Abstract
The papillomavirus life cycle is closely linked to the differentiation program of the host keratinocyte. Thus, late gene expression and viral maturation are restricted to terminally differentiated keratinocytes. A variety of cellular transcription factors including those of the C/EBP family are involved in the regulation of keratinocyte differentiation. In this study we show that the papillomavirus transcription factor E2 cooperates with C/EBPα and -β in transcriptional activation. This synergism was independent of an E2 binding site. E2 and C/EBP factors synergistically transactivated a synthetic promoter construct containing classical C/EBPβ sites and the C/EBPα-responsive proximal promoter of the involucrin gene, which is naturally expressed in differentiating keratinocytes. C/EBPα or -β coprecipitated with E2 proteins derived from human papillomavirus type 8 (HPV8), HPV16, HPV18, and bovine papillomavirus type 1 in vitro and in vivo, indicating complex formation by the cellular and viral factors. The interaction domains could be mapped to the C terminus of E2 and amino acids 261 to 302 located within the bZIP motif of C/EBPβ. Our data suggest that E2, via its interaction with C/EBP factors, may contribute to enhancing keratinocyte differentiation, which is suppressed by the viral oncoproteins E6 and E7 in HPV-induced lesions.
Human papillomaviruses (HPVs) are small, double-stranded DNA viruses causing proliferative lesions of the skin or mucosa. The viral life cycle is closely linked to the keratinocyte's differentiation program. According to the current model, HPVs infect keratinocytes of the basal cell layer via microtraumata. In these cells the early viral genes start to be weakly expressed and a maintenance copy number of the viral genome is established. Normally, keratinocytes have to exit the cell cycle in order to differentiate. As HPVs depend on the host cell's DNA-dependent DNA polymerase, the viral oncoproteins E6 and E7 maintain keratinocytes in a proliferative stage, thus enabling vegetative viral DNA replication in the upper layers of the epithelium. Furthermore, E6 and E7 inhibit terminal keratinocyte differentiation induced by serum or calcium (39, 49). The early protein E1 and the viral transcription factor E2 play a major role in the initiation of viral replication. All E2 proteins share a common modular structure containing an N-terminal transactivation domain and a C-terminal DNA binding and dimerization domain. These highly conserved domains are linked by the less conserved hinge region. The E2 protein of HPV type 16 (HPV16) was found to be expressed in the superficial layers of cervical intraepithelial neoplasia I (CINI) and CINII lesions and only at low levels in the intermediate and basal layers. In CINIII lesions, E2 is only weakly expressed, and in squamous cell carcinomas it is absent (44). In a benign cutaneous lesion of a patient suffering from epidermodysplasia verruciformis, E2-specific mRNA was detected in the upper two-thirds of the epidermis (15). E2 not only regulates E6 and E7 oncogene transcription but also induces G1 cell cycle arrest, apoptosis, or senescence in HPV-positive cells (7, 8, 20, 47). In the uppermost differentiated epithelial layers viral maturation and assembly take place. How E6/7 oncoprotein-mediated inhibition of keratinocyte differentiation is overcome to allow later stages of the viral life cycle is currently unknown.
Differentiation of keratinocytes is associated with changes in the expression patterns of a variety of transcription factors, i.e., the C/EBP (CCAAT/enhancer binding protein) and AP-1 (activator protein-1) families of transcription factors (for a review see reference 10). These factors belong to the basic leucine zipper (bZIP) transcription factors involved in multiple protein-protein interactions. The C/EBP family includes C/EBPα, C/EBPβ (variously named also LAP, NF-IL-6, NF-M, CRP2, or IL-6DBP), C/EBPγ, C/EBPδ (NF-IL-6β), C/EBPɛ, and C/EBPζ (C/EBP-homologous-protein CHOP-10, GADD 153). All C/EBP proteins form homo- and heterodimers via their C-terminal bZIP motifs (see reference 36 for a review). N-terminally, most C/EBP factors, including C/EBPα and C/EBPβ, possess transactivation domains. Through usage of three alternative initiation codons, the C/EBPβ gene gives rise to three proteins: the full-length protein, a protein starting from amino acid 24, and the shorter form, LIP. LIP lacks the complete transactivation domain and therefore acts as a dominant-negative inhibitor of the other forms. Transcription from C/EBP-dependent promoters is regulated by the expression levels and phosphorylation status of the different C/EBP isoforms. In the epidermis C/EBPβ is expressed at low levels in the basal cell layer, showing a cytoplasmic location. Starting from the middle to upper stratum spinosum, C/EBPβ is strongly expressed and localizes to the nuclei of keratinocytes. C/EBPα is expressed even later in keratinocyte differentiation (25, 31). In hyperplastic epidermis and squamous papillomas C/EBPα and C/EBPβ are present in all suprabasal layers, whereas their expression seems to be reduced in squamous cell carcinomas (31).
From various studies evidence is accumulating that C/EBP factors play a regulatory role in keratinocyte differentiation. Forced expression of C/EBPβ in murine keratinocytes results in growth inhibition, a more highly differentiated phenotype, and up-regulation of keratin 1 and keratin 10, two early markers of keratinocyte differentiation. Three C/EBP binding sites were found within the promoter of the murine keratin 10 gene conferring transcriptional activation (26, 52). The expression of involucrin, an integral component of the cornified envelope, starts in the upper spinous layers. The human involucrin promoter has been extensively studied. The distal regulatory region of the human involucrin promoter (−2473 to −1953) is required for expression in the epidermis of transgenic mice (5). The promoter-proximal regulatory region (−241 to −7) is responsible for almost half the activity, for which both a functional AP-1 and a C/EBP binding site are essential. The C/EBP binding site is strongly stimulated by C/EBPα (1, 48).
Here we show that the papillomavirus transcription factor E2 binds to and cooperates with C/EBP factors in transcriptional activation. This novel E2 activity may contribute to enhanced activation of differentiation-regulated genes, as exemplified by the hyperactivation of the proximal involucrin promoter.
MATERIALS AND METHODS
Plasmids.
Plasmids for eucaryotic expression and in vitro transcription.
Cytomegalovirus (CMV)-C/EBPβ (also known as CMV-NFIL6) was kindly provided by S. Kyo (2). C/EBPβ and its deletion mutants ΔN23, LIP, and ΔC (see Fig. 3) were amplified by PCR and inserted into the XhoI site of pET-14b (Novagen, Madison, Wis.). This vector allows in vitro transcription from the T7 promoter. For the construction of pcDNA-C/EBPα the coding region of C/EBPα was amplified from CMV-C/EBPα (kindly provided by G. J. Darlington) by PCR and subcloned in pCR-BluntII-TOPO (Invitrogen, Karlsruhe, Germany). The KpnI/XhoI fragment was inserted into the appropriate sites of pcDNA3.1+ (Invitrogen). pcDNA-HPV8-E2 was generated by inserting a BclI-StuI fragment, encoding the E2 open reading frame of the HPV8 genome, into the BamHI and EcoRV sites of pcDNA3.1+. To prepare pcDNA-HPV18-E2, the E2 gene of HPV18 was amplified by PCR and inserted into the EcoRV site of pcDNA3.1+. For the eucaryotic expression of fusion proteins consisting of the enhanced yellow fluorescent protein (EYFP) and HPV18 E2 or the C-terminal DNA binding and dimerization domain of HPV8 E2, the respective DNA fragments of the HPV18 E2 gene (nucleotides 4 to 1098) or the HPV8 E2 gene (nucleotides 1228 to 1497) were amplified by PCR. They were cloned into the EcoRI and BamHI sites of pEYFP-C1 (Becton Dickinson, Heidelberg, Germany) and designated EYFP-HPV18-E2 and EYFP-HPV8-E2 C, respectively. pXJ42-HPV8-E2 ΔC, encoding an E2 protein that lacks amino acids 409 to 489, was kindly provided by M. May (12).
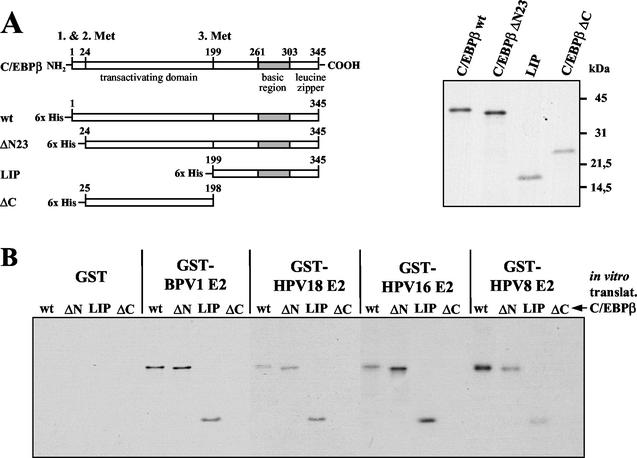
FIG. 3.
E2 binds to the C terminus of C/EBPβ. (A) Schematic presentation of C/EBPβ deletion mutants translated in vitro (left panel). Equal amounts of these proteins were resolved by SDS-PAGE (right panel). (B) In pull-down assays GST, GST-BPV1 E2, GST-HPV18 E2, GST-HPV16 E2, and GST-HPV8 E2 were bound to glutathione-Sepharose and incubated with in vitro-translated C/EBPβ and its deletion mutants. C/EBPβ proteins were visualized by SDS-PAGE and autoradiography.
Plasmids for procaryotic expression.
pET-HPV18-E2 was a gift from F. Thierry (6). For the bacterial expression of the fusion protein glutathione S-transferase (GST)-C/EBPα the coding region of C/EBPα was amplified by PCR and subcloned into pCR-BluntII-TOPO. After EcoRI/BamHI digestion an oligonucleotide linker containing a BamHI site in frame was ligated to the 5′ terminus of C/EBPα. This fragment was cloned into the BamHI site of pGEX-2T.
GST-C/EBPβ and its deletion mutants LIP, LIP ΔZ, LIP Z, LIP B, and LIP ΔC (see Fig. 4) were generated by cloning the respective PCR products of the C/EBPβ gene into the BamHI and EcoRI sites of pGEX-2T (Amersham Pharmacia, Freiburg, Germany). pGEX-HPV8-E2 and its deletion mutants ΔH205-409, ΔC410-499, ΔN1-329, and ΔN1-194 were kindly provided by M. May (12). pGEX-HPV16-E2 was a gift from H. Pajunk. pGEX-HPV18-E2 and pGEX-BPV1-E2 were kindly provided by G. Steger.
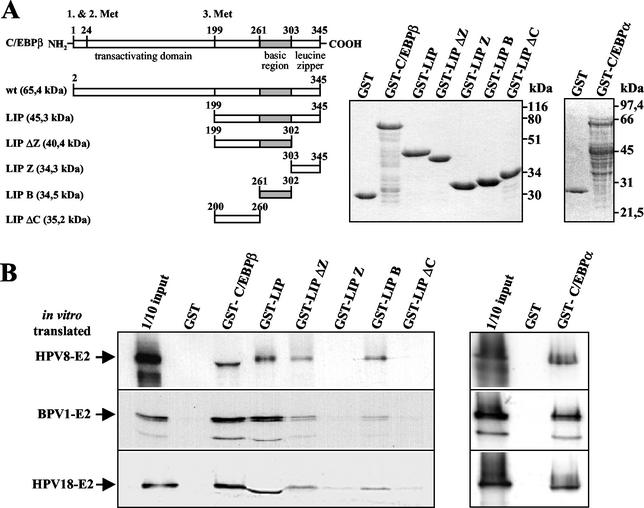
FIG. 4.
Fine mapping of the E2-interaction domain within C/EBPβ. (A) Schematic presentation of C/EBPβ deletion mutants fused to GST (left panel). Bacterially expressed and purified C/EBPβ or C/EBPα GST fusion proteins (middle and right panels, respectively) were used to precipitate in vitro-translated E2 proteins from HPV8, HPV18, and BPV1 (B). After washing, the bound 35S-labeled E2 proteins were separated by SDS-PAGE and detected by autoradiography.
Luciferase reporter constructs.
The luciferase expression plasmid E2wt-C/EBPwt-LUC is a derivative of C18-LUC (42), which contains four E2 binding sites and the adenovirus major late promoter upstream of the luciferase gene of the plasmid pALUC (9). To construct E2wt-C/EBPwt-LUC, two oligonucleotide adapters harboring the C/EBPβ binding site of the interleukin-6 promoter (in boldface) were inserted into the BamHI site (lowercase) of C18-LUC:
5′-gatctGTCACATTGCACAATCTTAg-3′
3′-aCAGTGTAACGTGTTAGAATcctag-5′
For the construction of E2mut-C/EBPwt-LUC, a fragment (−44 to +15) consisting of the adenovirus major late promoter and the two C/EBP sites of E2wt-C/EBPwt-LUC was amplified by PCR and cloned into the BamHI site of pALUC. Finally two preligated oligonucleotide adapters containing four mutated E2 binding sites (in boldface; mutations are underlined) were inserted within the BamHI and SalI sites (lowercase) of this construct:
5′-gatccTCTAGTCTGAAAACGATCGGGTCTGAAAACGAT-3′
3′-gAGATCAGACTTTTGCTAGCCCAGACTTTTGCTAGATTAGAC-5′
5′-CTAATCTGAAAACGATCGGGTCTGAAAACGATCTAGAg-3′
3′-TTTTGCTAGCCCAGACTTTTGCTAGATCTcagct-5′ pINV241 and pINV241-C/EBPmut, containing a mutated C/EBP binding site at positions −144 to −135, were gifts from R. L. Eckert (1, 48).
Cell culture, transfection, nuclear extracts, and luciferase assays.
The HPV-negative keratinocyte cell line RTS3b, derived from a human squamous cell carcinoma (35), was maintained in Dulbecco modified Eagle medium containing 25% Ham's F-12 medium, 8% fetal calf serum, 50 μg of gentamicin/ml, 10 ng of epidermal growth factor/ml (all from Invitrogen), 0.4 μg of hydrocortisone/ml, 10 ng of cholera toxin/ml, 5 μg of transferrin/ml, 2 × 10−11 M triiodothyronine, 1.8 × 10−4 M adenine, and 5 μg of insulin/ml (all from Sigma-Aldrich, Taufkirchen, Germany).
The HPV-negative cell line C33A, derived from a human cervical carcinoma, and 293T cells (32) were grown in Dulbecco modified Eagle medium (with Glutamax) supplemented with 8% fetal calf serum, 100 U of penicillin/ml, 0.1 mg of streptomycin/ml, and 1 mM sodium pyruvate (all from Invitrogen). C33A cells were transfected by the calcium phosphate method according to standard procedures. One day before transfection C33A cells were seeded in six-well plates at a density of 260,000 cells per well. The DNA was precipitated by calcium phosphate, added to the cells, and removed after 18 h.
RTS3b cells were seeded in six-well plates at a density of 170,000 cells/well. After 24 h cells were transfected with Fugene (Roche, Mannheim, Germany) according to the manufacturer's guidelines. The medium was changed after 6 h of incubation.
In the various experiments the total amount of DNA (2 μg) was kept constant by adding pcDNA3.1+. Cells were harvested 48 h after transfection, and luciferase extracts were prepared. The luciferase activity was measured and normalized with the protein concentration of the respective luciferase extract. Transfections were conducted in duplicate, and the indicated values were averaged from at least three independent experiments.
Nuclear extracts were prepared from transiently transfected C33A and 293T cells according to the protocol of Schreiber et al. (40).
In vitro transcription and translation.
In vitro transcription and translation were performed in the presence of [35S]methionine (Amersham Pharmacia) with the TNT-T7-coupled reticulocyte lysate system from Promega (Mannheim, Germany). To quantify the incorporation of [35S]methionine, 1 μl of the translation product was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were fixed and treated with Amplify (Amersham Pharmacia) for 20 min. Subsequently, the gels were dried and subjected to autoradiography.
Expression and purification of His and GST fusion proteins.
The E2 protein of HPV18 was expressed in bacteria as an N-terminally His-tagged fusion protein and purified as described in the work of Hoffmann and Roeder (18) with nickel-nitrilotriacetic acid-agarose (Qiagen, Hilden, Germany) columns.
GST fusion proteins were expressed in Escherichia coli and purified with glutathione-Sepharose according to the work of Enzenauer et al. (12). To semiquantify GST and GST fusion proteins, they were immobilized on glutathione-Sepharose. The beads were boiled in Laemmli buffer, and eluted proteins were analyzed by SDS-PAGE and Coomassie blue staining. GST-BPV1 E2, GST-HPV16 E2, GST-HPV8 E2, ΔH205-409, and ΔN1-329 were expressed only at low levels in bacteria or yielded partially truncated products.
In vitro binding assays.
For interaction assays proteins from nuclear extracts, purified His-tagged proteins, or in vitro-translated proteins were incubated with GST fusion proteins bound to glutathione-Sepharose beads for 2 h at 4°C. Subsequently the beads were washed three times for 3 min at 4°C with LSDB buffer (50 mM Tris-HCl [pH 7.9], 20% glycerol, 1 mM dithiothreitol, and 0.1% NP-40) containing 500 mM KCl and once with the same buffer containing 100 mM KCl. The beads were boiled in Laemmli buffer, and eluted proteins were analyzed by SDS-PAGE. In the case of [35S]methionine-labeled proteins, gels were fixed, treated with Amplify, dried, and subjected to autoradiography.
His-tagged proteins and proteins from nuclear extracts were blotted onto nitrocellulose membranes. After the membranes were blocked with 5% dry milk in 0.1% Tween-0.01% NaN3-phosphate-buffered saline (PBS) for 1 h, they were incubated with primary antibodies overnight: mouse anti-Flag (1:50; M5; Sigma-Aldrich), goat anti-C/EBPα (1 μg/ml; N-19; Santa Cruz Biotechnology, Heidelberg, Germany), or rabbit anti-C/EBPβ (1.5 μg/ml; C-19; Santa Cruz Biotechnology). After the membranes were washed three times with 0.1% Tween-PBS, they were incubated with peroxidase-coupled secondary antibody for 1 h. Goat anti-mouse, rabbit anti-goat, and goat anti-rabbit antibodies (all from Dianova, Hamburg, Germany) were used at a concentration of 160 ng/ml in 5% dry milk-PBS-0.1% Tween. The immunoblots were developed with enhanced chemiluminescence as described by the manufacturer (Roche).
Coimmunoprecipitation assays.
293T cells were seeded at a density of 8.9 × 106 cells per 150-cm2 dish. One day later they were transfected with 30 μg of expression plasmids by using Fugene. After 24 h cells were harvested by using trypsin and washed two times with PBS (4°C). The cells were incubated for 30 min in lysis buffer (1× PBS, 5 mM EDTA, 0.5% Triton X-100, 25 μg of aprotinin/ml, 10 μg of pepstatin A/ml, 10 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride) at 4°C in a rotating apparatus. Cell debris was pelleted by centrifugation at maximum speed and 4°C for 30 min. The protein content of the lysates was determined by using the Bradford reagent (Bio-Rad, Munich, Germany) according to the manufacturer's instructions. To preclear the lysates, equal protein amounts were incubated for 30 min with 30 μl of protein G-Sepharose (Amersham Pharmacia) at 4°C in a rotating apparatus. Protein G-Sepharose was pelleted by centrifugation, and the cleared lysates were supplemented with polyclonal rabbit anti-full-length EYFP antibody (1:100; Becton Dickinson) or rabbit anti-C/EBPβ (1:100; C-19) and 30 μl of protein G-Sepharose and were rotated overnight at 4°C. The Sepharose beads were washed five times for 2 min each with ice-cold PBS. Bound proteins were eluted with Laemmli buffer containing β-mercaptoethanol at 94°C for 4 min. After SDS-PAGE Western blotting was performed as described above. EYFPs or C/EBPs were identified in Western blots by using the antibody rabbit anti-EYFP peptide (1:100; Becton Dickinson) or mouse anti-EYFP (1:1,000; Becton Dickinson), mouse anti-C/EBPβ (2 μg/ml; H-7; Santa Cruz Biotechnology), or goat anti-C/EBPα (as above), respectively.
EMSA.
293T cells were transfected with CMV-C/EBPβ or pcDNA-C/EBPα as described above. Twenty-four hours later nuclear extracts were prepared. Sixty micrograms of these extracts was incubated with 2.5 ng of either 32P-labeled wild-type or mutated C/EBP oligonucleotides (positions −165 to −139 of the human interleukin-6 gene) according to the work of Akira et al. (2) for 40 min at 4°C. Ten percent of these mixtures were analyzed by electrophoretic mobility shift assay (EMSA) as described in reference 41 with C/EBP EMSA buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mg of bovine serum albumin/ml, 25 μg of dI-dC/ml, 10 mM dithiothreitol, and 0.5 mg of single-stranded DNA/ml). DNA-bound complexes were visualized by autoradiography. Ninety percent of the reaction mixtures were further incubated with GST or GST-HPV8 E2 immobilized on glutathione-Sepharose for 2 h at 4°C. The beads were washed three times with LSDB buffer containing 500 mM KCl and once with the same buffer containing 100 mM KCl at 4°C. Subsequently, bound complexes were eluted with C/EBP-EMSA buffer containing 25 mM glutathione for 1 h at 4°C. Eluates were resolved by EMSA, and DNA-bound complexes were visualized by autoradiography.
RESULTS
The HPV8 E2 protein synergizes with C/EBPβ in transcriptional activation.
C/EBP factors are expressed in the suprabasal layers of the epithelium in a similar pattern as the viral E2 protein, suggesting colocalization in lesional skin. We were therefore interested in whether these transcription factors might influence each other's activity. A luciferase reporter plasmid containing four E2 and two C/EBP binding sites upstream of the adenovirus major late promoter was constructed (E2wt-C/EBPwt-LUC [Fig. 1A ]). This reporter plasmid was cotransfected into the HPV-negative cervical carcinoma cell line C33A in combination with expression plasmids for HPV8 E2 and C/EBPβ. In preceding experiments each expression plasmid was titrated to yield approximately 10-fold stimulation of the reporter construct. Thus, 0.8 μg of CMV-C/EBPβ alone activated the reporter construct 13-fold, and 0.4 μg of pcDNA-HPV8 E2 activated the construct 8-fold (Fig. 1B). Together the two factors showed a strong synergism in transactivation (37-fold). These data indicate that the viral and the cellular transcription factors may cooperate in transcriptional activation if binding sites for both factors are present.
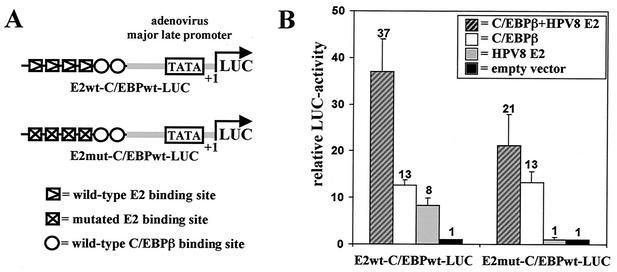
FIG. 1.
HPV8 E2 and C/EBPβ synergize in transcriptional activation. (A) Schematic presentation of reporter plasmids containing the adenovirus major late promoter and two C/EBP and four intact or mutated E2 binding sites upstream of the luciferase gene. (B) C33A cells were seeded in six-well plates. The next day the reporter plasmid E2wt-C/EBPwt-LUC or E2mut-C/EBPwt-LUC (0.5 μg each) was transfected with or without plasmids expressing C/EBPβ (0.8 μg) and HPV8 E2 (0.4 μg). The total amount of DNA was adjusted with empty pcDNA3.1+ vector. After 48 h luciferase activities were determined and normalized with the protein concentration of the respective luciferase extract. The value of the control (empty pcDNA3.1+ vector only) was set for 1. Transfections were conducted in duplicate, and the indicated values were averaged from at least three independent experiments.
The transcriptional synergism of E2 and C/EBPβ is independent of E2 binding sites.
Most cellular genes do not contain binding sites for the viral E2 protein. Therefore, we investigated whether E2 is still able to enhance the C/EBP activity on such promoters. To test this hypothesis, the E2 binding sites of the reporter construct were mutated (E2mut-C/EBPwt-LUC [Fig. 1A]). E2 proteins were unable to bind to these mutated sites as tested by EMSA (data not shown), and this reporter construct was not activated by HPV8 E2 anymore (Fig. 1B). In contrast, C/EBPβ transactivation was unchanged compared to that for the parental reporter construct, E2wt-C/EBPwt-LUC (13-fold). Coexpression of HPV8 E2 and C/EBPβ, however, resulted in a much stronger activation than that seen for C/EBPβ alone (21-fold), indicating that the two factors together synergistically activated E2mut-C/EBPwt-LUC. Western blot analysis confirmed that E2 and C/EBPβ coexpression (both driven by the CMV-immediate-early promoter) did not alter the C/EBPβ protein expression level or pattern (data not shown). These data demonstrate for the first time that the viral E2 protein enhances C/EBPβ-dependent transcription and that E2 binding sites are dispensable for this effect.
E2 proteins from different HPV types interact with C/EBPα and -β.
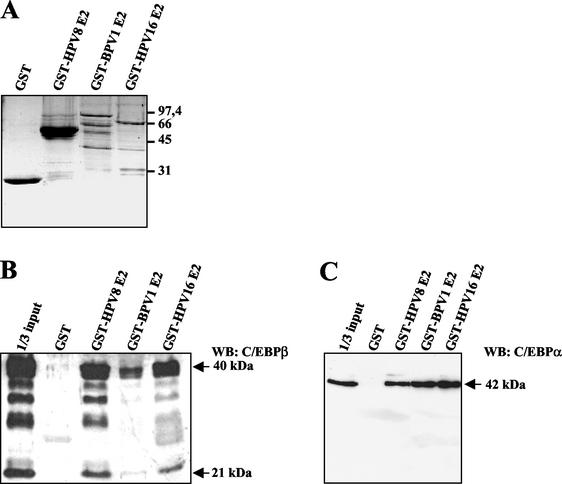
To investigate whether C/EBP factors and E2 proteins associated with each other, coprecipitation experiments were performed. Fusion proteins consisting of GST and E2 proteins from HPV8, HPV16, or bovine papillomavirus type 1 (BPV1) were expressed in E. coli and purified. Analysis by SDS-PAGE and Coomassie blue staining demonstrated that GST-BPV1 E2 and -HPV16 E2 could be expressed only at low levels and were used at lower amounts than those of GST and GST-HPV8 E2 (Fig. 2A). C/EBPβ was overexpressed in C33A cells, and nuclear extracts were prepared. Western blot analysis revealed that C/EBPβ was expressed in multiple forms in C33A cells, the 40-kDa band representing full-length C/EBPβ, whereas the 21-kDa band corresponded to LIP, the C-terminal portion of the transcription factor. GST-E2 fusion proteins from HPV8, HPV16, and to a lesser extent BPV1 but not GST alone precipitated C/EBPβ from nuclear extracts (Fig. 2B). Similarly, C/EBPα expressed in C33A cells was precipitated by the E2 proteins from all investigated papillomavirus types (Fig. 2C).
FIG. 2.
E2 physically interacts with C/EBPα and C/EBPβ. Nuclear extracts were prepared from C33A cells overexpressing C/EBPα or C/EBPβ. In coprecipitation assays GST or E2 proteins from different papillomavirus types fused to GST (A) were bound to glutathione-Sepharose and incubated with these extracts. After washing, the bound proteins were subjected to SDS-PAGE. C/EBPβ (B) and C/EBPα (C) were identified by Western blotting with anti-C/EBPβ or anti-C/EBPα antibodies. In panel B the larger (40-kDa) and the shorter (21-kDa) forms of C/EBPβ were detected. WB, Western blot. Numbers at right of panel A are molecular masses in kilodaltons.
Since E2 and C/EBP factors are DNA binding proteins, interaction between these proteins might have been mediated by DNA contamination. Several control experiments were performed to exclude this possibility. Addition of oligonucleotides containing binding sites for E2 or C/EBPβ or both factors, nonspecific salmon sperm DNA, or ethidium bromide did not affect the interaction. Similarly, incubation with DNase I and RNase A did not influence the interaction between HPV8 E2 and C/EBPβ (data not shown), suggesting that this association was due to protein-protein interaction rather than interaction of the transcription factors with contaminating nucleic acids. Furthermore, GST-E2 proteins from BPV1, HPV18, HPV16, and HPV8 precipitated not only C/EBP factors from nuclear extracts but also in vitro-translated C/EBPβ (Fig. 3).
To examine whether C/EBP factors were conversely able to pull down E2 proteins, C/EBPα and C/EBPβ were fused to GST and expressed in E. coli. GST-C/EBPα and GST-C/EBPβ were both able to precipitate in vitro-translated E2 proteins (Fig. 4). To confirm that the interaction between C/EBPβ and E2 was not mediated by other cellular proteins present in nuclear extracts and in vitro translations, it was important to show that the two factors could directly bind to each other. Therefore, His-tagged HPV18 E2 was expressed in E. coli and purified on nickel-nitrilotriacetic acid-agarose. We failed to purify His-HPV8 E2 or His-BPV1 E2 proteins. GST-C/EBPβ but not GST alone precipitated His-HPV18 E2, demonstrating a direct interaction between a representative E2 protein and C/EBPβ (data not shown).
Thus, in various kinds of interaction experiments a specific association of C/EBP factors and the papillomavirus E2 protein was confirmed. The interaction was not restricted to HPV8 E2 but was seen also for the E2 proteins from the genital HPV16 and HPV18 and even for the BPV1 E2 protein.
Identification of the C/EBPβ region involved in the interaction with E2.
As shown in Fig. 2B, the HPV E2 proteins associated not only with the larger 40-kDa form but also with a shorter 21-kDa form of C/EBPβ, corresponding to LIP. This suggested that E2 binds to the C-terminal part of C/EBPβ. To further investigate this, a number of C/EBPβ deletion mutants (Fig. 3A) were translated in vitro and tested for their ability to interact with GST-E2 proteins from different papillomavirus types. C/EBPβ, ΔN23, and LIP were precipitated by E2 proteins from BPV1, HPV18, HPV16, and HPV8 (Fig. 3B). Only the deletion mutant C/EBPβ ΔC did not associate with any of the E2 proteins, reconfirming that the E2 proteins in fact interacted with the C terminus of C/EBPβ.
This part of the protein was analyzed in more detail (Fig. 4) by using GST-LIP deletion mutants (Fig. 4A). Whereas GST-LIP, ΔZ, and B bound to all E2 proteins, GST-LIP ΔC and GST-LIP Z failed to associate with or only weakly bound to in vitro-translated E2 proteins from HPV8, HPV18, and BPV1 (Fig. 4B). These data indicated that the region interacting with E2 is located between amino acids 261 and 302 within the bZIP motif of C/EBPβ.
C/EBP factors bind to the carboxy terminus of HPV8 E2.
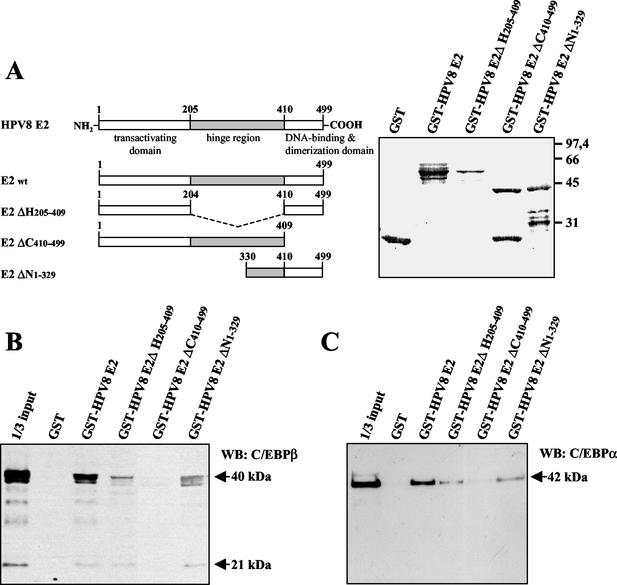
To determine which part of the E2 protein interacts with C/EBPα and C/EBPβ, deletion mutants of HPV8 E2 each lacking different functional domains were fused to GST (Fig. 5A). These GST-E2 proteins were purified from E. coli and incubated with C/EBPα or C/EBPβ from nuclear extracts. Both the E2 ΔH and E2 ΔN mutants seemed to precipitate the C/EBP factors less effectively. For both E2 deletion mutants the most likely explanation is that less protein was used for precipitation (Fig. 5A). Only the E2 deletion mutant lacking the C-terminal DNA binding and dimerization domain ΔC 410-499 did not precipitate C/EBPβ (Fig. 5B) or C/EBPα (Fig. 5C), despite higher amounts of input protein compared with E2 ΔH (Fig. 5A). This indicates that the C-terminal domain of E2 is important for the binding of C/EBP factors.
FIG. 5.
C/EBP factors interact with the C-terminal part of HPV8 E2. (A) Schematic presentation of HPV8 E2 deletion mutants fused to GST (left panel). The hinge region is marked as a gray box, and all amino acid positions are indicated. In pull-down assays GST and the GST-HPV8 E2 fusion proteins (right panel) were coupled to glutathione-Sepharose and incubated with nuclear extracts from C33A cells overexpressing C/EBPβ (B) or C/EBPα (C). All bound proteins were separated by SDS-PAGE. The C/EBP factors were identified by Western blot (WB) analysis with the respective antibodies. Numbers at right of panel A are molecular masses in kilodaltons.
In vivo association of HPV8 E2 and C/EBPβ.
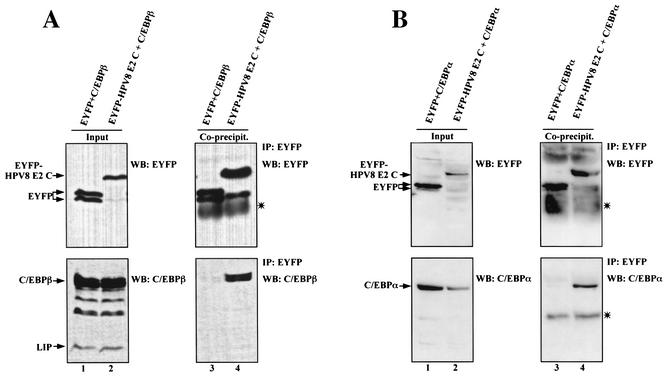
To confirm that E2 interacts with C/EBP factors in vivo, coimmunoprecipitations were performed. Since HPV E2-specific antibodies were not available, HPV E2 proteins and the C-terminal DNA binding and dimerization domain of HPV8 E2 were fused to the EYFP. Whereas full-length EYFP-HPV8 E2 expressed in 293T cells was not extractable from the nuclear matrix (unpublished observation), EYFP-HPV8 E2 C and full-length EYFP-HPV18 E2 could be recovered in nuclear extracts and used for subsequent experiments. Expression plasmids for this EYFP-HPV8 E2 C fusion protein or wild-type EYFP were transfected in 293T cells together with a plasmid expressing C/EBPβ or C/EBPα. After 24 h cells were harvested and cell lysates were prepared.
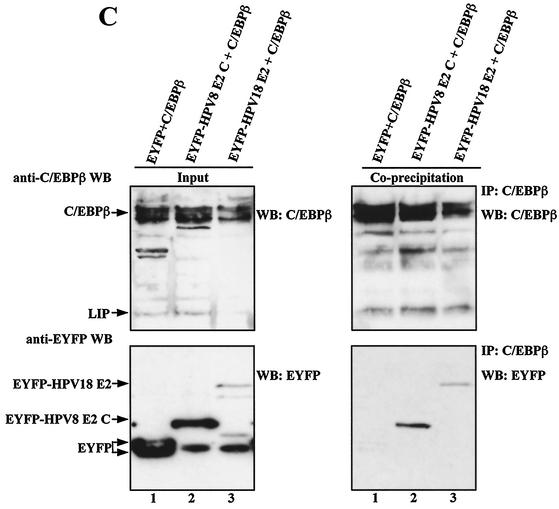
In coprecipitation experiments input levels of C/EBP factors or of EYFP and EYFP-HPV8 E2 C were virtually the same (Fig. 6A, lanes 1 and 2) or even less in the case of EYFP-HPV8 E2 C (Fig. 6B, lanes 1 and 2). EYFP-HPV18 E2 was expressed only at low levels (Fig. 6C, lower left panel, lane 3). EYFP and EYFP-HPV8 E2 C were precipitated by an EYFP-specific antibody with similar efficiencies (Fig. 6A and B, top panels, lanes 3 and 4). However, C/EBPβ and C/EBPα were precipitated only by EYFP-HPV8 E2 C and not by EYFP alone (Fig. 6A and B, bottom panels, lanes 3 and 4), confirming that E2 and C/EBP factors indeed specifically associate in mammalian cells. Similarly, when a C/EBPβ-specific antibody was used for precipitation, EYFP-HPV8 E2 C and EYFP-HPV18 E2 were coprecipitated (Fig. 6C, lower right panel, lanes 2 and 3) but not EYFP (lane 1).
FIG. 6.
Coimmunoprecipitation of C/EBPβ or C/EBPα and the HPV E2 protein in vivo. Plasmids encoding EYFP-HPV8 E2 C (10 μg) or equimolar amounts of EYFP vector were cotransfected with C/EBPβ (A) or C/EBPα (B) expression plasmids (20 μg) into 293T cells (150-cm2 dishes). For panel C 20 μg of EYFP-HPV8 E2 C or EYFP-HPV18 E2 or equimolar amounts of EYFP vector and 10 μg of C/EBPβ plasmid were used. The amount of DNA was adjusted with empty pBluescript SKII(+) vector. After 24 h extracts were incubated with rabbit polyclonal EYFP-specific antibodies (A and B) or rabbit polyclonal C/EBPβ-specific antibodies (C) and protein G-Sepharose. The precipitates were washed and resolved by SDS-PAGE (lanes 3 and 4 in panels A and B and right panels in panel C). Left panels of each panel show 1/33 (A, lanes 1 and 2), 1/70 (B, lanes 1 and 2), or 1/100 (C, lanes 1 to 3) of the input extracts used for coprecipitation. EYFP fusion proteins, C/EBPβ, or C/EBPα was detected by Western blotting with rabbit anti-EYFP peptide antibody (A and B), mouse anti-EYFP antibody (C), mouse anti-C/EBPβ antibody, or goat anti-C/EBPα antibody. The asterisks indicate the light chains of immunoglobulins used for immunoprecipitation which were detected by the secondary antibodies in Western blot analysis. WB, Western blot; IP, immunoprecipitation.
Formation of a ternary complex of HPV8 E2, C/EBP factors, and the C/EBP-specific DNA binding site.
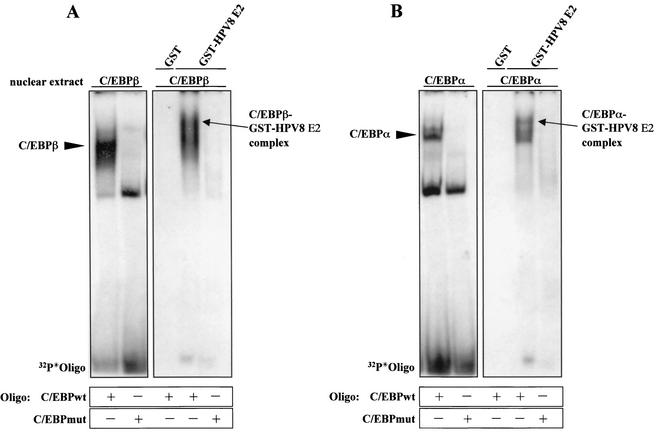
So far the data demonstrated the interaction of E2 and C/EBP factors in vitro and in vivo. The question arose whether complexes between the viral and cellular factors were formed also at the C/EBP binding site. To investigate this, C/EBPα and C/EBPβ were expressed in 293T cells and recovered from nuclear extracts. Incubation with radiolabeled oligonucleotides and subsequent EMSA demonstrated strong binding of both factors to the wild-type but not the mutated C/EBP DNA binding site (Fig. 7A and B, left panels). To test whether E2 was recruited by C/EBP factors bound to DNA, GST-HPV8 E2 or GST immobilized on glutathione-Sepharose was added and precipitated. After stringent washing and elution with glutathione, the complexes were analyzed by EMSA. In autoradiography, retarded complexes indicating a higher mass were detected only in the case of GST-HPV8 E2 and not GST (Fig. 7A and B, right panels). In fact, GST did not precipitate DNA-bound C/EBP factors, nor did GST-HPV8 E2 precipitate the radiolabeled mutated oligonucleotides, which were unable to bind C/EBP factors. From this we concluded that the E2 fusion protein is specifically incorporated into the complexes of C/EBPβ or C/EBPα when bound to DNA, leading to ternary complex formation.
FIG. 7.
HPV8 E2 forms complexes with C/EBPβ and C/EBPα in a DNA-bound state. Nuclear extracts were prepared from 293T cells overexpressing C/EBPβ (A) or C/EBPα (B). Sixty micrograms of each extract was incubated with wild-type or mutated C/EBP oligonucleotides for 40 min at 4°C. Ten percent of these reaction mixtures were investigated by EMSA (left panels). GST and the GST-HPV8 E2 fusion proteins (Fig. 2C) were coupled to glutathione-Sepharose and added to 90% of these mixtures for 2 h in LSDB buffer containing 100 mM KCl at 4°C. After washing, complexes were eluted in 25 μl of EMSA buffer containing 25 mM glutathione for 1 h at 4°C and then resolved by EMSA (right panels). Oligo, oligonucleotide.
HPV8 E2 and C/EBP factors synergistically activate the proximal involucrin promoter.
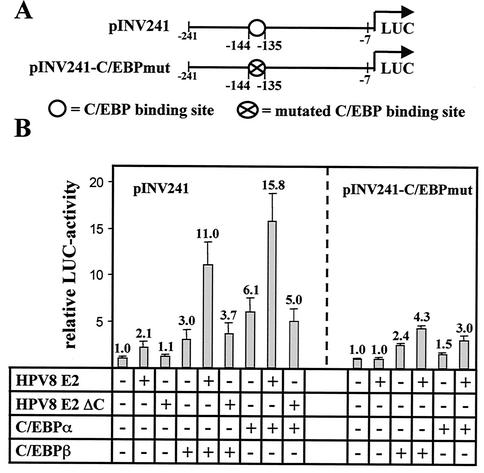
In the epithelium C/EBP factors are regulators of keratinocyte differentiation. They are directly involved in the differentiation-specific expression of proteins. In fact, Agarwal et al. showed that a C/EBP binding site (−135 to −144) within the proximal part of the involucrin promoter is essential for its basal and phorbol ester-mediated activation in normal foreskin keratinocytes (1). A luciferase reporter construct harboring the proximal −7 to −241 bp of the involucrin promoter (pINV241 [Fig. 8A ]) was strongly induced by transfection of very low amounts of C/EBPα expression plasmid (0.01 μg) into the skin-derived keratinocyte cell line RTS3b used here (sixfold, Fig. 8B). C/EBPβ or HPV8 E2 expression vectors alone (applied at the same concentrations as used for Fig. 1) activated this promoter construct threefold or twofold, respectively. Coexpression of C/EBPα and HPV8 E2 led to a synergistic induction (16-fold), although E2 binding sites were lacking in this promoter construct. A strong synergistic transactivation (11-fold) of pINV241 was observed when HPV8 E2 was combined with the weak activator C/EBPβ. Thus, the presence of E2 seems to convert even C/EBPβ into a strong inducer of the involucrin promoter. This strong synergism was not observed when C/EBP factors were combined with equimolar amounts of an expression plasmid encoding an E2 protein lacking the C terminus (3.7-fold induction in combination with C/EBPβ and 5.0-fold induction in combination with C/EBPα). This further underlined the importance of the C-terminal portion of the E2 protein not only for physical but also for functional interaction with C/EBP factors. To investigate the impact of the C/EBP binding site on the E2-C/EBP synergism in the context of a natural promoter, a pINV241 construct containing a mutated C/EBP binding site (pINV241-C/EBPmut) was used (Fig. 8B, right panel). E2 alone did not activate this promoter, indicating that the twofold activation of the natural pINV promoter might have been due to synergistic activation of E2 with low levels of endogenous C/EBP factors present in the RTS3b cell line. Whereas the capability of C/EBPα to transactivate this mutated promoter construct was strongly impaired (1.5-fold transactivation), C/EBPβ still led to a 2.4-fold activation. However, coexpression of HPV8 E2 and C/EBP factors led to a much weaker transactivation of this mutated construct than that of the wild-type promoter construct (4.3-fold in the case of C/EBPβ and 3.0-fold in the case of C/EBPα).
FIG. 8.
HPV8 E2 and C/EBP factors synergistically transactivate the involucrin promoter. (A) Schematic presentation of the reporter plasmid pINV241 comprising the proximal promoter fragment of the involucrin promoter upstream of the luciferase gene with wild-type or mutated C/EBP binding sites. (B) pINV241 (left panel) or pINV241-C/EBPmut (right panel) (0.5 μg each) was transfected in RTS3b cells with or without plasmids expressing C/EBPβ (0.8 μg), C/EBPα (0.01 μg), and HPV8 E2 full-length (0.4 μg) or equimolar amounts of HPV8 E2 ΔC lacking the C terminus of E2. The total amount of DNA was adjusted with empty pcDNA3.1+ vector. After 48 h luciferase activities were determined and normalized with the protein concentration of the respective luciferase extract. The value of the control (empty pcDNA3.1+ vector only) was set for 1. Transfections were conducted in duplicate. The indicated values were averaged from three independent experiments.
These experiments revealed that HPV8 E2 and C/EBP factors strongly synergize on different promoters (pINV241 and E2mut-C/EBPwt-LUC) in two different epithelial cell lines (RTS3b and C33A) when a C/EBP binding site is present. However, this synergism is independent of sequence-specific DNA binding of the viral transcription factor E2.
DISCUSSION
In this study we show that the papillomavirus-encoded E2 protein strongly synergizes with cellular transcription factors from the C/EBP family in transcriptional activation. The classical E2 binding site 5′-ACCN6GGT-3′ is dispensable for this synergism. Coprecipitation assays demonstrated that E2 proteins and C/EBP factors interact with each other in vivo and in vitro at the C/EBP binding site. The cooperative transactivation of C/EBP factors and E2 was detected in two different promoter contexts and with two different C/EBP factors (α and β). Furthermore this effect was seen in two HPV-negative epithelial cell lines, derived from skin (RTS3b) or cervix (C33A).
C/EBP-responsive promoter constructs with different features were employed in this study. E2 synergized with C/EBPβ on a synthetic promoter construct with classical C/EBPβ sites in the presence or absence of E2 binding sites. A similar synergistic activation by E2 was obtained when C/EBPα and the C/EBPα-responsive proximal involucrin promoter were used. In contrast, C/EBPβ does not activate or only weakly activates the involucrin promoter (1; this study). In combination with E2, however, C/EBPβ was converted into a strong activator. Synergistic transactivation between C/EBPα and E2 was strongly impaired when the C/EBP binding site was mutated in the pINV promoter. In the cell line used here, C/EBPβ weakly transactivated also this mutated promoter construct, almost to a similar extent as the wild-type promoter. Interestingly, this promoter fragment still contains a functional AP-1 binding site (48). C/EBPβ had been shown elsewhere to form functional complexes with other bZIP proteins, e.g., of the AP-1 family, or non-bZIP transcription factors, which may alter its binding specificity (19, 36). It may thus weakly transactivate the pINV promoter also through heterodimerization with other transcription factors in the cell line used here. However, in the case of a C/EBP binding site-independent transactivation the synergism with the viral E2 protein was strongly reduced. Synergistic transactivation did not depend on E2 binding sites. This was investigated systematically for the synthetic C/EBPβ-dependent promoter construct. This synthetic promoter construct was also synergistically transactivated if functional E2 binding sites were lacking, albeit somewhat less efficiently. From these experiments showing synergistic transactivation on the wild-type synthetic promoter, two mechanisms could be hypothesized. First, viral and cellular transcription factors bound to their individual binding sites could cooperate. Second, physical interaction of E2 and C/EBP factors could lead to hyperactivation via the C/EBP binding site. With use of the E2 binding site-mutated synthetic promoter, only the second mechanism could take place. Computer-based sequence analysis of the pINV promoter with the MatInspector program revealed no E2 binding sites. Thus, this study adds a novel way of E2 binding site-independent transactivation by E2. Similar mechanisms of E2 transactivation had been described previously with different promoters and cellular transcription factors (16, 17, 43).
The question arose by which mechanism E2 might synergize with C/EBP factors. Several possibilities were investigated. E2 enhanced neither C/EBP expression levels nor its expression pattern (data not shown). In various coprecipitation experiments using nuclear extracts and in vitro-translated and bacterially expressed proteins, E2 coprecipitated with C/EBP factors, confirming direct protein-protein interactions. In both molecules the interaction domains were mapped to their C termini. For C/EBPβ the domain was further reduced to amino acids 261 to 302 within the basic region. This region is highly conserved among members of the C/EBP family, possibly explaining why E2 interacts not only with C/EBPα and -β as shown here but also with C/EBPδ, rat C/EBPβ, and rat LIP (data not shown). The importance of the E2 C terminus was further supported by functional experiments on the pINV promoter. In contrast to the wild-type E2 protein, a C-terminally truncated E2 protein no longer synergized with C/EBP factors in transcriptional activation. Finally, we could demonstrate ternary complex formation of the viral E2 protein with C/EBPα and -β bound to the specific C/EBP binding site. Thus, our data indicate that C/EBP factors can recruit E2 proteins to promoters lacking E2 binding sites via protein-protein interaction.
In vivo, it is likely that E2 associates not only with C/EBP factors but also with a variety of other important regulatory molecules, such as Sp1, YY-1, AMF-1, and p53 (4, 22-24). In this study, both cell lines employed were devoid of functional p53. C33A cells derived from a cervical carcinoma with mutated p53 (38). RTS3b cells, derived from a cutaneous carcinoma (35), did not give rise to detectable p53 expression (S. Smola-Hess, unpublished data). Thus, the effects observed here were independent not only of other HPV proteins but also of p53. E2 also associates with the transcriptional coactivator CBP/p300 (21, 30, 33) and factors of the preinitiation complex (3, 12, 37, 51). Therefore, E2 might rather form complexes involving several of these known and potentially also yet unknown factors. An intriguing hypothesis is that E2 and C/EBP factors, which all associate with CBP/p300 (13, 27), would also recruit this transcriptional coactivator into a complex.
What might be the physiological role of the synergism between E2 and C/EBP factors, which are both mainly expressed in suprabasal layers of the epithelium? As E2 and C/EBPβ are both complex regulators of HPV oncogene expression, they may also synergize in this respect. No synergistic effects were seen in transient-transfection experiments on the HPV18 early promoter, which is suppressed by C/EBPβ (data not shown). However, a similar synergism in transactivation as that observed with the pINV promoter was obtained when an HPV late promoter construct was investigated, which was activated by C/EBP factors (S. Smola-Hess, unpublished).
It is tempting to speculate that one function of E2 might be to enhance keratinocyte differentiation, which is inhibited by the viral oncoproteins E6 and E7 in HPV-induced lesions. A differentiated cellular phenotype is required for the late gene expression and viral maturation. Interestingly, C/EBPα, similarly to other E2-interacting cellular factors mentioned above, is involved in the activation of the p21 promoter (11, 28, 34, 45, 50). This cell cycle inhibitor plays an important role in the regulation of terminal keratinocyte differentiation (29). Thus, E2 may overcome oncoprotein-mediated repression of differentiation via its interaction with members of the C/EBP transcription factor family. In fact, our data show that E2 is able to hyperactivate the C/EBP-responsive promoter fragment of the involucrin gene, which might be regarded as representative for other cellular genes regulated during keratinocyte differentiation. The expression levels of keratin 1 and keratin 10, two further structural proteins, which are differentially expressed in stratified epithelia, are increased by C/EBPβ (52). Therefore, it would be interesting to investigate whether E2 can also enhance the activity of their promoters.
Previously, it was shown that the papillomavirus E2 protein induces apoptosis in HPV-positive keratinocytes (7, 8, 46). Differentiating keratinocytes show common, but also distinct, features compared with cells undergoing apoptosis (14). In this regard, it was recently hypothesized that E2 might activate a program that helps the cell to terminally differentiate (7). To exert this function in vivo, E2 might employ a variety of strategies and molecules including Sp1, p53, p300, and, as shown here, C/EBP factors.
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to S.S.-H. (Az. FOR 265/2-2). S.S.-H. is supported by the Innovationsprogramm Forschung of the MSWWF Nordrhein-Westfalen.
We thank Corinna Pick and Ute Sandaradura de Silva for excellent technical assistance. We are grateful to G. Steger, M. May, H. Pajunk, R. L. Eckert, S. Kyo, G. Darlington, and F. Thierry for generously providing plasmids used in this study.
REFERENCES
- 1.Agarwal, C., T. Efimova, J. F. Welter, J. F. Crish, and R. L. Eckert. 1999. CCAAT/enhancer-binding proteins. A role in regulation of human involucrin promoter response to phorbol ester. J. Biol. Chem. 274:6190-6194. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., H. Isshiki, T. Sugita, O. Tanabe, S. Kinoshita, Y. Nishio, T. Nakajima, T. Hirano, and T. Kishimoto. 1990. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, J. D., R. Lawande, and P. M. Howley. 1997. Conserved interaction of the papillomavirus E2 transcriptional activator proteins with human and yeast TFIIB proteins. J. Virol. 71:8041-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breiding, D. E., F. Sverdrup, M. J. Grossel, N. Moscufo, W. Boonchai, and E. J. Androphy. 1997. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol. Cell. Biol. 17:7208-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crish, J. F., T. M. Zaim, and R. L. Eckert. 1998. The distal regulatory region of the human involucrin promoter is required for expression in epidermis. J. Biol. Chem. 273:30460-30465. [DOI] [PubMed] [Google Scholar]
- 6.Demeret, C., M. Yaniv, and F. Thierry. 1994. The E2 transcriptional repressor can compensate for Sp1 activation of the human papillomavirus type 18 early promoter. J. Virol. 68:7075-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desaintes, C., C. Demeret, S. Goyat, M. Yaniv, and F. Thierry. 1997. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 16:504-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desaintes, C., S. Goyat, S. Garbay, M. Yaniv, and F. Thierry. 1999. Papillomavirus E2 induces p53-independent apoptosis in HeLa cells. Oncogene 18:4538-4545. [DOI] [PubMed] [Google Scholar]
- 9.Dong, X. P., F. Stubenrauch, E. Beyer-Finkler, and H. Pfister. 1994. Prevalence of deletions of YY1-binding sites in episomal HPV 16 DNA from cervical cancers. Int. J. Cancer 58:803-808. [DOI] [PubMed] [Google Scholar]
- 10.Eckert, R. L., J. F. Crish, and N. A. Robinson. 1997. The epidermal keratinocyte as a model for the study of gene regulation and cell differentiation. Physiol. Rev. 77:397-424. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 12.Enzenauer, C., G. Mengus, A. Lavigne, I. Davidson, H. Pfister, and M. May. 1998. Interaction of human papillomavirus 8 regulatory proteins E2, E6 and E7 with components of the TFIID complex. Intervirology 41:80-90. [DOI] [PubMed] [Google Scholar]
- 13.Erickson, R. L., N. Hemati, S. E. Ross, and O. A. MacDougald. 2001. p300 coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 276:16348-16355. [DOI] [PubMed] [Google Scholar]
- 14.Gandarillas, A., L. A. Goldsmith, S. Gschmeissner, I. M. Leigh, and F. M. Watt. 1999. Evidence that apoptosis and terminal differentiation of epidermal keratinocytes are distinct processes. Exp. Dermatol. 8:71-79. [DOI] [PubMed] [Google Scholar]
- 15.Haller, K., F. Stubenrauch, and H. Pfister. 1995. Differentiation-dependent transcription of the epidermodysplasia verruciformis-associated human papillomavirus type 5 in benign lesions. Virology 214:245-255. [DOI] [PubMed] [Google Scholar]
- 16.Haugen, T. H., L. P. Turek, F. M. Mercurio, T. P. Cripe, B. J. Olson, R. D. Anderson, D. Seidl, M. Karin, and J. Schiller. 1988. Sequence-specific and general transcriptional activation by the bovine papillomavirus-1 E2 trans-activator require an N-terminal amphipathic helix-containing E2 domain. EMBO J. 7:4245-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heike, T., S. Miyatake, M. Yoshida, K. Arai, and N. Arai. 1989. Bovine papilloma virus encoded E2 protein activates lymphokine genes through DNA elements, distinct from the consensus motif, in the long control region of its own genome. EMBO J. 8:1411-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann, A., and R. G. Roeder. 1991. Purification of His-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 19:6337-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu, W., T. K. Kerppola, P. L. Chen, T. Curran, and S. Chen-Kiang. 1994. Fos and Jun repress transcription activation by NF-IL6 through association at the basic zipper region. Mol. Cell. Biol. 14:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang, E. S., D. J. Riese II, J. Settleman, L. A. Nilson, J. Honig, S. Flynn, and D. DiMaio. 1993. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J. Virol. 67:3720-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, D., B. Lee, J. Kim, D. W. Kim, and J. Choe. 2000. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J. Biol. Chem. 275:7045-7051. [DOI] [PubMed] [Google Scholar]
- 22.Lee, K. Y., T. R. Broker, and L. T. Chow. 1998. Transcription factor YY1 represses cell-free replication from human papillomavirus origins. J. Virol. 72:4911-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, R., J. D. Knight, S. P. Jackson, R. Tjian, and M. R. Botchan. 1991. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell 65:493-505. [DOI] [PubMed] [Google Scholar]
- 24.Massimi, P., D. Pim, C. Bertoli, V. Bouvard, and L. Banks. 1999. Interaction between the HPV-16 E2 transcriptional activator and p53. Oncogene 18:7748-7754. [DOI] [PubMed] [Google Scholar]
- 25.Maytin, E. V., and J. F. Habener. 1998. Transcription factors C/EBP alpha, C/EBP beta, and CHOP (Gadd153) expressed during the differentiation program of keratinocytes in vitro and in vivo. J. Investig. Dermatol. 110:238-246. [DOI] [PubMed] [Google Scholar]
- 26.Maytin, E. V., J. C. Lin, R. Krishnamurthy, N. Batchvarova, D. Ron, P. J. Mitchell, and J. F. Habener. 1999. Keratin 10 gene expression during differentiation of mouse epidermis requires transcription factors C/EBP and AP-2. Dev. Biol. 216:164-181. [DOI] [PubMed] [Google Scholar]
- 27.Mink, S., B. Haenig, and K. H. Klempnauer. 1997. Interaction and functional collaboration of p300 and C/EBPβ. Mol. Cell. Biol. 17:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Missero, C., E. Calautti, R. Eckner, J. Chin, L. H. Tsai, D. M. Livingston, and G. P. Dotto. 1995. Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc. Natl. Acad. Sci. USA 92:5451-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Missero, C., F. Di Cunto, H. Kiyokawa, A. Koff, and G. P. Dotto. 1996. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev. 10:3065-3075. [DOI] [PubMed] [Google Scholar]
- 30.Müller, A., A. Ritzkowsky, and G. Steger. 2002. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J. Virol. 76:11042-11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh, H. S., and R. C. Smart. 1998. Expression of CCAAT/enhancer binding proteins (C/EBP) is associated with squamous differentiation in epidermis and isolated primary keratinocytes and is altered in skin neoplasms. J. Investig. Dermatol. 110:939-945. [DOI] [PubMed] [Google Scholar]
- 32.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng, Y. C., D. E. Breiding, F. Sverdrup, J. Richard, and E. J. Androphy. 2000. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J. Virol. 74:5872-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prowse, D. M., L. Bolgan, A. Molnar, and G. P. Dotto. 1997. Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J. Biol. Chem. 272:1308-1314. [DOI] [PubMed] [Google Scholar]
- 35.Purdie, K. J., C. J. Sexton, C. M. Proby, M. T. Glover, A. T. Williams, J. N. Stables, and I. M. Leigh. 1993. Malignant transformation of cutaneous lesions in renal allograft patients: a role for human papillomavirus. Cancer Res. 53:5328-5333. [PubMed] [Google Scholar]
- 36.Ramji, D. P., and P. Foka. 2002. CCAAT/enhancer binding proteins: structure, function and regulation. Biochem. J. 10:561-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rank, N. M., and P. F. Lambert. 1995. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TFIIB. J. Virol. 69:6323-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffner, M., K. Munger, J. C. Byrne, and P. M. Howley. 1991. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc. Natl. Acad. Sci. USA 88:5523-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlegel, R., W. C. Phelps, Y. L. Zhang, and M. Barbosa. 1988. Quantitative keratinocyte assay detects two biological activities of human papillomavirus DNA and identifies viral types associated with cervical carcinoma. EMBO J. 7:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ′mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 17:6419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smola-Hess, S., U. S. de Silva, D. Hadaschik, and H. J. Pfister. 2001. Soluble interleukin-6 receptor activates the human papillomavirus type 18 long control region in SW756 cervical carcinoma cells in a STAT3-dependent manner. J. Gen. Virol. 82:2335-2339. [DOI] [PubMed] [Google Scholar]
- 42.Steger, G., J. Ham, O. Lefebvre, and M. Yaniv. 1995. The bovine papillomavirus 1 E2 protein contains two activation domains: one that interacts with TBP and another that functions after TBP binding. EMBO J. 14:329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steger, G., C. Schnabel, and H. M. Schmidt. 2002. The hinge region of the human papillomavirus type 8 E2 protein activates the human p21(WAF1/CIP1) promoter via interaction with Sp1. J. Gen. Virol. 83:503-510. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson, M., L. C. Hudson, J. E. Burns, R. L. Stewart, M. Wells, and N. J. Maitland. 2000. Inverse relationship between the expression of the human papillomavirus type 16 transcription factor E2 and virus DNA copy number during the progression of cervical intraepithelial neoplasia. J. Gen. Virol. 81:1825-1832. [DOI] [PubMed] [Google Scholar]
- 45.Timchenko, N. A., M. Wilde, M. Nakanishi, J. R. Smith, and G. J. Darlington. 1996. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 10:804-815. [DOI] [PubMed] [Google Scholar]
- 46.Webster, K., J. Parish, M. Pandya, P. L. Stern, A. R. Clarke, and K. Gaston. 2000. The human papillomavirus (HPV) 16 E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. J. Biol. Chem. 275:87-94. [DOI] [PubMed] [Google Scholar]
- 47.Wells, S. I., D. A. Francis, A. Y. Karpova, J. J. Dowhanick, J. D. Benson, and P. M. Howley. 2000. Papillomavirus E2 induces senescence in HPV-positive cells via pRB- and p21(CIP)-dependent pathways. EMBO J. 19:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welter, J. F., J. F. Crish, C. Agarwal, and R. L. Eckert. 1995. Fos-related antigen (Fra-1), junB, and junD activate human involucrin promoter transcription by binding to proximal and distal AP1 sites to mediate phorbol ester effects on promoter activity. J. Biol. Chem. 270:12614-12622. [DOI] [PubMed] [Google Scholar]
- 49.Woodworth, C. D., S. Cheng, S. Simpson, L. Hamacher, L. T. Chow, T. R. Broker, and J. A. DiPaolo. 1992. Recombinant retroviruses encoding human papillomavirus type 18 E6 and E7 genes stimulate proliferation and delay differentiation of human keratinocytes early after infection. Oncogene 7:619-626. [PubMed] [Google Scholar]
- 50.Xiao, H., T. Hasegawa, and K. Isobe. 2000. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J. Biol. Chem. 275:1371-1376. [DOI] [PubMed] [Google Scholar]
- 51.Yao, J. M., D. E. Breiding, and E. J. Androphy. 1998. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J. Virol. 72:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, S., H. S. Oh, M. Shim, E. Sterneck, P. F. Johnson, and R. C. Smart. 1999. C/EBPβ modulates the early events of keratinocyte differentiation involving growth arrest and keratin 1 and keratin 10 expression. Mol. Cell. Biol. 19:7181-7190. [DOI] [PMC free article] [PubMed] [Google Scholar]