Abstract
The role of the cis replication element (cre) in the 2CATPase coding region of the poliovirus (PV) genome has been studied with a series of mutants derived from either a PV1 full-length genome or a replicon (P/L) containing the firefly luciferase reporter gene in place of the capsid region. Using the P/L replicon we have inserted cre elements at three different locations in the genome including the 5′ nontranslated region and within the open reading frame. The successful recovery of replication of a nonviable P/L (A5C) mutant replicon with an artificial cre element as “rescuer,” in addition to the results of site-directed mutagenesis and experiments with truncated forms of PV-cre(2C), indicated that (i) the sequence within the upper stem and loop regions contains the minimal cre RNA required for VPg uridylylation in vitro, (ii) the location of the cre RNA in the poliovirus genome is not relevant to RNA infectivity, and (iii) specific binding of 3CDpro to PV-cre(2C) occurs within the upper stem region and probably involves several contact residues. The role of a 14-nucleotide conserved “core” sequence among known cre structures in picornaviruses was examined by site-directed mutagenesis of individual nucleotides. In addition to a conserved AAA (4472 to 4474) triplet previously shown to be the primary RNA template for VPg uridylylation by the PV RNA polymerase 3Dpol (E. Rieder, A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer, J. Virol. 74:10371-10380, 2000), we have now shown that important residues (G4468 and A4481) are contained in a predicted internal bulge at the upper stem-loop of PV-cre(2C). We have further demonstrated that the viral proteins 3CDpro and 3Cpro form stable complexes with a transcript PV-cre(2C) RNA that can be considered critical for VPg uridylylation.
The interplay of viral proteins and host factors in virus replication within the Picornaviridae has been the subject of studies for 2 decades. Although important discoveries emerged from these studies, the steps required to successfully replicate picornavirus RNA in host cells are not yet fully understood.
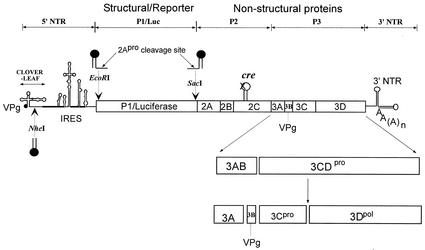
During the course of infection poliovirus RNA serves as mRNA and directs the synthesis of a large polyprotein, which is processed by virus-encoded proteinases into structural and nonstructural proteins. The genome of poliovirus possesses a 5′ end characteristic of picornaviruses, consisting of a long nontranslated region (5′ NTR) that is linked to a small viral peptide, VPg. The linkage is between the 5′ phosphate of the terminal UMP and the hydroxyl group of a tyrosine in VPg (2, 32). The 5′ NTR contains two highly structured RNA signals: the cloverleaf, which is required for genome replication (3, 4), and the internal ribosome entry site (IRES; (12, 28), which directs translation initiation (Fig. 1). The cloverleaf has been shown to interact with viral protein 3CDpro and either 3AB or cellular poly(rC) binding protein (PCBP2) to perform its function in RNA synthesis (4, 6, 7, 10, 21, 31, 39). More recently, a model of the genome in a circular form aided by the interaction between CL, 3CDpro, and PCBP2 at the 5′ NTR and poly(A) and poly(A) binding protein on the 3′ NTR has been proposed (5, 11).
FIG. 1.
Schematic diagram of replicon and full-length poliovirus genomes used in this study. Open circle, wt PV-cre(2C). Arrows, positions of unique restriction endonuclease sites engineered for the insertion of the second copy of cre of various origins. In rescue experiments, the native cre(2C) was inactivated through a point mutation (A5C) in the conserved AAA sequence of the PV-cre loop (X) and a second copy of cre (filled hairpin loop) from poliovirus, HRV2, or HRV14 or an artificial cre was inserted at locations indicated by arrows.
The poly(A) tail and the heteropolymeric region of the 3′ NTR (Fig. 1) contain signals for replication of the poliovirus genome. Despite the observed deleterious effects of mutations within the heteropolymeric region, the role of this highly structured segment of the poliovirus genome in virus replication remains unsolved (1). Truncated forms of the genome lacking the 3′ NTR have been shown to replicate, albeit not as efficiently as the wild-type genome (35).
A new structural element, cre (cis replication element, oriI), has been identified in the genomes of several picornaviruses. cres have been described for human rhinovirus 14 (HRV14) (18), cardioviruses (Theiler's murine encephalomyelitis virus 15), poliovirus types 3 and 1 (9, 26, 30), HRV2 (8) (Fig. 2), and foot-and-mouth disease virus (17). Interestingly, the locations of these elements in the genomes of different picornaviruses vary, although all seem to perform the same function. In assays where the uridylylation partners are reconstituted from purified viral proteins, the cre elements act entirely independently of other RNA signals of the genomic RNA (26, 30) (E. Rieder et al., submitted for publication). This extends to a variant of poliovirus RNA, whose cloverleaf cannot bind 3CDpro yet functions with wild-type (wt) efficiency in uridylylation (Rieder et al., submitted). In contrast, in studies of the translation/replication system reported by Lyons et al. (16), the cloverleaf was found to be essential for uridylylation of VPg. This apparent conflict in the results remains to be resolved and may be due to differences in the experimental approach.
FIG. 2.
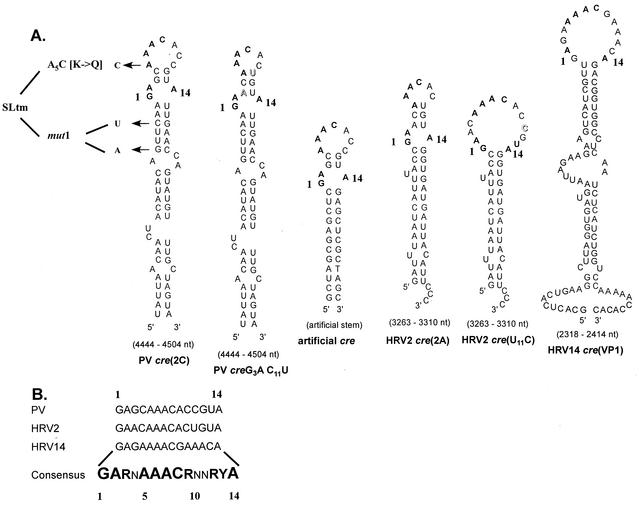
Comparison of predicted cre structures of various picornaviruses used in this study. (A) Hairpin structures predicted by the MFOLD RNA program (Michael Zuker) for wt PV-, HRV14-, HRV2-cre, HRV2-cre carrying a U11C mutation which enlarges the top loop structure, and a cre constructed to contain 14 nt from the PV-cre top loop and an unrelated artificial stem. Arrows, locations of the substitutions in mutant PV-cres corresponding to A5C, G4462A C4465U, and SLtm. Letters in shadow style, mutations introduced in PV-cre and HRV2-cre to alter their loop sizes. (B) Sequences of the top 14 nt of three naturally occurring picornavirus cres. The consensus sequence referred to in the text as the core sequence is in boldface. The degree of conservation at each nucleotide position is indicated by the size of the letter. N, any ribonucleotide; R, purine; Y, pyrimidine.
cre RNAs so far described for picornaviruses consist of a hairpin structure composed of a stem that is heterogeneous in length and that includes internal bulges and a terminal loop of variable size (Fig. 2). Recently, we have shown that poliovirus RNA containing a mutation (underlined) of AAA to CAA (4472 to 4474) in the loop of PV-cre(2C) is nonviable (30). Moreover, we have demonstrated that the PV-cre(2C), in particular the first two As of the AAA triplet, serves as an efficient template for VPg-pUpU synthesis in vitro, which, in turn, is the primer for RNA synthesis (26, 30). Uridylylation of VPg occurs via catalysis by the RNA-dependent RNA polymerase 3Dpol in a reaction that is greatly stimulated by 3CDpro (26).
Our previous studies have also suggested that a slide-back mechanism is involved in the nucleotidylylation of VPg, where the first UMP in VPg-pUpU is linked to VPg to form VPg-pU by using A4472 as the template. This is followed by a translocation of the uridylylation complex to, and base pairing with, A4473 and the addition of a second UMP to form VPg-pUpU (8, 30; A. V. Paul et al., unpublished data).
Available biochemical and genetic evidence has led to the suggestion that cre RNAs in picornaviruses are required in the sense strand for negative-strand RNA synthesis, acting in cis (9, 19, 30). A minigenome transcript RNA of poliovirus was used to show that the sense, but not the antisense, PV-cre(2C) is the functional template for the in vitro VPg uridylylation reaction (26). Given these observations, it appeared likely that poliovirus and probably all picornaviruses have acquired a mechanism to efficiently replicate their own genomic RNA via an exquisite interplay of regulatory mechanisms involving virally encoded proteins (precursors and their end products), a number of host factors, and at least three structural RNA elements.
To determine the critical structural features required for cre function, we have generated dual-cre poliovirus genomic constructs as well as a dual-cre (luciferase) replicon system. This strategy was based on previous observations that cre elements can be moved within replicons of HRV14 (18) or poliovirus (9). In our experiments, we placed second cres in three different locations of the poliovirus genome. Moreover, we used cres from two different picornaviruses (HRV14 and HRV2) to assess their ability to rescue the lethal phenotype caused by mutations in the native cre (nucleotides [nt] 4445 to 4504), mapping to the coding region of poliovirus 2CATPase. In addition, we have analyzed the effect on in vitro VPg uridylylation of partial truncations in PV-cre(2C) and mutations within the core sequence in the upper stem and terminal loop of this hairpin. Our data confirm and extend recent data obtained with the HRV14 cre by Yang et al. (40). We also present evidence for the specific interaction between PV-cre(2C) and 3CDpro (or its cleavage product 3Cpro).
MATERIALS AND METHODS
Enzymes.
Poliovirus RNA polymerase was expressed in Escherichia coli from plasmid pT5T-3Dpol, which encodes the wt sequence of 3Dpol preceded by an ATG, and was purified by the method of Pata et al. (22).
For His tag expression of 3CDpro, we used the plasmid pET21b/3CDpro (3Cpro/H40A). The protein was expressed in E. coli and purified by Ni2+ chromatography as described elsewhere (26). The untagged 3CDpro (H40A) and expression vectors for His-tagged 3Cpro were gifts from C. E. Cameron (23).
Cellular PCBP2 containing a His tag was expressed in E. coli from plasmid pET21b/PCBP2 (this is a derivative of the plasmid pQE30-PCBP2, a gift from B. L. Semler) (21) and purified as previously described (26).
The expression of 3AB in E. coli and the purification of the protein were described by Lama et al. (13).
Construction of plasmids and DNA manipulation.
pT7PV1(M) (36) refers to the full-length PV1(M) cDNA. Site-directed mutagenesis was performed with the QuikChange mutagenesis kit from Stratagene, and mutagenic oligonucleotides are described in Tables 1 and 2. Mutations and final constructs were verified through sequencing using the ABI Prism DNA sequencing kit.
TABLE 1.
Oligonucleotides used for mutagenesis and cloning (involving PCR amplification)
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| PVcre (SacI) forward | GCC GAG CTC ACA ACC TAT GGG TTC GGA ACT ATT AAC AAC TAC ATA C |
| PVcre (SacI) reverse | GC GAG CTC TAC TAG CAA ACA TAC TGG |
| HRV14cre (SacI) forward | GCC GAG CTC ACA ACC TAT GGG TTC GGA GCA CTC ACT GAA GGC TTA G |
| HRV14cre (SacI) reverse | GGC GAG CTC TTT TTG TGT GTG TTT TGG |
| HRV2cre (SacI) forward | GCC GAG CTC ACA ACC TAT GGG TTC GGA GAT TTA ATC ATT TAC CGA AC |
| HRV2cre (SacI) reverse | GGC GAG CTC GGG AAT GTA ATC ATC ACC |
| HRV2cre U11C forw. (QM)a | ATT TAC CGA ACA AAC ACC GTA GGT GAT GAT TAC |
| PVcre (EcoRI) forward | CGG AAT TCA CTA TTA ACA ACT ACA TAC AG |
| PVcre (EcoRI) reverse | CGG AAT TCC CCA AAG CCG TAA GTC GTT ACT AGC AAA CAT ACT GG |
| PVcre G3A C11U forw. (QM) | CAT ACA GTT CAA GAA CAA ACA CTG TAT TGA ACC AGT ATG |
| PVcre A14C forward (QM) | CAA GAG CAA ACA CCG TCT TGA ACC AGT ATG TTT G |
| PVcre G1A forward (QM) | CTA CAT ACA GTT CAA AAG CAA ACA CCG TAT TG |
| PVcre G1C forward (QM) | CTA CAT ACA GTT CAA CAG CAA ACA CCG TAT TG |
| PVcre G1U forward (QM) | CTA CAT ACA GTT CAA TAG CAA ACA CCG TAT TG |
| PVcre A14G forward (QM) | CAA GAG CAA ACA CCG TGT TGA ACC AGT ATG TTT G |
| PVcre A14U forward (QM) | CAA GAG CAA ACA CCG TTT TGA ACC AGT ATG TTT G |
| PV/Luc+(NheI) forw. (QM) | GTT TTA TAC TCC CTT CCC GCT AGC TTA GAC GCA CAA AAC CAA G |
| PVcre (NheI) forward | CTA GCT AGC ACT ATT AAC AAC TAC |
| PVcre (NheI) reverse | CTA GCT AGC TAC TAG CAA ACA TAC |
| Artificial cre (NheI) forward | CTA GCT AGC GAG CTC GAG CAA ACA CCG TAG AGC |
| Artificial cre (NheI) reverse | GAT GCT AGC GAG CTC TAC GGT GTT TGC TCG AGC |
| Artificial cre A5C(NheI) forw. | CTA GCT AGC GAG CTC GAG CCA ACA CCG TAG AGC |
| Artificial cre A5C(NheI) rev. | GAT GCT AGC GAG CTC TAC GGT GTT GGC TCG AGC |
| PVcre (SLtm) forward (QM) | CTA TTA ACA ACT ACA TAC AAT TTA AGA GCC AAC ACC GTA TCG AAC |
QM, oligonucleotide used for QuikChange mutagenesis.
TABLE 2.
Oligonucleotides used for the synthesis of truncated PV-cre(2C)
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| T7 Δ1cre forward | GGT GCC GGC TAA TAC GAC TCA CTA TAG GCT ACA TAC AGT TCA AGA GCA AAC |
| Δ1cre reverse | AAG AAT TCC AAA CAT ACT GGT TCA ATA CGG TGT TTG CTC TTG AAC TGT |
| T7 Δ2cre forward | GGT GCC GGC TAA TAC GAC TCA CTA TAG GTT CAA GAG CAA AC |
| Δ1cre reverse | CAA GAA TTC CTA GGG TTC AAT ACG GTG TTT GCT CTT GAA CCT ATA G |
| T7 Δ3cre forward | GTG CCG GCT AAT ACG ACT CAC TAT AGG CAA ACA CCG TAT TG |
| Δ3cre reverse | AAG AAT TCC TAG GGT TCA ATA CGG TGT TTG CCT ATA GT |
For simplification of the nomenclature, we have numbered a functionally important sequence (consensus sequence, see below) of the cre element as depicted in Fig. 2. Thus, the highly conserved AAA triplet is numbered A5A6A7. The A5C (AAA to CAA) mutation (underlined) in PV-cre(2C) has been characterized previously and is referred to by Rieder et al. (30) as mut7. The nomenclature for dual-cre constructs is as follows: PV or P/L (poliovirus-luciferase) refers to the parental (full-length or replicon) plasmid, followed by the genotype of the native PV-cre (A5C or SLtm), followed by the genotype of the “rescuer” and its insertion site in the genome (N, NheI; E, EcoRI; S, SacI).
P/L (A5C) is a derivative of replicon P/L (wt) (14) carrying a lethal AAA to CAA (A5C) mutation (underlined) in the cognate PV-cre(2C) (see above) (30) and three unique restriction sites that are used in this study: an NheI site between the cloverleaf and IRES, a SacI site at the beginning of the viral 2Apro protein, and an EcoRI site next to the initiating AUG (Fig. 1). Dual-cre plasmids were constructed by ligation of PCR-amplified DNA fragments carrying either a point mutant PV-cre, an artificial cre, or foreign cre elements to the inactivated P/L (A5C) or triple mutant G4462A A4472C C4465U (named SLtm for stem and loop triple mutant) replicons at either SacI, EcoRI, or NheI restriction enzyme sites. To introduce a point mutation (U11C) in the native HRV2-cre(2A), site-directed mutagenesis was performed using pT7HRV2 full-length cDNA (33) as the template and oligonucleotides described in Table 1. HRV14-cre was amplified with pT7HRV14 cDNA (34) as the template.
pT7PVM(S) is a full-length poliovirus cDNA clone used as a parental construct that contains an engineered restriction site (SacI) between P1 and 2Apro coding sequences (14). DNA fragments carrying the mutations in PV-, HRV2-, or HRV14-cre described above were generated by PCR amplification from the corresponding templates and inserted at the SacI site. To assemble a dual-cre PV1(M) with a rescuer cre inserted between the cloverleaf and the IRES, a fragment between PmlI and AgeI in pT7PV(M) was replaced with that of the plasmid P/L (A5C)/PVwt(N).
cDNA templates corresponding to the PV-cre(2C) sequence (nt 4445 to 4504; Fig. 2) were obtained by PCR amplification from plasmid pT7PVM as described previously (30).
In vitro transcription, transfection, and plaque assay.
All plasmids were linearized with DraI. Truncated derivatives of PV-cre RNA used in in vitro uridylylation experiments were generated by PCR amplification using oligonucleotides described in Table 2. RNAs were synthesized with phage T7 RNA polymerase, and the RNA transcripts were transfected into HeLa R19 cell monolayers by the DEAE-dextran method as described previously (36). The incubation time was 12 h for luciferase replicons and up to 3 days for full-length viral constructs. Virus titer was determined by a plaque assay (20).
Analysis of recombinant viruses.
To measure one-step growth kinetics, we used HeLa R19 monolayers (5 × 106 cells) infected with virus at a multiplicity of infection of 5 to 10. The plates were incubated at 37°C, and the infected cells were harvested at 0, 1, 2.5, 3, 5, 8, and 12 h postinfection. The virus yield of the cell lysates was determined by plaque assay.
VPg uridylylation assay.
The reaction mixture contained 50 mM HEPES, pH 7.5, 3.5 mM magnesium acetate, 1 μg of purified 3Dpol, 2 μg of synthetic poliovirus VPg, ∼0.75 μCi of [32P]UTP (3,000 Ci/mmol; DuPont NEN), 10 μM unlabeled UTP, and 0.5 μg of template RNA transcribed in vitro from a PCR product containing cre stem-loop RNA or various PV1(M) constructs (26). Unless otherwise indicated either 1 μg of 3CDpro or 0.5 μg of 3Cpro was added to the reaction mixture. After incubation at 34°C for 30 min, the reaction was stopped by the addition of 5 μl of gel loading buffer. The samples were then analyzed on Tris-Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (Bio-Rad) with 13.5% polyacrylamide. Gels were dried without fixing and autoradiographed. The incorporation of [32P]UMP into VPg-pU and VPg-pUpU was measured on a phosphorimager (Molecular Dynamics; Storm 860).
Luciferase assay.
After being transfected with replicon RNA, the monolayer cultures (35-mm-diameter dishes) of HeLa R19 cells were incubated at 37°C in standard tissue culture medium. After 12 h, the growth medium was removed from the dishes, and the cells were washed gently with 2 ml of phosphate-buffered saline. HeLa cell dishes were overlaid with 300 μl of “passive” lysis buffer supplied by Promega (catalog no. E194A) and rocked at room temperature for 15 min, after which the lysate was transferred to a tube. Fifty microliters of luciferase assay reagent (Promega; luciferase assay system catalog no. E1501) was mixed with 20 μl of lysate, and the firefly luciferase activity was measured in an Optocomp I luminometer (MGM Instruments, Inc.).
Electrophoretic mobility shift and filter binding assays.
Assays were performed as described previously with some modifications (10, 39). The radiolabeled RNA cre probes were generated by in vitro transcription in the presence of [32P]UTP by using templates generated through PCR with primers encoding the T7 polymerase promoter. To prevent nonspecific binding of the proteins to the RNA, an excess of nonspecific RNA (10 μg of tRNA) was added to all reaction mixtures. Radiolabeled PV-cre(2C) RNA (7 nM; nt 4445 to 4504; Fig. 2) was added to the binding buffer (5 mM MOPS [pH 7.4], 25 mM KCl, 2 mM MgCl2, 20 mM dithiothreitol) containing either HeLa S10 extract or various amounts of recombinant protein, i.e., His-tagged 3Cpro, 3CDpro with or without the His tag, 3Dpol, 3AB, or PCBP2), and incubated for 10 min at 30°C. Reactions were stopped by the addition of glycerol to a final concentration of 10%, and reaction products were loaded on a native 0.5× Tris-borate-EDTA-5% polyacrylamide (40:1) gel containing 5% glycerol. The cellular extracts (cytoplasmic S10 extract) were prepared as described previously (20). In filter binding assays, the reactions were performed in duplicate. A 20-μl reaction mixture containing various amounts of proteins plus 1 μl of radiolabeled RNA (7 nM) was incubated for 15 min at room temperature, after which aliquots were filtered at low negative pressure through prewetted nitrocellulose membranes (pore size, 0.45 μm; Schleicher & Schuell). Beneath the nitrocellulose membrane a nylon membrane (Boehringer Mannheim) and then a 3MM cellulose membrane were placed (23). Filters were washed once with wash buffer (20 mM HEPES-KOH [pH 6.8], 1 mM magnesium acetate, 10 mM 2-mercaptoethanol) and dried, and the radioactivity was measured by liquid scintillation counting in the presence of EcoLite scintillation cocktail (ICN).
RNA structure prediction.
We used the MFOLD program designed by Michael Zuker (http:/www.bioinfo.rpi.edu/applications/mfold).
RESULTS
Rescue of a replication-incompetent P/L (A5C) replicon with a second copy of cre of different picornavirus origin.
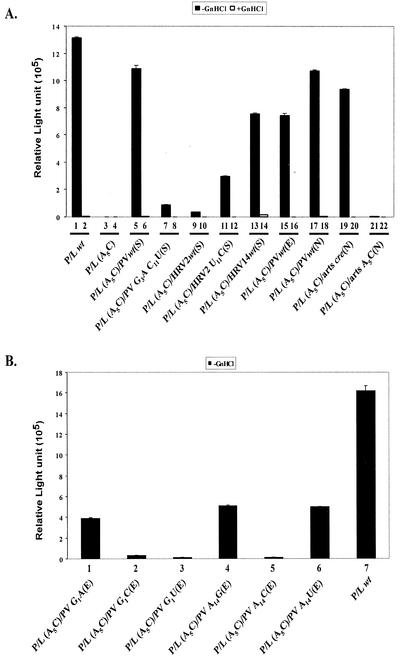
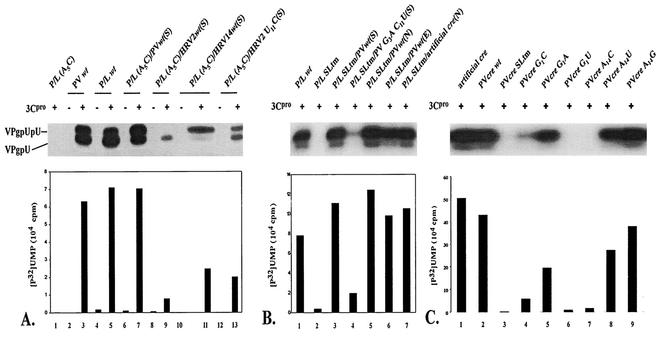
Previously, we demonstrated that the conserved A5A6A7 sequence of the PV-cre(2C) plays a role in the replication of poliovirus RNA (30). For example, the A5C mutation in the loop of PV-cre(2C) rendered the resulting virus nonviable, and no revertants could be recovered even after five blind passages in HeLa R19 cells. In agreement with these findings a P/L (A5C) replicon carrying the same mutation in the endogenous PV-cre(2C) showed little luciferase activity after transfection (see below). The A5C mutation leads to amino acid change K117Q in 2CATPase. Available evidence suggests that this change does not significantly interfere with the 2CATPase activity (29, 30) and that the effect of the mutation is due to the inactivation of the cre signal. To provide experimental evidence for this assumption and to allow for extensive genetic analysis of the cre element, we introduced a second copy of wt PV-cre (63 nt from 4444 to 4506) at different locations (Fig. 1). For example, the wt cre was inserted between the coding sequence for the C terminus of P1/luciferase and that for the N terminus of 2Apro (Fig. 1, SacI site inserts). To facilitate the processing of the modified polyprotein, a 2Apro cleavage site was introduced between the translation product of the new cre and the P1/luciferase so that after translation the extra 21 amino acids encoded by the cre would be cleaved out of the polyprotein. As shown in Fig. 3A, the second copy of wt PV-cre at this position can restore the replication of an inactive P/L (A5C) replicon to that of the wt P/L (Fig. 3A, compare lanes 3 and 5 with lane 1). Furthermore, a dual-cre replicon carrying two mutant copies of A5C cre (Fig. 3A, lanes 21 and 22) failed to show any luciferase activity above the background signal. The background signal was determined by repeating the experiments in the presence of 2 mM guanidine-HCl, an inhibitor of poliovirus RNA synthesis (Fig. 3A, even-numbered lanes). Taken together, the data, in agreement with previous results, suggest that (i) the 63-nt PV-cre is a functional signal for the replication of poliovirus RNA in vivo (9, 26, 30), (ii) the function of PV-cre is independent of the cognate amino acid sequence of the viral 2CATPase protein, and (iii) a functional cre element can be moved to a different location within the poliovirus genome (9).
FIG. 3.
Luciferase activities of various P/L replicons as a measure of RNA replication. (A) RNAs from dual-cre P/L replicons with second rescuer cre elements derived from poliovirus, HRV2, or HRV14. Transfection of RNA and luciferase activity measurements are described in Materials and Methods. Even-numbered lanes, transfections repeated in the presence of 2 mM guanidine-HCl (GnHCl). arts, artificial. (B) Dual-cre luciferase replicons with the rescuer cre from poliovirus carrying point mutations in G1, A2, and A14 in the core cre sequence were tested for their ability to replicate in HeLa R19 cells. The nomenclature is as follows. P/L refers to the parental replicon plasmid followed by A5C when it carries an inactivating mutation in the 2CATPase coding sequence. This is followed by the sequences of the rescuer cre and the insertion site (N, NheI; E, EcoRI; S, SacI).
We have recently shown that cre elements from HRV14 and poliovirus can be functionally exchanged in our in vitro uridylylation assays using poliovirus 3Dpol (26). Therefore, we were interested in determining whether heterologous cre elements could rescue the replication of a P/L (A5C) replicon carrying a lethal mutation in the native PV-cre(2C) by engineering the foreign cre elements into dual-cre RNAs. As shown in Fig. 3A (lane 13) HRV14-cre restored the replication level of this replicon to 70% of that of wt dual-cre (lane 5). HRV2-cre, on the other hand, barely (3%) rescued the replication of the dual-cre replicon (Fig. 3A, compare lanes 5 and 9). This is in agreement with our findings that HRV2-cre(2A) is a very poor template for poliovirus 3Dpol in VPg uridylylation (A. V. Paul and E. Wimmer, unpublished data).
Rescue by genetically altered PV-cre(2C), HRV2-cre(2A), or artificial cre's.
As shown in Fig. 2A, PV-, HRV2-, and HRV14-cre differ significantly from each other in their primary nucleotide sequences. However, we have highlighted a consensus sequence of 14 nt (named core in this paper; Fig. 2B) that is highly conserved among all cre structures. Curiously, although HRV2-cre appears to be more similar to PV-cre than to HRV14-cre in its core sequence, HRV14-cre was much more efficient in rescuing a debilitated (A5C) dual-cre replicon than HRV2-cre (Fig. 3A, compare lane 5 with lanes 9 and 13). A major difference between the upper portions of HRV2-cre and HRV14-cre is the size of the loop. We were interested to test the hypothesis that a predicted “tight” structure in the core sequence of HRV2-cre that leads to base pairing of A5 prevented it from being an efficient rescuer in the context of the poliovirus genome. Therefore, a point mutation (U11C) was engineered into the core sequence of wt HRV2-cre so that the predicted secondary structure for the altered HRV2-cre resembles that of the HRV14-cre top loop. Indeed, the altered HRV2-cre was about 10-fold more efficient in rescuing a debilitated (A5C) PV-cre than the wt HRV2-cre (Fig. 3A, compare lane 5 with lanes 9 and 11). The reason why the recovery of luciferase activity is still low compared to that for PV-cre or HRV14-cre is not known but is undoubtedly related to the cre structure and its interaction with the proteins involved in uridylylation. Perhaps the presence of an unpaired U residue in the upper region of the stem is responsible for the low activity in the context of a poliovirus genome (30). In some cases, a relationship between higher-order structure and function can be predicted. For example, a mutant PV-cre carrying two point mutations (G3A and C11U) in its core sequence has a predicted loop structure identical to that of wt HRV2-cre. Indeed, this variant PV-cre produced only 8% luciferase activity when used as a rescuer cre in the dual-cre replicon (Fig. 3A; compare lanes 5 and 7).
The core sequences of PV-, HRV2-, and HRV14-cre are relatively more conserved than the rest of the hairpin sequence (Fig. 2B). Along with the A5A6A7 sequence, the first two (G1 and A2) nucleotides and the last (A14) nucleotide were among the most conserved. Mutations A2C and A2U did not influence the ability of the second cre element to rescue a lethal mutation in the cognate cre (data not shown). On the other hand, some G1 and A14 mutants [P/L (A5C)/PV G1A(E), P/L (A5C)/PV A14G(E), and P/L (A5C)/PV A14U(E)] displayed significant reduction in the level of replication compared to wt P/L (Fig. 3B, compare lane 7 with lanes 1, 4, and 6). Interestingly, other G1 mutants [P/L (A5C)/PV G1C(E), P/L (A5C)/PV G1U(E), and A14(P/L) A5C/PV A14C(E)] exhibited very little if any replication signal (Fig. 3B, compare lane 7 with lanes 2, 3, and 5). Currently, we have no explanation for the remarkable intolerance for transversions at the G1 position, but it is likely related to the reduced ability of this mutant to bind 3CDpro (see Fig. 9C). A possible explanation for the relatively high activity of the A14U replicon (lane 6), compared to that of the A14C replicon (lane 5) will be given in Discussion.
FIG. 9.
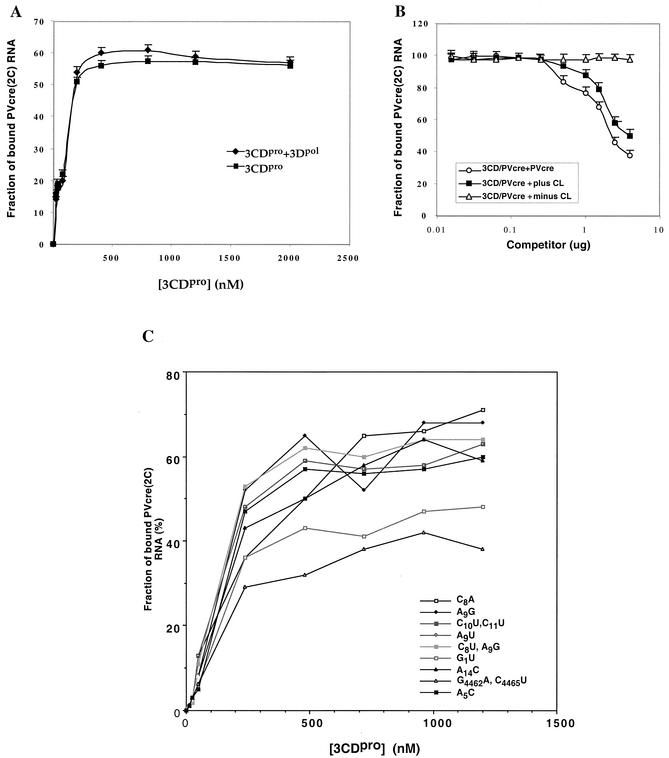
Filter binding assays to assess the effect of addition of 3Dpol or competitor (mutant PV-cre or cloverleaf) RNAs on the binding of 3CDpro to PV-cre(2C). (A) Binding reaction between PV-cre(2C) (63 nt, 7 nM) and 3CDpro (0.01 to 0.8 μM, untagged) performed in the absence (squares) or in the presence (diamonds) of 1 μM 3Dpol. 3CDpro used in this experiment and that described in the panel B legend is a recombinant (3Cpro/H40A) protein that does not carry additional tag sequences (23). The filter binding data collected are shown as the means of three measurements at each data point. (B) Effect of plus and minus strand poliovirus cloverleaf and wt PV-cre on the binding of 3CDpro to PV-cre. Competition was performed in the presence of either PV-cre (circles) or plus strand cloverleaf (squares) or minus strand cloverleaf (triangles) RNA competitors. Quantitative analysis of the 3CDpro/PV-cre complexes was carried out by using a filter-binding assay as described in Materials and Methods. (C) Comparison of the relative binding affinities of wt and mutant PV-cre RNAs for the site within 3CDpro in a filter-binding assay as described in Materials and Methods.
The stem structures beneath the core sequence display the biggest diversity among the cre structures studied. To evaluate the role of sequences located in the stem region beneath the core sequence, we constructed an artificial cre consisting of an authentic PV-cre core (G4468AGCAAACACCGUA4481) sequence and an artificial stem of unrelated nucleotide sequence (Fig. 2A). This stem was designed to accommodate an NheI restriction site for easy insertion between the cloverleaf structure and the IRES element in the 5′ NTR (Fig. 1) and will be discussed below. As seen in Fig. 3A, this artificial cre, when inserted into the P/L (A5C) replicon genome, efficiently rescued the lethal phenotype caused by an A5C mutation in the native PV-cre. This observation suggests that the structure (i.e., scaffolding) of the stem beneath the core sequence of various cres is more important than its nucleotide sequence.
Rescue of replication by wt PV-cre(2C) placed at different locations in the P/L replicon.
To determine if the genomic position of PV-cre affects its role in poliovirus replication, we tested the effect of a rescuing a cre element at three locations in the dual-cre replicon genome: inside the polyprotein open reading frame [ORF; P/L (A5C)/PVwt(S)], in the beginning of the ORF [P/L (A5C)/PVwt(E)], and in the noncoding region in the 5′ NTR between the cloverleaf and the IRES element [P/L (A5C)/PVwt(N)]. The result showed that the location of the cre elements has very little effect on the replication of the genome (Fig. 3A, compare lane 1 with lanes 5, 15, and 17).
The efficient rescuing function of the poliovirus cre element outside the ORF was surprising. As mentioned above, even an artificial cre element inserted into the NheI site allowed replication of the RNA carrying a lethal A5C mutation in the cognate cre element (Fig. 3A, compare lanes 17 and 19). In contrast, an artificial cre with an A5C mutation in the core sequence did not rescue replication (Fig. 3A, compare lanes 17 and 21), an observation proving that RNA synthesis strictly depended on a functional cre, regardless of its locus in the genome.
In vitro uridylylation of VPg using dual-cre poliovirus RNAs as templates.
We have previously developed an in vitro assay in which the viral RNA-dependent RNA polymerase 3Dpol transfers a UMP to the tyrosine residue in the small viral protein VPg, by using poly(A) as the template (27). We further made the discovery that such a reaction was stimulated (by 1 order of magnitude) by the presence of viral protein 3CDpro when genome RNA or the cre sequence was used as the template (26, 30). Enhancement of uridylylation was observed also with 3Cpro (23). Since the stimulatory activity of 3Cpro is more reproducible than that of the much larger (His-tagged) 3CDpro, we used 3Cpro in uridylylation reactions catalyzed by dual-cre replicon RNAs. As shown in Fig. 4A, in the absence of 3Cpro, there was little uridylylation of VPg by 3Dpol when any of the different RNAs were used as the template (Fig. 4A, lanes 2, 4, 6, 8, 10, and 12). The P/L (A5C) replicon RNA carrying the lethal A5C mutation in cre(2C) failed to function as a template for the uridylylation of VPg (Fig. 4A, lane 1), as expected (30). In the presence of 3Cpro, only RNAs with a functional cre were able to direct significant uridylylation of VPg above the background level (Fig. 4A, lanes 3, 5, 7, 9, 11, and 13). This is in good agreement with the observed replication levels of these replicons in tissue culture, as determined by measuring luciferase activity (Fig. 3A, lanes 1, 5, 9, 11, and 13). This was also observed for the mutated HRV2-cre (U11C), which is a better template for uridylylation of VPg in vitro than the dual-cre replicon carrying wt HRV2-cre(2A) (Fig. 4A, compare lanes 9 and 13). Reciprocally, when the loop (14 nt) of PV-cre(2C) was converted to that of HRV2-cre(2A), the hybrid cre (PV G3A U11C) not only displayed a much-reduced ability to replicate (Fig. 3A, lane 7) but also was a poor template for the uridylylation of VPg in vitro (Fig. 4B, lane 4).
FIG. 4.
In vitro uridylylation of VPg using either a dual-cre P/L replicon or mutated PV-cre RNAs as the template. Uridylylation reactions were performed as described in Materials and Methods. (A) Dual-cre replicons carrying a lethal (A5C) mutation in the 2CATPase coding sequence with a rescuer cre from HRV14 or HRV2 or a mutant U11C HRV2-cre (predicted by the Zuker program to enlarge the terminal loop of HRV2-cre to resemble that of HRV14-cre). (B) VPg uridylylation using inactivated dual-cre (SLtm; triple mutant) replicons as templates and the rescuer wt PV-cre inserted at different locations. (C) Mutant or wt cre RNAs as templates in VPg uridylylation.
The different location of the rescuer copy of PV-cre(2C) did not influence significantly its template activity for the uridylylation of VPg in vitro, as shown in Fig. 4B. The dual-cre replicons that displayed wt P/L luciferase activity (Fig. 3A, lanes 1, 5, 15, and 17) in HeLa cells also exhibited VPg uridylylation levels similar to that of the wt construct (Fig. 4B, lanes 1, 3, 5, and 6). This further suggests that the cre(2C) RNA sequence functions as a replication signal independently of the function of the viral 2CATPase and local RNA-RNA interactions. Not surprisingly, the dual-cre replicon carrying the artificial cre as the rescuer is a good template for both the uridylylation of VPg in vitro (Fig. 4B, lane 7) and replication in HeLa R19 cells (Fig. 3A, lane 19).
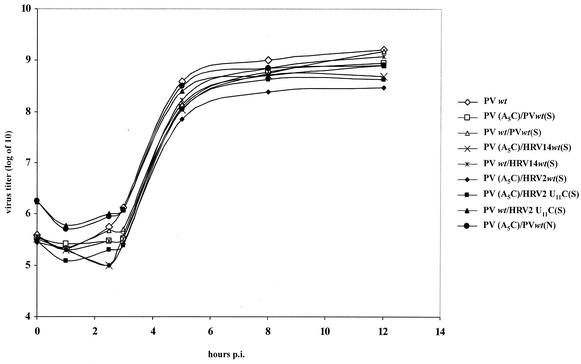
One-step growth curve of dual-cre polioviruses.
To examine to what extent rescue by cognate or foreign cres influences virus yield, we tested virus production with genomes carrying a lethal A5C mutation in PV-cre(2C) as well as a second cre (PV-cre, HRV2-cre, HRV14-cre, or HRV2-cre U11C) element. Two locations of the second cre were used for these experiments: (i) between the P1 and P2 coding sequences and (ii) between the cloverleaf and the IRES element in the 5′ NTR. As shown in Fig. 5, the dual-cre viruses with a second copy of functional viral cre displayed similar growth kinetics. These results suggest that, in vivo, the uridylylation of VPg might not be the rate-limiting step of poliovirus RNA replication. Note that HRV2-cre, a weaker template for in vitro VPg uridylylation by poliovirus 3Dpol (Fig. 4A, lane 9), still stimulated the dual-cre virus to amplify by over 2 orders of magnitude at 12 h postinfection (Fig. 5). The results also suggested that the location of the rescuer cre has little effect on the recovery of virus, as PV (A5C)/PVwt(N) has growth kinetics similar to those of PV (A5C)/PVwt(S) and both proliferated at a rate comparable to that for wt PV (Fig. 5).
FIG. 5.
One-step growth curves for various dual-cre polioviruses. HeLa R19 cells were infected with virus derived from wt and mutant poliovirus cDNA plasmids, and the virus yield was determined at the indicated time points (see Materials and Methods).
To exclude the possibility that the insertion of a second cre impairs replication of the resulting virus, we also inserted the above cre elements into the wt poliovirus genome and studied the resulting viruses in one-step growth experiments. As shown in Fig. 5, a dual-cre virus carrying wt cre in the 2C coding region grew just as well as wt poliovirus, regardless of the content of the second cre, and slightly better than the dual-cre viruses carrying the A5C mutation.
The growth phenotypes of the constructs carrying mutations in the cre element could also be due to rapid mutation to wt sequences or second-site suppressor mutations. Sequence analysis of the genomic RNAs recovered after two passages in HeLa cells did not reveal changes in the cre sequences. This result excludes direct reversion, but it does not eliminate second-site suppressor mutations. This interesting possibility is currently under investigation.
In vitro uridylylation of VPg using PV-cre G1 and A14 mutants and artificial cre RNAs as templates.
We were also interested in the ability of PV-cre's to direct VPg uridylylation when either the G1 or A14 nucleotides were altered. As shown in Fig. 4C, transversions at these positions caused a strong defect in the protein priming reaction. Mutant PV-cres carrying either G1C or G1U or A14C mutations in the core cre sequence lost more than 90% of VPg uridylylation activity compared to the wt PV-cre (Fig. 4C, compare lane 2 with lanes 4, 6, and 7), whereas PV-cre A14U is about 50% as functional as wt PV-cre (Fig. 4C, compare lanes 8 and 2). On the other hand, transitions are better tolerated at both nucleotide positions: PV-cre G1A and PV-cre A14G retained 50 and 90% of wt PV-cre activity for the uridylylation of VPg in vitro, respectively (Fig. 4C, compare lane 2 with lanes 5 and 9). As a negative control, mutant PV (SLtm) (Fig. 2) was constructed to carry the debilitating A5C change and two point mutations in the upper stem region (G4462A and C4465U); these three mutations are predicted to alter the secondary structure of cre drastically. As expected, PV (SLtm) showed only a background level signal for the in vitro uridylylation of VPg (Fig. 4C, lane 3), which is in accordance with previous findings with full-length viral RNA and poliovirus replicons (9, 30). These results suggest a preference for purine at the N1 and N14 nucleotide positions in the core sequence of PV-cre(2C), specifically a G1 and an A14.
We also tested our artificial cre, carrying the wt core sequence from PV-cre(2C) and a stem of unrelated nucleotide sequence, in the in vitro VPg uridylylation assay. As shown in Fig. 4C (compare lanes 1 and 2), the artificial cre is as functional as the wt PV-cre for directing the synthesis of VPg-pU and VPg-pUpU under the experimental conditions, in agreement with our data for the dual-cre replicon shown in Fig. 3A (lane 19).
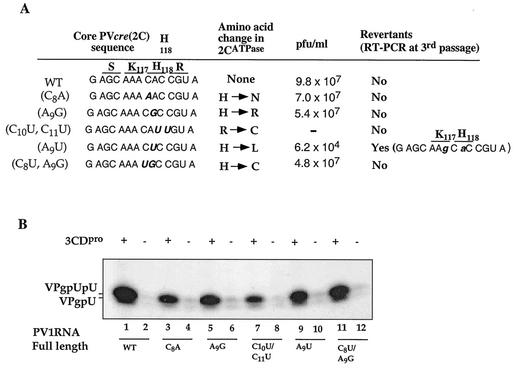
Analysis of full-length poliovirus genomes carrying mutations within the loop of PV-cre(2C).
Further characterization of the effect of mutations within the core PV-cre sequence was carried out by using mutant full-length polioviruses. The growth phenotype and uridylylation activities of these full-length constructs are shown in Fig. 6A and B, respectively. Similar to the wt, C8A, A9G, and C8U A9G mutants developed complete cytopathic effect (CPE) after 24 h of incubation at 37°C posttransfection. A delay in the appearance of CPE was observed with the A9U mutant, an observation suggesting that this mutant RNA may be impaired in replication. We suspected that the inefficient replication may lead to a reversion or other genetic alterations. This was confirmed by sequence analysis (reverse transcription-PCR) of RNA extracted from the virus that was collected after three passages following the original transfection. It reveals a reversion at the site of mutagenesis (U9A) to the original codon (resulting in a change from Leu118 to His). In addition, we detected a change at position A7G in the core sequence (synonymous mutation). No CPE was detected with C10U C11U mutant RNA even after three additional blind passages in HeLa R19 cells. This result is not unexpected since the substitutions introduced into this double mutant are likely to lead to base pairing with the critical A5 and A6 nucleotides in the core cre sequence. This is supported only by computer-aided folding; no biochemical evidence has been generated. The C10U C11U mutation also greatly reduces the yield of VPg-pU and VPg-pUpU products in in vitro uridylylation (Fig. 6B). The results are also consistent with the data from dual-cre replicons [P/L (A5C)/PV G3A C11U(S)] shown in Fig. 3A, where a mutation in the PV-cre loop which reduces the size of the terminal loop reduced luciferase activity 10-fold relative to that for the P/L carrying wt PV-cre. However, since the double mutation changes the amino acid sequence from H118R119 to H118C119, we cannot rule out the possibility that this may critically impair 2CATPase function. The C10U and C11U mutations have not yet been tested in a dual-cre construct.
FIG. 6.
Summary of the effect of nucleotide substitutions within the PV-cre(2C) loop in full-length PV1(M) genome RNA on genome replication and uridylylation. (A) Phenotypic properties of viruses derived from transfection experiments with HeLa R19 cells. Following transfection into HeLa cells the cell lysates were subjected to three additional passages and the viral RNAs were sequenced following reverse transcription-PCR, as described in Materials and Methods. The letters above the nucleotides indicate the deduced amino acid sequence and its location within 2CATPase. Mutations or reversions in nucleotides are in boldface and italics. Nucleotide reversions are shown with lowercase letters. (B) VPg uridylylation was assayed as described in Materials and Methods with full-length wt or mutant RNAs as templates. Where indicated, poliovirus 3CDpro was added to the reaction mixtures.
All mutant versions of the full-length RNAs tested above are functional templates in the in vitro VPg uridylylation reaction (Fig. 6B). However, smaller amounts of VPg-pU and VPg-pUpU products were produced than were produced with wt PV1 RNA (Fig. 6B, compare lane 1 with lanes 3, 5, 7, 9, and 11). Among them, only one (C10U C11U mutant; 10% of wt activity in uridylylation; Fig. 6B, compare lane 1 with lane 7) rendered the genome nonviable. Polypeptide 3CDpro significantly stimulated uridylylation in all cases, as its absence greatly reduced synthesis of the VPg-pU and VPg-pUpU products (Fig. 6B, lanes 2, 4, 6, 8, 10, and 12). The existence of viable mutants containing amino acid changes at His118 in 2CATPase suggests that this His residue is not essential for 2CATPase function in the replication of poliovirus (Fig. 6A). However, whereas the His118Arg change was tolerated, a the His118Leu change produced a replication phenotype that led to reversion to the original codon and amino acid. Since this reversion is accompanied by a synonymous change in the preceding codon, it is impossible to decide whether the growth phenotype of the A9U mutant resulted from the amino acid change, from the structure of the cre, or both. Finally, a G1U mutation in cognate PV-cre(2C) of poliovirus RNA rendered the genome nonviable, even after three blind passages in HeLa cells (data not shown). Taken together, the data support the hypothesis that critical features of the cre required for replication are contained in a 14-nt core sequence in the apical portion of the RNA stem-loop.
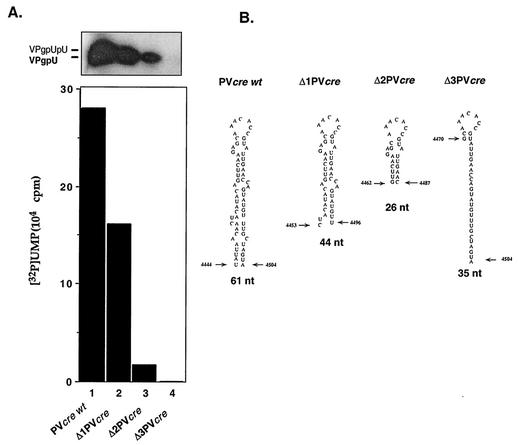
Minimal length of PV-cre(2C) required for uridylylation.
We have performed experiments to analyze the relationship between the length of the stem in PV-cre(2C) and VPg uridylylation. Two truncated PV-cre RNAs were synthesized in vitro and tested as templates in the in vitro uridylylation assays. As shown in Fig. 7, both RNAs were able to serve as templates, but the yield was reduced to 70 to 80% of that with the wt PV-cre [Δ1 PV-cre(2C)] and to 25% of that with Δ2 PV-cre(2C) (compare lane 1 with lanes 2 and 3). The data indicate that the upper stem and loop of PV-cre(2C) carry the minimal structural determinants for the assembly of a uridylylation-competent complex. On the other hand, an RNA spanning the sequence 4470 to 4504 [Δ3 PV-cre(2C)] of the poliovirus genome is totally inactive as a template in the reaction (Fig. 7, compare lanes 1 and 4), suggesting that the sequence determinants contained in this region must be contained within a certain spatial arrangement to be recognized by the proteins of the uridylylation complex.
FIG. 7.
In vitro uridylylation using shortened PV-cre(2C) RNAs as templates. (A) Effect of truncation on the ability of PV-cre(2C) RNA to serve as the template in VPg uridylylation. The uridylylation of VPg was measured by the standard method described in Materials and Methods. The yield of the reaction (graph) was shown by autoradiography. (B) Schematic drawing of the secondary structure of complete and truncated PV-cre RNAs utilized in this experiment. The first and last nucleotides as well as the length of each RNA template are indicated.
Proteins binding to the poliovirus oriI [PV-cre(2C)].
The function of PV-cre(2C) is likely to involve an RNP complex consisting of more than one protein. As shown previously, PV-cre(2C) serves as an efficient template for 3Dpol-catalyzed VPg uridylylation only if 3CDpro or 3Cpro is present (23, 26) (see our results above). We therefore used a binding assay to test interactions between viral and possibly cellular polypeptides and PV-cre(2C).
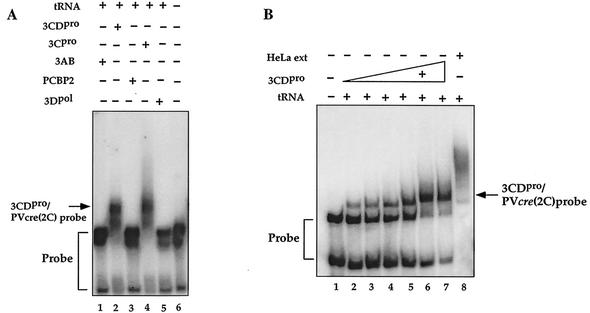
We first investigated if nonstructural proteins that originated from the P3 region of the polyprotein (3CDpro, 3AB, and 3Dpol) and the cellular protein PCBP2 possess specific affinity to PV-cre(2C) by gel shift assays, carried out in the presence of a 1,000-fold excess of nonspecific competitor tRNA (10). Purified polypeptide 3AB, known to be a nonspecific RNA-binding protein (25, 38), does not bind to PV-cre(2C) under these conditions (Fig. 8A, compare lane 1 with lane 6). In contrast, 3CDpro and its cleavage product 3Cpro retarded the mobility of the labeled probe in the presence of the 1,000-fold excess of tRNA (Fig. 8A, lanes 2 and 4, respectively). Gel shift assays carried out with increasing amounts of 3CDpro-His showed that the amount of RNP complex formed is dependent on the concentration of the protein in the reaction mixture up to about 400 nM 3CDpro (Fig. 8B, lanes 2 to 7, and 9A). This is true regardless of whether the recombinant 3CDpro is attached to a His tag at the C terminus (Fig. 8) or is free of any extra tag sequence (Fig. 9A). The formation of an RNP complex between 3CDpro and PV-cre(2C) could be competed by the addition of a 200-fold excess of nonlabeled PV-cre(2C) RNA (Fig. 8C). Moreover, this RNP complex is similarly susceptible to competition by the addition of an unlabeled plus sense cloverleaf RNA but not by its cRNA sequence (referred to as a minus sense cloverleaf; Fig. 9B). This competition experiment supports our claim of specificity since plus strand cloverleaf RNA is known to bind 3CDpro (4, 10, 41). As expected, a 100-fold excess of domain II RNA of the poliovirus IRES, a hairpin containing an AAACCA sequence in the loop region, did not compete in 3CDpro/PV-cre(2C) complex formation (data not shown).
FIG. 8.
Relative abilities of poliovirus and HeLa cell proteins to form complexes with PV-cre(2C) RNA. The electrophoretic mobility shift assays were carried out with a 32P-labeled PV-cre(2C) probe in the presence of a 1,000-fold excess of tRNA as described in Materials and Methods. (A) Lane 6, radiolabeled PV-cre (63-nt, 7 nM) probe in binding buffer; lane 1, 3AB (0.8 μM); lane 2, 3CDpro (His tagged, 0.5 μM); lane 3, PCBP (1.1 μM); lane 4, 3Cpro (1.2 μM); lane 5, 3Dpol (1 μM). (B) Binding curve for PV-cre/3CDpro with increasing amounts of 3CDpro. Lane 1, PV-cre probe in binding buffer; lanes 2 to 7, probe in binding buffer with increasing amounts (0.05, 0.13, 0.2, 0.4, 0.5, and 0.6 μM) of 3CDpro (His tagged); lane 8, PV-cre probe in binding buffer and HeLa cell extract proteins (15 μg). (C) Competition for the binding of 3CDpro to the radiolabeled PV-cre probe (63 nt, 7 nM) with unlabeled PV-cre. Binding reactions were performed in the absence (lane 1) or in the presence (lanes 2 to 7) of recombinant 3CDpro (His tagged). Lanes 3 to 7 contain radiolabeled PV-cre probes in binding buffer, purified 3CDpro (0.5 μM; His tagged), and increasing concentrations (from 10 to 500 M excess) of unlabeled PV-cre RNA as the competitor.
As 3AB did not bind to PV-cre(2C), it also did not supershift a 3CDpro/PV-cre(2C) complex (data not shown). This is in contrast to the formation of a 3AB/3CDpro/cloverleaf complex, which we have shown previously to be essential for plus strand RNA synthesis (10, 38, 39). Similarly, the cellular RNA binding protein PCBP2 can form a complex with 3CDpro and the 5′ cloverleaf (7, 21). Just like 3AB, purified PCBP2 did not bind PV-cre(2C) RNA (Fig. 8A, lane 3), nor did it supershift the 3CDpro/PV-cre(2C) complex (data not shown). These results are consistent with our previous report showing that the addition of purified 3AB or PCBP2 had no effect on the uridylylation of VPg in the presence of 3Dpol, 3CDpro, and PV-cre(2C) (26). It is likely therefore, that 3AB and PCBP2 do not play a direct role in VPg uridylylation.
The viral RNA polymerase 3Dpol did not bind PV-cre(2C) RNA under the conditions of the gel shift assay (Fig. 8A, lane 5) and was also unable to influence the formation of the 3CDpro/PV-cre(2C) complex (Fig. 9A) in filter binding experiments. Notably, 1 μg of 3Dpol, the amount of protein normally used in uridylylation experiments, did not affect the dose-response curve resulting from the binding of increasing amounts of 3CDpro to the PV-cre probe (Fig. 9A).
Components of an S10 HeLa cell extract formed one or more RNP complexes with PV-cre(2C), even in the presence of competitor tRNA (Fig. 8B, lane 8). Preliminary data suggested that a 50-kDa protein present in HeLa cell extracts is the major component that binds PV-cre(2C) (data not shown). However, early studies have indicated that the addition of an extract of uninfected HeLa cells did not stimulate the uridylylation reaction under the condition of the experiments (26). Currently we do not know the identity of the cellular protein, nor do we know its function.
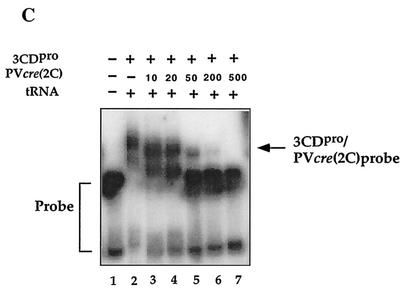
To test whether mutations in the loop or stem of PV-cre(2C) affect the binding to 3CDpro, we performed filter-binding assays with selected mutant PV-cre RNA probes. As shown in Fig. 9C, use of C8A, A9G, C10U C11U, A9U, C8U A9G, or A5C loop mutant RNA or A14C stem mutant RNA resulted in the retention of amounts of 3CDpro on the nitrocellulose membrane similar to that resulting from the use of the wt PV-cre probe. Interestingly, quantification of the binding of the PV-cre G1U and G4462A C4465U (mut1 in reference 30) mutant RNAs revealed an 8- to 15-fold reduction in binding to 3CDpro, as judged by a decreased amount of RNP complex retained on the nitrocellulose membranes. The diminution in binding of PV-cre G1U and G4462A C4465U RNAs by 3CDpro is in agreement with our uridylylation studies and the phenotypic properties of either the full-length viral genome (G4462A C4465U) (30) or a replicon RNA (G1U) (Fig. 3B).
DISCUSSION
Originally, the highly structured 5′ and 3′ NTR sequences of entero- and rhinovirus genomes were considered necessary and sufficient for the initiation of RNA replication (1). The discovery of internal cre elements, first described by McKnight and Lemon (18) for HRV14, which are also essential for viral RNA replication, came as a surprise. Similarly surprising was the subsequent observation that these elements can direct 3Dpol-catalyzed uridylylation of VPg, thereby providing the primer for the initiation of RNA synthesis (8, 26, 30). A suggestion that the poliovirus cloverleaf is involved in VPg uridylylation was provided in a recent report by Lyons et al. (16). This finding, together with the recent model of the circularization of the poliovirus genome prior to RNA synthesis (11), suggests that at least three RNA domains (5′-terminal cloverleaf, cre, and 3′-terminal poly[A]) must interact with viral and cellular polypeptides to orchestrate the initiation of minus strand synthesis. The details of these events are not yet understood.
We have focused on the cre elements of poliovirus, HRV14, and HRV2 to decipher structural parameters by genetic and biochemical analyses. With the exception of those of foot-and-mouth disease virus (17), all cre elements described so far are located in the ORFs encoding components of polyproteins (8, 9, 15, 18). Genetic analysis requires site-directed mutagenesis of the cognate PV-cre(2C) without destroying 2CATPase function, a strategy that is severely limited. Therefore, in this study, the cognate cre was inactivated by a single point mutation (A5C) and a second cre was inserted to rescue genome replication. Since McKnight and Lemon (18) and Goodfellow et al. (9) had shown previously that HRV14-cre and PV3-cre, respectively, could be moved from their original locations, our strategy was likely to work. Indeed, the data presented here show that a second cre element, even if it was derived from a different picornavirus, was efficient in rescuing replication. As any genetic alteration at the second cre does not complicate the proper processing and functioning of viral proteins, the dual-cre system was suitable for genetic analyses. Remarkably, the location of the second cre had no detectable effect on its rescuing function, even when it mapped to the region between the cloverleaf and the IRES.
Our genetic and biochemical analyses have led to the designation of a core sequence (Fig. 2B) essential for cre function. A similar core sequence has been proposed very recently also by Yang et al. (40) based on an analysis of HRV14-cre. Mutations within the conserved 14-nt core sequence (Fig. 2) produce phenotypes that suggest that several purines within this core sequence, i.e., G1, A5, A6, and A14, are necessary for cre function. Interestingly, transition mutations in the loop (C10U and C11U) that may lead to base pairing with A5 and A6 abolished cre function in uridylylation and rendered the genome nonviable even after four passages. This observation indicates that a degree of structural integrity in the core sequence is essential for its function. The preferred nucleotide in position 1 of the core is the purine G, which is highly sensitive to base pairing since a mutant carrying an A14C transversion reduces the luciferase signal dramatically in the dual-cre replicon system and nearly abolishes cre-dependent uridylylation of VPg.
Although PV-cres with mutation in G1 or A14 of the core sequence produced a reduction in cre function when used as a rescuer in the dual-cre replicon system, the role of G1 appears to be more important than that of A14. Transversions are not tolerated at the G1 position, whereas the A14U mutant is still active in VPg uridylylation in vitro and in the rescue function in vivo. Initially, we were surprised about the functional activity of the A14U mutant since either a C or a U at nt 14 of the core sequence could base pair with G1. However, since U14 has the option to base pair with A2 and since A2 is not essential for cre function (J. Yin and E. Wimmer, unpublished data), an A2::U14 base pairing may not be debilitating. Perhaps the observed conservation of an A at position 14 is solely due to the fact that this A is the least favorable nucleotide to base pair with G1.
Experiments with truncated PV-cre elements (Fig. 7) support our previous finding that important structural determinants for cre function are contained in the upper stem and loop region. Δ2 PV-cre(2C) RNA, consisting of only 26 nt, can still function as a template for VPg uridylylation although the template activity is greatly reduced. The artificial cre has even fewer virus-specific nucleotides, yet it is not only an excellent template for uridylylation in vitro but also an efficient rescue element in vivo (Fig. 3 and 4). These results suggest that the sequences contained in the middle and bottom regions of the cre stem might be required only for proper structural presentation of the core sequence. This conclusion is in agreement with evidence derived from mutational analyses of cre elements of poliovirus and HRVs (8, 9, 18, 19, 30, 40).
The flexibility by which the poliovirus genome tolerates different locations of the cre element is surprising. Even when placed between two highly structured entities, the cloverleaf and the IRES, nearly wt function was retained. These results suggest that sequences in the vicinity of the cognate cre play little if any role in cre function. Mason et al. (17) have recently shown that the cre element of foot-and-mouth disease virus, an aphthovirus, is located in the 5′-terminal region of its genome and could be moved to the 3′ NTR. This result, together with the other known locations of cre elements in entero-, rhino-, and cardiovirus genomes, supports the notion that cre-mediated uridylylation is location independent.
We have previously reported that cre RNAs of HRV14 and HRV2 can serve as templates in the uridylylation of poliovirus VPg in vitro in the presence of poliovirus 3Dpol and 3CDpro (8, 26). We now show that these heterologous cre elements are capable also of rescuing RNA synthesis in dual-cre genomes of replicons in vivo. The efficiency of the rescuing ability covaries with the efficiency of in vitro uridylylation: HRV14-cre is far superior to HRV2-cre (Fig. 3A). Of the three cres that we have analyzed, that of HRV2 has the smallest loop domain. Whereas this configuration must be optimal for HRV2 replication, it is suboptimal for poliovirus polypeptides such as VPg, 3Dpol, and 3CDpro that must recognize it. It appears that the poliovirus polypeptides, as well as those of HRV14, prefer a larger loop since a U11C mutation in HRV2-cre enhances significantly both the rescue function and template function in uridylylation.
Our dual-cre viruses showed less-than-expected differences when analyzed in one-step growth curve experiments. Compared to the results from in vitro uridylylation assays using dual-cre P/L replicon RNAs, the differences in viral growth kinetics in vivo are far less pronounced. The reason for this unexpected result is unknown. One possible explanation is that in infected cells VPg-pUpU is synthesized in excess of what is needed to produce progeny RNAs. Another possibility is that reversions in the mutated PV-cre(2C) emerge rapidly to obscure the original replication phenotype. However, we have sequenced both cre regions in plaque-purified dual-cre viruses isolated after the second passage and have found that the A5C mutation in the 2CATPase locus was always retained. However, in two out of four progeny viruses that were plaque purified after two passages of PV (A5C)/HRV2-wt(S) virus, we found an identical U11C transition in HRV2-cre, a mutation that enhances HRV2-cre function in the context of the poliovirus genome. In any event, we cannot exclude the possibility that some of the isolates after the second passage harbor second-site suppressor mutations.
Nothing has been known about the mechanism of interaction between viral polypeptides and cre elements. Equally obscure is the possible participation of cellular polypeptides in uridylylation. For this reason, we have conducted protein binding studies using gel mobility shift and filter binding assays. We found that 3CDpro and its cleavage product 3Cpro, but not 3Dpol, recognize PV-cre(2C) with high specificity. Other RNA binding proteins known to be involved in poliovirus RNA replication, e.g., 3AB and PCBP2, did not display affinity to cre under the conditions of the experiment. Available evidence suggests that it is the 3Cpro moiety of 3CDpol that is responsible for the binding of the polypeptide to cre (26). Moreover, since 3Cpro can stimulate uridylylation, first reported by Pathak et al. (23) and confirmed here, 3Dpol may directly bind to an as yet undefined region in 3Cpro (23). Preliminary experiments support this hypothesis (J. Yin, E. Wimmer, and E. Rieder, unpublished results). On the other hand, no binding between 3Dpol and 3Cpro was observed in a yeast two-hybrid system (37), which may be a false-negative result.
We have not yet been able to determine those nucleotides in PV-cre(2C) making contact with 3CDpro or 3Cpro. Our data suggest that recognition of the RNA may involve more than 1 nt located in the upper bulge and upper terminal stem. This hypothesis is based on the observation that the G1U (G4468U and G4462A C4465U) mutants were less efficient not only in replication but also in the formation of the RNP complex. In contrast, all mutants with mutations in the terminal loop (including the one altering the first critical A5C nucleotide) bind to 3CDpro or 3Cpro with efficiencies similar to that of wt PV-cre. Whether the formation of an RNP complex stabilizes the configuration of cre or whether this event is necessary for putative structural changes in the 3Cpro moiety for 3Dpol/VPg recruitment remains to be seen.
All available evidence suggests that the first step in poliovirus RNA synthesis is cre-dependent uridylylation of VPg by 3Dpol, yielding the primer for minus strand synthesis. Because of its internal location, Paul has designated the function of the cre locus oriI (24). The observation that cre elements can be moved to different genome loci supports this designation. However, the mechanism by which the VPg-pUpU/3D/3CD initiation complex is subsequently moved to the 3′ terminus for the initiation of minus strand RNA synthesis remains a puzzle. It has been proposed that the initiation of minus strand synthesis requires the poliovirus genome to circularize (5, 11). As has been proposed by Paul (24) and Pathak et al. (23), we suggest that an initiation complex formed at any oriI locus within a circular RNA could bind to the 3-terminal poly(A) by template displacement in cis.
Acknowledgments
We thank Anna Niewiadomska and JoAnn Mugavero for expert assistance. We thank C. E. Cameron for sharing unpublished data and for supplying recombinant non-His-tagged 3CDpro and 3Cpro-His expression vectors and proteins. Our extended gratitude goes to B. Semler for providing a PCBP-expression vector, X. Li for providing the P/L replicon plasmid, and K. Kirkergaard for providing the 3Dpol vector.
This work was supported by grant AI15122 of the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. [DOI] [PubMed] [Google Scholar]
- 2.Ambros, V., and D. Baltimore. 1978. Protein is linked to the 5′ end of poliovirus RNA by phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263-5266. [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 5.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 93:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2Apro. J. Virol. 75:10979-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 11.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang, S. K., H.-G. Kräusslich, M. J. H. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lama, J., A. V. Paul, K. S. Harris, and E. Wimmer. 1994. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as co-factor for viral polymerase 3Dpol. J. Biol. Chem. 269:66-70. [PubMed] [Google Scholar]
- 14.Li, X., H. H. Lu, S. Mueller, and E. Wimmer. 2001. The C-terminal residues of poliovirus proteinase 2A(pro) are critical for viral RNA replication but not for cis- or trans-proteolytic cleavage. J. Gen. Virol. 82:397-408. [DOI] [PubMed] [Google Scholar]
- 15.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason, P. W., S. V. Bezborodova, and T. M. Henry. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the IRES of foot-and-mouth disease virus. J. Virol. 76:9686-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 21.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 22.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 23.Pathak, H. B., S. K. Ghosh, A. W. Roberts, S. D. Sharma, J. D. Yoder, J. J. Arnold, D. W. Gohara, D. J. Barton, A. V. Paul, and C. E. Cameron. 2002. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol): a surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J. Biol. Chem. 277:31551-31562. [DOI] [PubMed] [Google Scholar]
- 24.Paul, A. V. 2002. Possible unifying mechanism of picornavirus genome replication, p. P. 227-246. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 25.Paul, A. V., X. Cao, K. S. Harris, J. Lama, and E. Wimmer. 1994. Studies with poliovirus polymerase 3Dpol. Stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 269:29173-29181. [PubMed] [Google Scholar]
- 26.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 28.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 29.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleotide triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits replication of poliovirus. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 30.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roehl, H. H., T. B. Parsley, T. V. Ho, and B. L. Semler. 1997. Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation. J. Virol. 71:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothberg, P., T. Harris, A. Nomoto, and E. Wimmer. 1978. 04-(5′-Uridylyl)-tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc. Natl. Acad. Sci. USA 75:4868-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skern, T., W. Sommergruber, D. Blaas, P. Gruendler, F. Fraundorfer, C. H. Pieler, I. Fogy, and E. Kuechler. 1985. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 13:2111-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanway, G., P. J. Hughes, R. C. Mountford, P. D. Minor, and J. W. Almond. 1984. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 12:7859-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Werf, S., J. Bradley, E. Wimmer, F. W. Studier, and J. J. Dunn. 1986. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang, W., A. Cuconati, D. Hope, K. Kirkegaard, and E. Wimmer. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 72:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang, W., A. Cuconati, A. V. Paul, X. Cao, and E. Wimmer. 1995. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1:892-904. [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, Y., R. Rijnbrand, K. L. McKnight, E. Wimmer, A. Paul, A. Martin, and S. M. Lemon. 2002. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J. Virol. 76:7485-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ypma-Wong, M.-F., P. G. Dewalt, V. H. Johnson, J. G. Lamb, and B. L. Semler. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265-270. [DOI] [PubMed] [Google Scholar]