjugated secondary antibodies to detect endogenous lamin B and delineate the nucleus (row 1). To detect CTIP1 and CTIP2, cells were incubated with anti-HA (row 2) and anti-Flag (rows 3, 4, and 5) antibodies and stained with cyanine 3-conjugated secondary antibodies. To detect Tat, cells were incubated with monoclonal Tat antibodies and stained with cyanine 2-conjugated secondary antibodies (row 5). Masks were obtained after selection of the double-labeled pixels in the two-dimensional scatter histograms of gray values constructed from red and green images. Bars: 10 μm. HA-CTIP1 exhibited a diffuse staining pattern, with did not alter the Tat-GFP nucleolar and nuclear fluorescence (row 2). Flag-CTIP2 exhibited a ball-like staining in most of the nuclei. Tat-GFP and Tat were located within these ball-like structures that were distributed randomly in 80 to 90% of the nuclei (rows 3 and 5) or localized at the periphery of the inner nuclear membrane in 10 to 20% of nuclei (row 4). Tat-GFP and Tat colocalize with CTIP2 at the periphery of CTIP2-induced structures (columns 3 and 4).
Interestingly, Flag-CTIP2 exhibited an unusual staining pattern of ball-like structures filling the perinuclear space in 80 to 90% of the nuclei, as shown in a representative nucleus (row 3). In about 10 to 20% of cells, the CTIP2 structures were more peripheral to the inner nuclear membrane (row 4). CTIP2 was detected not inside the structures but more in juxtaposition with the outside face. The number of these typical structures varied between 10 and 30, and their size ranged between 1 and 5 μm, with an average size of 2.5 μm.
Surprisingly, in the presence of CTIP2, nucleoplasmic and nucleolar Tat-GFP were recruited inside the ball-like structures, clearly showing a dramatic nuclear relocation (rows 3 and 4). Overlaying of the red and green images revealed that the two proteins colocalize at the periphery of the ball-like structures (columns 3 and 4). When HA-CTIP1 was examined as a control, it exhibited a more diffuse staining pattern, with no alteration of the Tat-GFP distribution (row 2). These images confirm that CTIP2 and Tat-GFP are able to interact within the nuclei of cotransfected cells, as evidenced by their peripheral colocalization and the ability of the former protein to dramatically disrupt the nuclear localization of Tat-GFP
We next investigated whether the localization of the Tat-GFP fusion protein within the ball-like structures was similar to that of the Tat protein. Thus, cells expressing Tat were stained with monoclonal Tat antibodies revealed with Cy2 anti-mouse antibodies. We used conditions in which Tat was not detected in nucleoli (results not shown). Under these conditions, in cells expressing low levels of Tat, Tat was redistributed from the nucleoplasm within the ball-like structures (row 5), similar to the images observed with Tat-GFP. It is important to note that most of the Tat proteins present in the nucleoplasm (rows 1 and 2) are redistributed in the CTIP2-induced structures (rows 3, 4, and 5).
In some cells expressing higher levels of Tat, the green fluorescence was not detected within the balls but was concentrated at the periphery of these structures. To understand this observation, cells expressing Tat-GFP were stained with monoclonal Tat antibodies revealed with cyanine 3 anti-mouse antibodies. Again, in cells expressing high levels of Tat-GFP, the green fluorescence was detected inside the balls and the red staining was visible only at the periphery (results not shown). These observations suggest that monoclonal anti-Tat antibodies are impaired in their ability to contact high concentrations of Tat proteins packed within ball structures. This result confirmed that detection of the intrinsic Tat-GFP fluorescence represents the best tool to precisely visualize the subnuclear location of Tat.
CTIP2 colocalizes with HP1α in nuclei.
The observation that in some cells CTIP2 displayed a peripheral nuclear membrane location suggested a dynamic relocation next to the inner nuclear membrane. Recent data have shown the existence of dynamic associations between elements of the nuclear envelope and heterochromatin-associated proteins HP1, which are regulators of heterochromatin-mediated silencing (25, 38, 39). There are three HP1 protein family members in mammals, HP1α, HP1β, and HP1γ. The HP1α isoform is detected predominantly in transcriptionally repressed heterochromatic regions (29).
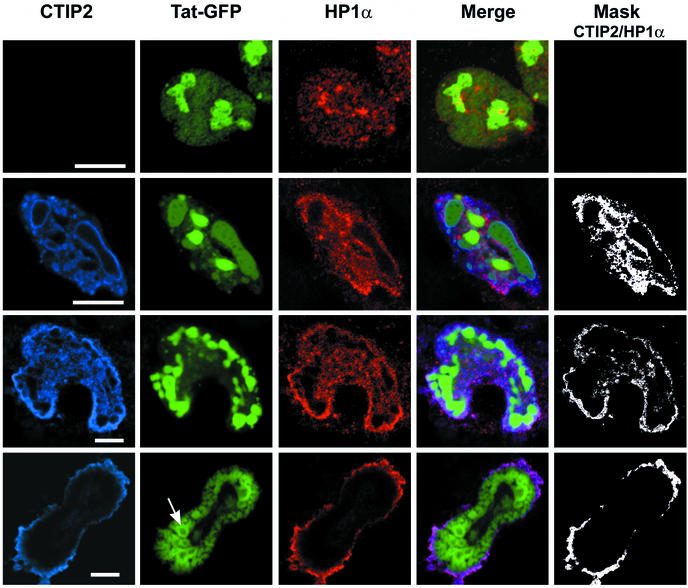
We therefore examined whether CTIP2 was associated with HP1α by visualizing their respective nuclear localization (Fig. 6). In the absence of CTIP2, endogenous HP1α (in red in Fig. 6) exhibited a random speckled distribution in all nuclei (row 1), in agreement with previous reports (25, 29). In the presence of transfected CTIP2 (in blue in Fig. 6), the distribution of HP1α was less random and in most spots was coincident with that of CTIP2 (rows 2 to 4). Overlaying of the confocal images (rows 4 and 5) revealed that these two proteins colocalize (in pink in Fig. 6) within the nucleoplasm of the vast majority of cells (rows 2 and 3) and next to the inner nuclear membrane of a minority of cells (row 4), suggesting that CTIP2 and HP1α are able to interact.
FIG. 6.
CTIP2 colocalizes with HP1α and redistributes Tat-GFP to distinct subnuclear structures within the nucleoplasm or next to the nuclear membrane. Microglial cells were transfected with vectors expressing Tat-GFP (500 ng) in the absence (row 1) or presence of Flag-CTIP2 (rows 2 to 4). Cells were incubated with anti-Flag antibodies and stained with cyanine 5-conjugated secondary antibodies to detect overexpressed CTIP2 (column 1; blue) or incubated with anti-HP1α and stained with Alexa Fluor 568-conjugated secondary antibodies to detect endogenous HP1α (column 3; red). For double immunofluorescence detection, cells were first treated to detect HP1 and then treated to detect CTIP2 (column 4). Masks representing the region of colocalization HP1α/CTIP2 were generated by selecting the double-labeled pixels. Bars: 10 μm. CTIP2 exhibited a staining pattern of distinct structures filled with Tat-GFP (rows 2 and 3). In some nuclei, CTIP2 was peripheral to the nuclear membrane and to ball-like structures of Tat-GFP (row 4). In these nuclei, Tat-GFP recruited at the inner nuclear membrane formed empty ball structures. An arrow points to one of these structures. Note that in all nuclei, HP1α colocalizes with CTIP2 (columns 4 and 5).
When CTIP2 and HP1α were distributed in the nucleoplasm, Tat-GFP filled the ball-like structures (Fig. 6) located randomly in the nucleoplasm (row 2) or next to the inner nuclear membrane (row 3). In contrast, in rare cells where CTIP2 and HP1α were adjacent to the nuclear membrane, Tat-GFP was also recruited to the inner membrane and formed empty ball structures, suggesting a dynamic process and changes in its interactions with CTIP2 (row 4). Of note, HP1α staining was excluded from the spots of Tat-GFP concentrations. On close inspection, HP1α appeared located at the outside of the CTIP2-induced structures. These observations suggest formation of sandwich structures with CTIP2 in the middle, HP1α on the outside, and Tat on the inside. This result further suggests that in a nuclear context CTIP2 interacts with an interface of HP1 different from that of Tat.
CTIP2 interacts with HP1α in vitro and harbors two HP1α interaction interfaces.
To precisely map the interaction domains of CTIP2 with HP1α, pull down assays were performed with GST-HP1α and different in vitro translated 35S-labeled deletion mutants of CTIP2 (Fig. 4B). Both full-length CTIP2 and the peptide 717-813 did mediate interaction with GST-HP1α and not with the control GST protein. This confirmed that CTIP2 interacts with HP1α in vitro and indicated that the 717-813 domain is involved in binding to HP1α. The 350-716 and 1-354 CTIP2 peptides were unable to bind to GST-HP1α, confirming that the C-terminal 717-813 domain mediates binding to HP1α. Similar to the results obtained with Tat, the central 145-434 region was also able to interact with GST-HP1α. These results demonstrate that CTIP2 harbors two independent HP1α interaction interfaces, regions 145-434 and 717-813. Interestingly, these same two domains are also involved in the in vitro association with Tat (Fig. 4A).
Analysis of HP1 binding to the CTIP2 interaction interfaces.
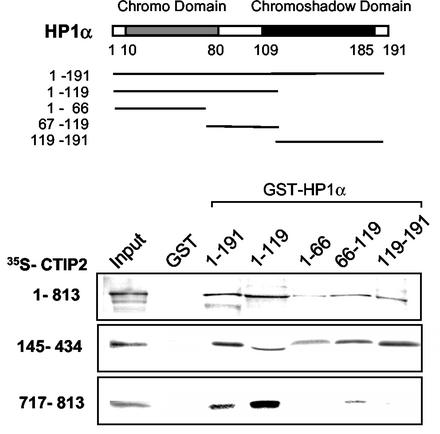
HP1-type proteins consist of two homologous but distinct globular domains, corresponding to the chromodomain (CD) and chromo shadow domain (CSD), separated by a hinge region (Fig. 7) (39). We next assessed which HP1α region was targeted by CTIP2. Different HP1α deletion mutants expressed as GST fusion proteins were used in pull down assays with full-length CTIP2, the 145-434 and 717-813 CTIP2 peptides (Fig. 7). Results confirmed that all three CTIP2 proteins interact with HP1α. CTIP2 peptide 717-813 bound specifically to the CD and hinge region. In contrast, full-length CTIP2 and CTIP2 peptide 145-434 bound more or less to each of the HP1α domains. Taken together, these results help understand how HP1α and CTIP2 associate in vitro.
FIG. 7.
Definition of the CTIP2 in vitro interaction domain of HP1α. The HP1α deletion mutants (indicated in the schematic diagram) were expressed as GST fusion proteins and bound to glutathione-Sepharose beads to examine their interaction with different domains of in vitro translated 35S-labeled full-length CTIP2 and the two indicated deletion mutants. GST and GST-HP1α mutants were incubated with the indicated 35S-CTIP2 proteins (25 μl). Input lane, 35S-CTIP2 (0.2 μl).
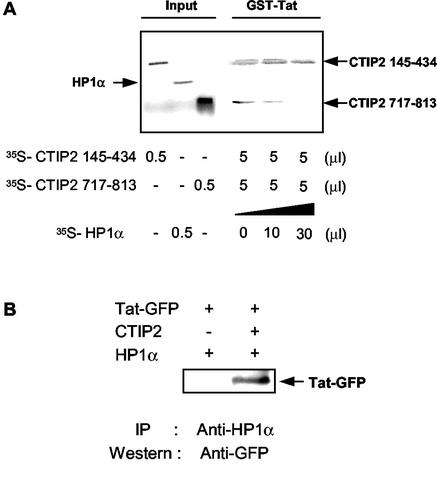
Tat and HP1α form a complex with CTIP2, by, respectively, binding to CTIP2 domains 145-434 and 717-813. Experiments performed with either HP1α or Tat indicated that each protein alone was able to bind to the two CTIP2 interaction interfaces. The question arises to what happens in the simultaneous presence of Tat and HP1α, as found in a cellular context. We therefore conducted competition pull down assays, using GST-Tat with both35S-labeled CTIP2 145-434 and CTIP2 717-813 in the presence of increasing amounts of 35S-HP1α (Fig. 8A). Results clearly showed that CTIP2 145-434 remained bound to GST-Tat, while CTIP2 717-813 was displaced by HP1α. This result shows that HP1 competes for Tat binding to the CTIP2 C-terminal domain. These findings indicate that the 145-434 domain has a higher binding affinity for Tat, and that the 717-813 domain preferentially binds HP1α. These results suggest that in the presence of Tat, HP1α is preferentially bound to the 717-813 domain and gets displaced from the 145-434 domain.
FIG. 8.
Analysis of the in vitro and in vivo interactions between CTIP2, Tat and HP1α. (A) Analysis of the preferential CTIP2 interaction domain of Tat by GST pull down competition assays. GST-Tat was immobilized on glutathione-Sepharose beads to examine the interaction with the two indicated 35S-labeled CTIP2 deletion mutants, in the absence and in the presence of increasing amounts of 35S-labeled HP1α. Note that HP1α does not bind directly to GST-Tat and that CTIP2 145-434 remains bound to GST-Tat and is not displaced by HP1α. (B) CTIP2 interacts with HP1 in cells only in the presence of CTIP2. Nuclear protein extracts from microglial cells, previously cotransfected with Tat-GFP and HP1 in the absence or presence of CTIP2 were immunoprecipitated with anti-HP1 antibodies and analyzed for Tat-GFP by Western blotting.
Interestingly, HP1α did not directly bind to GST-Tat in vitro (Fig. 8A). This result was confirmed by coimmunoprecipitation assays with extracts of microglial cells transfected with HP1α and Tat-GFP, in the absence or presence of CTIP2. Anti-HP1α antibodies were able to coimmunoprecipitate Tat-GFP, only in the presence of CTIP2 (Fig. 8B). This confirms the confocal microscopy observations and the HP1-CTIP2-Tat sandwich hypothesis, in which HP1 and Tat do not interact directly. Taken together, our data suggest a model of a three-protein complex in which (145-434) CTIP2 interacts with Tat, while (717-813) CTIP2 binds to HP1 via its chromo domain and hinge region.
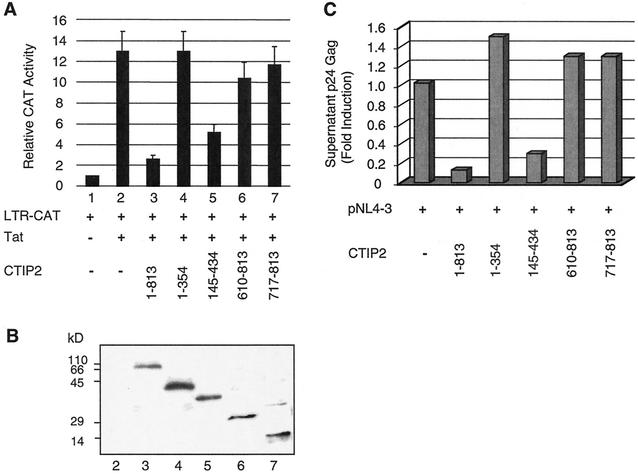
Effect of CTIP2 deletion mutants on HIV-1 gene transcription and on viral replication.
To test the validity of our in vitro model, we investigated the functional effect of different CTIP2 deletion constructs. Microglial cells were cotransfected with HIV-1 LTR-CAT and Tat in the presence of CTIP2 deletion mutants (Fig. 9A). As expected, the 1-354 peptide that harbors no Tat interaction domain did not affect the level of Tat-mediated transactivation, while 145-434 CTIP2 that contains a Tat binding site was able to inhibit Tat-induced stimulation. Interestingly, both 610-813 and 717-813 CTIP2 were unable to alter Tat transactivation. Western blot analysis was performed to control the expression levels of the CTIP2 peptides (Fig. 9B). Results showed that except for a higher expression of the 1-354 peptide, all other CTIP2 proteins were expressed at similar levels.
FIG. 9.
Full-length and 145-434 CTIP2 inhibit HIV-1 LTR-driven transcription and viral replication. (A) Microglial cells were cotransfected with vectors expressing HIV-1 LTR-CAT (1 μg) and Tat (2 ng) in the presence of the indicated CTIP2 deletion mutants (1 μg). CAT assays were performed after 2 days. CAT activities are expressed relative to the value obtained with LTR-CAT alone. Values correspond to an average of at least three independent experiments done in duplicate. (B) Western blot analysis of nuclear proteins extracted from cells transfected for 48 h with the indicated expression vectors used in panel A. The blot was probed with anti-Flag antibodies. Positions of size standards are indicated in kilodaltons. (C) Cells were cotransfected with vectors expressing HIV-1 pNL4-3 (1.5 μg) and the indicated CTIP2 deletion mutants (1 μg). Two days after transfection, samples of the culture supernatant were analyzed for p24 Gag content. Data represent an average of at least three independent experiments performed in duplicate. They are expressed relative to the value obtained with pNL4-3 alone taken as 1, which corresponds to an average of 2000 pg/ml of p24 Gag.
The CTIP2 mutants exhibited a similar behavior when tested in replication experiments, following cotransfection of pNL4-3 and CTIP2 vectors (Fig. 9C). Both full-length CTIP2 and CTIP2 145-434 appeared able to inhibit viral replication, in contrast with the other two mutants. Of note, the inhibitory effect of the mutant was less than that of full-length CTIP2.
These transcription and replication data support our in vitro model in which Tat preferentially associates with 145-434 CTIP2 and appears unable to interact with the 717-813 domain in the presence of HP1α. They confirm the importance of the 145-434 domain able to associate with both Tat and HP1α.
Localization of Tat-GFP in the presence of different CTIP2 mutants.
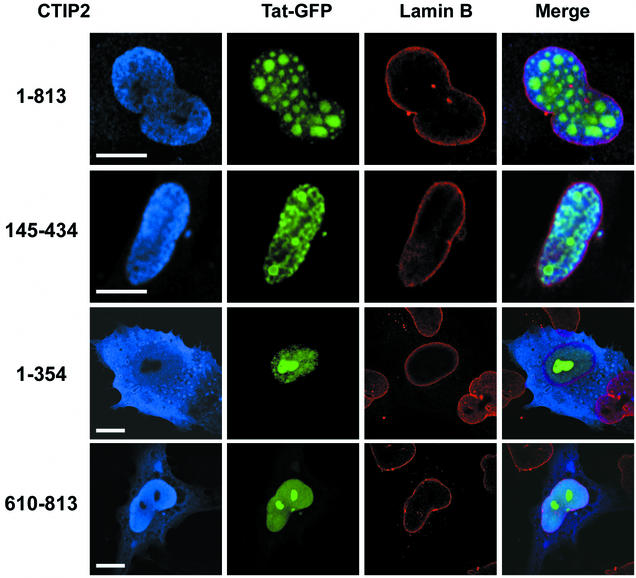
We next visualized the subcellular localization of Tat-GFP in the presence of the different CTIP2 mutants (Fig. 10). Nuclei were demarcated by immunolabeling with anti-lamin B antibodies (in red). The 145-434 CTIP2 mutant that was able to inhibit Tat function presented a distinct distribution (Fig. 10, row 2) that prefigured the typical ball-like structures formed in the presence of full-length CTIP2 (row 1). This mutant was expressed in the nucleus in most of the cells, formed a gradient toward the nuclear membrane and was structured in foci or small ball-like structures. The Tat-GFP distribution was clearly altered, and appeared concentrated in the vicinity of the foci-like structures. Although this pattern was distinct from that found in the presence of full-length CTIP2, it showed a clear recruitment of Tat next to the CTIP2 mutant-induced subnuclear structures (row 2).
FIG. 10.
Tat-GFP distribution is altered in the presence of full-length and 145-434 CTIP2. Microglial cells were transfected with vectors expressing Tat-GFP and the indicated full-length (residues 1 to 813) and deletion mutants of Flag-CTIP2. Nuclei are demarcated by immunolabeling with anti-lamin B antibodies (column 3, red). Cells were incubated with anti-Flag antibodies followed by staining with cyanine 5-conjugated antibodies to detect overexpressed CTIP2 proteins (columns 1 and 4, blue). Bars: 10 μm. Tat-GFP is redistributed within or next to subnuclear structures in nuclei overexpressing full-length and 145-434 CTIP2. In contrast, the nucleoplasmic and nucleolar distribution of Tat-GFP is not altered (compare with Fig. 5, row 1) in cells overexpressing 1-354 and 610-813 CTIP2 proteins in the cytoplasm and nucleus.
In contrast, the 1-354 and 610-813 CTIP2 truncated proteins that were unable to affect Tat activity exhibited a diffuse blue staining pattern in the cytoplasm and nucleus of all transfected cells. These mutants appeared clearly unable to alter Tat-GFP distribution (rows 3 and 4) similar to the pattern observed in the absence of CTIP2 (Fig. 5, row 1). These observations show a correlation between the ability of CTIP2 mutant proteins to alter Tat-GFP subnuclear location and their ability to affect Tat function. Interestingly, Tat relocation was less dramatic with the 145-434 CTIP2 mutant than with full-length CTIP2, correlating with a less potent effect of the mutant protein on Tat transactivation and viral replication.
DISCUSSION
We have investigated the functional effects of the recently described nuclear receptor cofactors CTIP1 and CTIP2 on HIV-1 gene expression and viral replication in human microglial cells. While the two related zinc finger proteins inhibit basal transcriptional activity, CTIP2 specifically acts as a potent inhibitor of HIV-1 Tat transactivation, leading to a potent repression of viral replication. This functional inhibition correlates with associations between the nuclear cofactor CTIP2, the viral transactivator Tat and the heterochromatin-associated protein HP1α, as discussed below. Preliminary results indicate that inhibition by CTIPs of the basal LTR-driven expression in the absence of Tat correlates with associations between the CTIP proteins with the cellular transcription factors COUP-TF and Sp1 (data not shown).
Association between CTIP2 and Tat.
CTIP2 was recently described as a cofactor of members of the COUP-TF subfamily of nuclear receptors (1). Our findings reveal that CTIP2 also directly associates with the N-terminal region (amino acids 1 to 48) of Tat. The inhibition of Tat function via this region correlates with the fact that it contains the short core domain (amino acids 21 to 40) sufficient to transactivate and induce HIV replication (4). The existence of a novel association between CTIP2 and Tat was also demonstrated by coimmunoprecipitation and confocal microscopy data in human microglial cells.
The CTIP2 protein harbors two independent Tat interaction interfaces, the 145-434 central domain and the 717-813 C-terminal domain. This bipartite interaction domain of CTIP2 is reminiscent of CTIP1, previously reported to contain two COUP-TF interaction domains (1). However, we found that in the presence of HP1, full-length CTIP2 interacts preferentially with Tat via its central 145-434 domain.
Within the nucleus, overexpressed CTIP2 exhibits a typical pattern of ball-like structures, mostly distributed throughout the nucleoplasm and in some cells concentrated at the periphery of the inner nuclear membrane. This expression pattern likely reflects a dynamic process during the cell cycle following CTIP2 overexpression. In a few cells, we could even detect a local disruption of the lamina leaking out CTIP2 within the cytoplasm (data not shown), likely associated with a late step of CTIP2 expression. Whether this process corresponds to CTIP2-induced apoptosis will be examined in future studies.
The nucleus is a highly dynamic organelle that contains distinct substructures. The best-studied nuclear compartments are the nucleolus and various nuclear bodies such as the splicing-factor compartments, the Cajal body, the promyelocytic oncogenic domains or nuclear domains 10 and a family of small dot-like nuclear speckles (12). With the exception of the nucleolus, none of the described nuclear substructures exceed 1.5 μm in diameter. On the basis of to their number, large size, and dynamic distribution, the CTIP2-induced ball-like structures likely represent a new class of nuclear substructures.
Overexpressed CTIP2 possesses the intriguing ability to dramatically disrupt the nuclear localization of viral Tat and to recruit Tat to the new ball-like structures. Interestingly, the related CTIP1 protein appears unable to form such distinct structures, to alter the localization of Tat, and to affect Tat function. These data show that sequestration of Tat by the CTIP2-created structures correlates with the inhibition of its transactivation ability. Besides a decrease of nucleolar Tat, most importantly, transcriptionally relevant Tat proteins present in the nucleoplasm are redistributed within the CTIP2-induced balls. This phenomenon is reminiscent of the capacity of CTIP1 to form punctate structures, to recruit COUP-TF to these structures and to repress COUP-TF activity (1). Similarly, the promyelocytic leukemia protein recruits the CREB-binding protein to subnuclear promyelocytic oncogenic domains and modulates CREB-binding protein function (11). This recruitment of transcription factors to nuclear subcompartments represents a novel mechanism whereby the function of nuclear factors is modulated.
How nuclear architecture affects gene expression remains largely unknown. There is growing evidence that nuclear compartmentalization influences the transcriptional activity of genes. Genes can be silenced through positioning into regions of chromosomes that are not accessible to chromatin remodeling factors or transcriptional activators (12, 15). A clear correlation exists between the silencing of a gene and its proximity to heterochromatin or to the nuclear periphery (6, 9). Our findings therefore suggest that transcription of the HIV-1 genome is inhibited through recruitment of Tat to distinct subnuclear structures associated with heterochromatin, as discussed below.
Association between CTIP2 and HP1α.
The HP1 family consists of small nonhistone proteins, primarily associated with heterochromatin, which contains mostly transcriptionally inactive or silent coding sequences (40). HP1 proteins are involved in gene silencing via the formation of heterochromatic structures and relocation of multiprotein complexes in heterochromatin (5, 14). Transcriptional repression requires the recognition of lysine 9-methylated histone H3 by the HP1 CD (2, 30).
Our findings reveal a novel direct association between HP1α and CTIP2. Full-length CTIP2 harbors two independent HP1α interaction interfaces, domains 145-434 and 717-813, that are the same two domains involved in the binding to Tat. The HP1 protein may bind as a monomer, with its CD binding to 717-813 CTIP2 and its CSD binding to 145-434 CTIP2. It is more likely that HP1 binds as a dimer and interacts with each of the two CTIP2 interfaces via its CD. Alternatively, the HP1 dimers may associate with two distinct CTIP2 proteins to form multiprotein complexes. Our confocal microscopy observations confirm that within the nucleus CTIP2 and HP1 are able to colocalize. These data support the idea that CTIP2, through its association with HP1, is relocated in transcriptionally inactive regions, as also shown by D. Avram and M. Leid (in preparation). Since HP1s self-associate via their CSD, bind to methylated histone H3 via their CD (residues 19 to 60) and interact with CTIP2 via part of the CD and hinge region (residues 60 to 119), it is likely that HP1 proteins link CTIP2 to histone H3 in repressed chromatin.
HP1-type proteins also serve as a linker, dynamically connecting transcriptionally silenced peripheral heterochromatin to the inner nuclear membrane, via cell-cycle regulated interactions with lamin B receptors (25, 38, 39). This ability of HP1 to associate with the nuclear membrane may account for the relocation of CTIP2 to the periphery of the nucleoplasm. The fact that HP1 appears located at the outside periphery of the CTIP2-induced substructures supports the hypothesis that HP1 is involved in relocating CTIP2 next to the inner nuclear membrane.
Association between HP1α, CTIP2 and Tat.
In a cellular context of HIV-1 infection that leads to viral Tat expression, HP1α and CTIP2 form a three-protein complex with Tat. The 145-434 CTIP2 domain interacts with Tat, while the 717-813 domain binds to HP1. Our functional data confirm the importance of the association between Tat and 145-434 CTIP2 and in contrast the inability of the 717-813 domain to affect the activity of Tat. No direct association could be detected in vitro and in cells between Tat and HP1. This perfectly correlates with confocal microscopy data showing the formation of an HP1-CTIP2-Tat sandwich, in which HP1 is located on the outside, CTIP2 in the middle and Tat on the inside.
The HP1-CTIP2-Tat association is involved in the dramatic recruitment of Tat within CTIP2-induced structures and relocation in distinct nuclear regions associated with heterochromatin. These associations correlate with CTIP2-mediated repression of Tat transactivation of the HIV-1 LTR. Interestingly, only full-length CTIP2 and the truncated 145-434 CTIP2 protein that possess the ability to create distinct subnuclear structures and to recruit Tat to these structures could function as Tat inhibitors. In agreement with studies showing that the availability of regulatory proteins influences regulation of gene expression during development and differentiation (12, 15) our data show that CTIP2, by reorganizing the subnuclear architecture, prevents Tat from playing its crucial role in viral gene expression.
While the vast majority of Tat-associated proteins function as positive factors (for review see (23) only a minority acts as repressors (28, 31). A recent report described a novel mechanism of Tat inactivation by overexpression of the host proteins YY1 and LSF, which recruit histone deacetylase 1 to the LTR and counteract the positive effect of histone acetyltransferases (20). Preliminary results show that CTIP2 is also able to interact with histone deacetylases (data not shown). Therefore, further investigations will assess whether inhibition of LTR transactivation by CTIP2 may also be linked to histone deacetylation and/or to Tat deacetylation. It will be interesting to examine the possibility of CTIP2 presenting a dual inhibitory action via recruitment of Tat to HP1 and to methylated histone H3 within heterochromatin, and via recruitment of deacetylases to modified histones and/or the acetylated Tat protein. Our findings present evidence of a correlation between Tat sequestration by CTIP2-created subnuclear structures, relocation in HP1-associated regions, and inhibition of Tat transactivating ability leading to repression of HIV-1 replication. These studies performed in human microglial cells and currently limited to this cell type reveal a novel mechanism of Tat inhibition through subnuclear relocalization in regions associated with heterochromatin. Further studies will be necessary to precisely examine the molecular mechanisms that underlie our observations. Moreover, future investigations need to be performed in various HIV-1 target cells to analyze whether and how the use of CTIP2-derived peptides could serve for a novel anti-HIV strategy and help establish latent HIV infection.
Acknowledgments
We thank George Pavlakis for providing pTat-GFP and Régine Losson for providing GST-HP1α vectors and anti-HP1α antibodies. We acknowledge the confocal microscopy facilities of IFR37.
This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM) and the Agence Nationale de Recherches sur le SIDA (ANRS). Work in the Leid laboratory was supported by NIH grants GM60852 (to M.L.) and AR02194 (to D.A.). Additional support was provided by the Oregon State University Environmental Health Sciences Center (under NIH grant ES00210).
REFERENCES
- 1.Avram, D., A. Fields, K. Pretty On Top, D. J. Nevrivy, J. E. Ishmael, and M. Leid. 2000. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 275:10315-10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Benkirane, M., R. F. Chun, H. Xiao, V. V. Ogryzko, B. H. Howard, Y. Nakatani, and K. T. Jeang. 1998. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem. 273:24898-24905. [DOI] [PubMed] [Google Scholar]
- 4.Boykins, R. A., R. Mahieux, U. T. Shankavaram, Y. S. Gho, S. F. Lee, I. K. Hewlett, L. M. Wahl, H. K. Kleinman, J. N. Brady, K. M. Yamada, and S. Dhawan. 1999. Cutting edge: a short polypeptide domain of HIV-1-Tat protein mediates pathogenesis. J. Immunol. 163:15-20. [PubMed] [Google Scholar]
- 5.Brasher, S. V., B. O. Smith, R. H. Fogh, D. Nietlispach, A. Thiru, P. R. Nielsen, R. W. Broadhurst, L. J. Ball, N. V. Murzina, and E. D. Laue. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19:1587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, K. E., S. S. Guest, S. T. Smale, K. Hahm, M. Merkenschlager, and A. G. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91:845-854. [DOI] [PubMed] [Google Scholar]
- 7.Burton, M., C. D. Upadhyaya, B. Maier, T. J. Hope, and O. J. Semmes. 2000. Human T-cell leukemia virus type 1 Tax shuttles between functionally discrete subcellular targets. J. Virol. 74:2351-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun, R. F., O. J. Semmes, C. Neuveut, and K. T. Jeang. 1998. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol. 72:2615-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cockell, M., and S. M. Gasser. 1999. Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9:199-205. [DOI] [PubMed] [Google Scholar]
- 10.Dingwall, C., I. Ernberg, M. J. Gait, S. M. Green, S. Heaphy, J. Karn, A. D. Lowe, M. Singh, M. A. Skinner, and R. Valerio. 1989. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. USA 86:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doucas, V., M. Tini, D. A. Egan, and R. M. Evans. 1999. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc. Natl. Acad. Sci. USA 96:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dundr, M., and T. Misteli. 2001. Functional architecture in the cell nucleus. Biochem. J. 356:297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efthymiadis, A., L. J. Briggs, and D. A. Jans. 1998. The HIV-1 Tat nuclear localization sequence confers novel nuclear import properties. J. Biol. Chem. 273:1623-1628. [DOI] [PubMed] [Google Scholar]
- 14.Eissenberg, J. C., and S. C. Elgin. 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10:204-210. [DOI] [PubMed] [Google Scholar]
- 15.Francastel, C., D. Schubeler, D. I. Martin, and M. Groudine. 2000. Nuclear compartmentalization and gene activity. Nat. Rev. Mol. Cell. Biol. 1:137-143. [DOI] [PubMed] [Google Scholar]
- 16.Gatignol, A., and K. T. Jeang. 2000. Tat as a transcriptional activator and a potential therapeutic target for HIV-1. Adv. Pharmacol. 48:209-227. [DOI] [PubMed] [Google Scholar]
- 17.Gaynor, R. B. 1995. Regulation of HIV-1 gene expression by the transactivator protein Tat. Curr. Top. Microbiol. Immunol. 193:51-77. [DOI] [PubMed] [Google Scholar]
- 18.Glass, J. D., and R. T. Johnson. 1996. Human immunodeficiency virus and the brain. Annu. Rev. Neurosci. 19:1-26. [DOI] [PubMed] [Google Scholar]
- 19.Goldman, R. D., Y. Gruenbaum, R. D. Moir, D. K. Shumaker, and T. P. Spann. 2002. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16:533-547. [DOI] [PubMed] [Google Scholar]
- 20.He, G., and D. M. Margolis. 2002. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol. Cell. Biol. 22:2965-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janabi, N., S. Peudenier, B. Heron, K. H. Ng, and M. Tardieu. 1995. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci. Lett. 195:105-108. [DOI] [PubMed] [Google Scholar]
- 23.Jeang, K. T., H. Xiao, and E. A. Rich. 1999. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274:28837-28840. [DOI] [PubMed] [Google Scholar]
- 24.Kingsman, S. M., and A. J. Kingsman. 1996. The regulation of human immunodeficiency virus type-1 gene expression. Eur. J. Biochem. 240:491-507. [DOI] [PubMed] [Google Scholar]
- 25.Kourmouli, N., P. A. Theodoropoulos, G. Dialynas, A. Bakou, A. S. Politou, I. G. Cowell, P. B. Singh, and S. D. Georgatos. 2000. Dynamic associations of heterochromatin protein 1 with the nuclear envelope. EMBO J. 19:6558-6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krebs, F. C., H. Ross, J. McAllister, and B. Wigdahl. 2000. HIV-1-associated central nervous system dysfunction. Adv. Pharmacol. 49:315-385. [DOI] [PubMed] [Google Scholar]
- 27.Moir, R. D., T. P. Spann, R. I. Lopez-Soler, M. Yoon, A. E. Goldman, S. Khuon, and R. D. Goldman. 2000. Review: the dynamics of the nuclear lamins during the cell cycle—relationship between structure and function. J. Struct. Biol. 129:324-334. [DOI] [PubMed] [Google Scholar]
- 28.Nelbock, P., P. J. Dillon, A. Perkins, and C. A. Rosen. 1990. A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator. Science 248:1650-1653. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen, A. L., M. Oulad-Abdelghani, J. A. Ortiz, E. Remboutsika, P. Chambon, and R. Losson. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7:729-739. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416:103-107. [DOI] [PubMed] [Google Scholar]
- 31.Ou, S. H., F. Wu, D. Harrich, L. F. Garcia-Martinez, and R. B. Gaynor. 1995. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 69:3584-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira, L. A., K. Bentley, A. Peeters, M. J. Churchill, and N. J. Deacon. 2000. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 28:663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peudenier, S., C. Hery, L. Montagnier, and M. Tardieu. 1991. Human microglial cells: characterization in cerebral tissue and in primary culture, and study of their susceptibility to HIV-1 infection. Ann. Neurol. 29:152-161. [DOI] [PubMed] [Google Scholar]
- 34.Rohr, O., D. Aunis, and E. Schaeffer. 1997. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J. Biol. Chem. 272:31149-31155. [DOI] [PubMed] [Google Scholar]
- 35.Rohr, O., C. Schwartz, C. Hery, D. Aunis, M. Tardieu, and E. Schaeffer. 2000. The nuclear receptor chicken ovalbumin upstream promoter transcription factor interacts with HIV-1 Tat and stimulates viral replication in human microglial cells. J. Biol. Chem. 275:2654-2660. [DOI] [PubMed] [Google Scholar]
- 36.Stauber, R. H., and G. N. Pavlakis. 1998. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology 252:126-136. [DOI] [PubMed] [Google Scholar]
- 37.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 38.Vlcek, S., T. Dechat, and R. Foisner. 2001. Nuclear envelope and nuclear matrix: interactions and dynamics. Cell. Mol. Life Sci. 58:1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye, Q., I. Callebaut, A. Pezhman, J. C. Courvalin, and H. J. Worman. 1997. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 272:14983-14989. [DOI] [PubMed] [Google Scholar]
- 40.Zhao, T., T. Heyduk, C. D. Allis, and J. C. Eissenberg. 2000. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem. 275:28332-28338. [DOI] [PubMed] [Google Scholar]