Abstract
Replication of the hepatitis C virus (HCV) genome has been proposed to take place close to the membrane of the endoplasmic reticulum in membrane-associated replicase complexes, as is the case with several other plus-strand RNA viruses, such as poliovirus and flaviviruses. The most obvious benefits of this property are the possibility of coupling functions residing in different polypeptidic chains and the sequestration of viral proteins and nucleic acids in a distinct cytoplasmic compartment with high local concentrations of viral components. Indeed, HCV nonstructural (NS) proteins were clearly colocalized in association with membranes derived from the endoplasmic reticulum. This observation, together with the demonstration of the existence of several physical interactions between HCV NS proteins, supports the idea of assembly of a highly ordered multisubunit protein complex(es) probably involved in the replication of the viral genome. The objective of this study, therefore, was to examine all potential interactions between HCV NS proteins which could result in the formation of a replication complex(es). We identified several interacting viral partners by using a glutathione S-transferase pull-down assay, by in vitro and ex vivo coimmunoprecipitation experiments in adenovirus-infected Huh-7 cells allowing the expression of HCV NS proteins, and, finally, by using the yeast two-hybrid system. In addition, by confocal laser scanning microscopy, NS proteins were clearly shown to colocalize when expressed together in Huh-7 cells. We have been able to demonstrate the existence of a complex network of interactions implicating all six NS proteins. Our observations confirm previously described associations and identify several novel homo- and heterodimerizations.
Hepatitis C virus (HCV) is the primary causative agent of parenterally transmitted non-A, non-B hepatitis, which affects a significant part of the world's population (14, 56, 76). Most infected individuals are unable to eliminate the virus, resulting in a persistent infection in about 80% of cases. Chronically infected patients often develop progressive liver disease, cirrhosis, hepatic failure, and hepatocellular carcinoma (71). There is no vaccine against HCV, and current therapies, including a combination of pegylated alpha-interferon (IFN-α) and ribavirin, are curative in only half of patients (25, 52, 97). The development of more effective anti-HCV therapeutic agents is hampered by the lack of an efficient cell culture system and an adequate animal model for HCV infection and replication.
HCV has been classified as belonging to the distinct genus Hepacivirus, in the family Flaviviridae, which also includes the classical flaviviruses with the prototype member Yellow fever virus and the animal pestiviruses, such as Bovine viral diarrhea virus (57, 85). These viruses are characterized by enveloped particles, which carry a single-stranded RNA genome of positive polarity. The HCV genome has a length of about 9,600 nucleotides (nt) and carries a single open reading frame flanked by nontranslated regions. The encoded polyprotein precursor of about 3,000 amino acids is translated via an internal ribosome entry site located in the 5′ nontranslated region (84, 89). The polyprotein is cleaved co- and posttranslationally by cellular and viral proteases into at least 10 different products (for recent reviews, see references 5 and 68). An additional HCV protein, F (for “frameshift protein”) or ARFP (for “alternate reading frame protein”), generated by an overlapping reading frame in the core (C) protein coding sequence, has been proposed (86, 88, 93). The structural proteins C, E1, E2, and p7, located in the amino-terminal region of the polyprotein, are processed by host signal peptidases. C is thought to be the nucleocapsid protein binding to the viral RNA genome (72), and E1 and E2 are the virion envelope glycoproteins (30). The short hydrophobic peptide p7, which separates the structural and nonstructural (NS) proteins, has an unknown function (49).
The NS proteins NS2, NS3, NS4A, NS4B, NS5A and NS5B are thought to be required for replication of the viral genome. Autocleavage at the NS2/3 junction is mediated by a protease activity encoded by the NS2 region and the amino-terminal NS3 protease domain flanking the cleavage site (29, 32). This autocleavage appears to be essential in vivo, as suggested by results obtained with an HCV clone lacking the NS2/3 protease activity which is unable to infect chimpanzees (47). It has been suggested that NS2/3 might be a cysteine protease with His and Cys residues essential for the autocleavage activity (26) or a metalloprotease stimulated by metal ions and inhibited by EDTA (28, 32). NS3 harbors three different enzymatic activities. The 180 amino-terminal residues constitute a chymotrypsin-like serine protease, which mediates the proteolytic release of mature NS4A, NS4B, NS5A, and NS5B (3, 28). The carboxy-terminal remainder possesses nucleoside triphosphatase and helicase activities (43, 78). The NS4A polypeptide functions as an essential cofactor for the NS3 serine protease (4, 19, 50) which is incorporated as an integral component into the amino-terminal β-barrel of the enzyme core (44, 96). The function of the hydrophobic NS4B protein is unknown. However, NS4B seems to be directly involved in the modulation of NS5A hyperphosphorylation (45, 59). Most HCV isolates contain two phosphoprotein variants of NS5A with apparent molecular masses of 56 and 58 kDa, corresponding to the basic and the hyperphosphorylated forms, respectively (40, 69, 82). A major serine phosphoacceptor site has been mapped for a genotype 1a isolate, but this site is not conserved in NS5A proteins of other HCV genotypes (67). The cellular kinase, which mediates NS5A phosphorylation, remains unknown, as is the importance of NS5A phosphorylation for its functions. NS5A protein appears to be involved in resistance of infected cells to the antiviral activity of IFN-α (16, 17, 23, 24). Indeed, at least for some HCV isolates, NS5A is able to bind to PKR, inhibiting the reduction of translation in IFN-treated cells (23, 24). Recently, Shirota et al. (75) showed that NS5A directly interacts with NS5B, the RNA-dependent RNA polymerase (RDRP), and modulates its enzymatic activity in a dose-dependent manner. This is, so far, the first observation supporting the idea that NS5A modulates HCV replication as a component of a replication complex. HCV RDRP NS5B has been extensively characterized biochemically (7, 20, 53, 95) and structurally (1, 10, 48). The protein contains motifs shared by all RDRPs and possesses the classical finger, palm, and thumb subdomains. NS5B has also an intrinsic ability to oligomerize or dimerize, and this oligomerization is prerequisite for RDRP activity (65).
Replication of the HCV genome has been proposed to take place close to the membrane of the endoplasmic reticulum (ER) in membrane-associated replicase complexes (36), as is the case with several other plus-strand RNA viruses, such as poliovirus and flaviviruses (8, 91). In support of this hypothesis, HCV NS proteins have been clearly shown to colocalize within membranes derived from the ER (7, 9, 15, 36, 92). The NS3 protein is directed to the ER via an interaction with its cofactor, NS4A (92), whereas NS2 (73), NS4A, NS4B, NS5A, and NS5B are targeted to the ER even if expressed individually (9, 35, 74, 92). These observations, together with the demonstration of the existence of several physical interactions between HCV NS proteins (22, 37, 51), support the hypothesis of the formation of a highly ordered multisubunit protein complex(es) probably involved in the replication of the viral genome. Indeed, in an in vitro immunoprecipitation experiment, Hijikata et al. (32) coprecipitated NS3 with NS4A, NS4B, NS5A, and NS5B and also NS2 with NS5A and with NS5B. NS4A, acting as an activating cofactor for NS3 protease/helicase, forms a tight complex with NS3 (6, 44, 61, 96) and was shown to associate with NS2 in the yeast two-hybrid system (22). RDRP NS5B also interacts with NS3 as well as with NS4A (37). NS4A, NS4B, and NS5A have been found to form a complex (51). Recently, Shirota et al. (75) have described binding of phosphoprotein NS5A to NS5B and modulation of NS5B activity. Finally, dimerization and/or oligomerization of viral NS proteins NS3 (41) and NS5B (65, 90), which harbor enzymatic activities, were shown as being critical for their functions.
The objective of this study, therefore, was to examine all possible interactions between HCV NS proteins, which are potentially involved in the assembly of a replication complex(es). The experiments were performed with in vitro-transcribed and -translated NS proteins, as well as in and ex vivo, in Saccharomyces cerevisiae, and in hepatic cell lines expressing the NS proteins. We observed a complex network of interactions implicating all six NS proteins, which confirms already described associations and identifies two novel homoassociation and four new heterodimerization events.
MATERIALS AND METHODS
Cell culture and transfection.
Cells of the human hepatoma cell line Huh-7 (58) were kindly provided by R. Bartenschlager (University of Heidelberg, Heidelberg, Germany). Huh-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS), 50 μg of gentamicin per ml, 0.1 mM nonessential amino acids, 2 mM l-glutamine, and 1 mM sodium pyruvate.
Human embryonic kidney cells (HEK 293), obtained from the American Type Culture Collection, were grown in DMEM supplemented with 10% FCS, 50 μg of gentamicin per ml, 0,1 mM nonessential amino acids, and 2 mM l-glutamine. All medium components were purchased from GIBCO-Invitrogen Corporation.
Monolayers of HEK 293 cells were cultured in 60-mm petri dishes and transfected with 10 μg of DNA by using a Profection mammalian transfection system with calcium phosphate (Promega).
Antibodies.
Polyclonal rabbit antiserum LaIV73 against NS2 was produced by immunization of rabbits with recombinant Escherichia coli-expressed fusion protein GST-NS2. Polyclonal rabbit antibodies (PAbs) to NS3, NS4B, and NS5B were kindly provided by R. Bartenschlager (University of Heidelberg, Heidelberg, Germany). The NS5A-specific mouse monoclonal antibody (MAb) 2D9F4 and MAb 2E3C2 against NS4A were kindly provided by C. Jolivet (BioMérieux, Lyon, France). Mouse MAb to NS4B (NS4B-52) was a gift from K. Mishinori (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan), and mouse MAbs against NS3 (1B6.14.2) and NS5B (12B7.54.1) were kindly provided by D. Moradpour (University of Freiburg, Freiburg, Germany). Rabbit polyclonal immunoglobulin G (IgG) to glutathione S-transferase (GST) was purchased from Santa Cruz Biotechnology.
Recombinant adenoviruses. (i) Construction.
The recombinant adenoviral genomes were generated as infectious plasmids by homologous recombination in E. coli as described previously (11). In brief, a ClaI Klenow blunt-ended/MluI DNA fragment from pIV1056 (described in “Plasmid constructions”) encoding the six HCV NS proteins (NS2 to NS5B) (nt 2769 to 9374) was inserted into the adenoviral shuttle plasmid pTG13387. In the resulting vector, pIV1052, NS polyprotein expression is under the control of a cytomegalovirus promoter and its sequence is surrounded by adenoviral sequences (nt 1 to 458 and nt 3328 to 5788 of the adenovirus type 5 [Ad5] genome). PIV1052 was used for homologous recombination with adenoviral sequences of the backbone vector pTG6624 (11). The resulting plasmid, pIV1053, contains the full-length adenoviral genome with a deletion in E3 (nt 28592 to 30470), whereas the E1 region (nt 459 to 3327) is replaced by the sequence encoding the HCV NS proteins. Recombinant adenovirus AdIV1053, containing the viral NS sequence, was generated by transfection of pIV1053 into the 293 complementation cell line after PacI digestion. AdIV1043, used as a negative control, is the same recombinant adenovirus as AdIV1053 in which a single nucleotide insertion in the beginning of the NS2 sequence results in a frameshift mutation.
(ii) Preparation of adenovirus and titration.
Virus propagation and titration by indirect immunofluorescence detection of the adenoviral DNA binding protein were carried out as described by Lusky et al. (55). Briefly, nearly confluent 293 cell monolayers in 75-cm2 flasks were infected with 500 μl of transfected 293 cell supernatants. After 48 h, the nearly completely detached cells and supernatants were collected. Recombinant adenoviruses were recovered by three cycles of flash freeze-thawing. Lysates were cleared by centrifugation at 1,000 × g for 5 min, and virus was stored in complete culture medium at −80°C. Titers of the infectious viral progeny were determined as infectious units by quantitative DNA-binding protein (DBP) immunofluorescence. To determine the IU titer, 293 cells were infected with serial dilutions of virus. This was followed by immunofluorescence staining at 16 h postinfection with B6α72K, an anti-DBP MAb, and quantitation (70).
Plasmid constructions.
All nucleotide numbers given in parentheses refer to the complete HCV-H genome sequence (genotype 1a) encoded by p90/HCV FL-Long pU (46), which was used as a template for the PCRs.
GST fusion protein expression vectors.
EcoRI restriction fragments encoding NS2 (nt 2769 to 3419), NS4B (nt 5475 to 6257), and NS5A (nt 6258 to 7601) were amplified by PCR. Each PCR product was inserted into pGEX-4T-1 (Amersham Pharmacia Biotech), in frame with the GST-encoding sequence, to generate pIV1020, pIV1021 and pIV1022, respectively. pIV320, encoding the GST-CD81 fusion protein, was constructed by ligation of a BamHI/EcoRI restriction fragment which encodes the large extracellular loop of hCD81 (amino acids 116 to 202) into pGEX-4T-1 (42).
Yeast two-hybrid system expression plasmids.
EcoRI/BamHI restriction fragments encoding NS2 (nt 2769 to 3419), NS3 (nt 3420 to 5312), and NS4A (nt 5313 to 6474, where three methionine AUG codons were added at the 5′ terminus and two were added at the 3′ terminus) were obtained by PCR and cloned into pGBKT7 (Clontech), generating pIV1054, pIV1051, and pIV1055. The expression constructs encode fusion proteins between the yeast GAL4 DNA-binding domain (BD) and the NS proteins. EcoRI PCR products encoding NS4B (nt 5475 to 6257) and NS5A (nt 6258 to 7601) were inserted into the EcoRI site of pGBKT7, and the resulting plasmids were designated pIV1024 and pIV1025. A ClaI/BamHI PCR fragment that was blunt-ended with Klenow fragment and that encoded NS5BΔ21, which harbors a deletion of the 21 carboxy-terminal amino acids (nt 7602 to 9311), was ligated into the BamHI-restricted and Klenow blunt-ended pGBKT7 to generate pIV1026. The same NS protein-encoding PCR products were also cloned into pGADT7 (Clontech), in phase with the GAL4 activation domain (AD) sequence, as follows. EcoRI NS2-, NS4B-, and NS5A-encoding PCR products were inserted into the EcoRI site of the cloning vector, generating pIV1027, pIV1028, and pIV1029, respectively. The ClaI/BamHI NS3, NS4A, and NS5BΔ21 PCR fragments were ligated into the ClaI/BamHI sites of pGADT7 to yield expression constructs pIV1030, pIV1035, and pIV1032, respectively.
pIV1056 NS polyprotein expression plasmid used for the construction of the recombinant adenovirus was constructed in three steps. First, a MluI/NheI PCR product, which contains the 114 amino-terminal NS2 residues (nt 2769 to 3419), was cloned into the MluI/NheI sites of pBI-L (Clontech) to give pIV1002. Then, a ClaI PCR product encoding the entire NS5B protein (nt 7602 to 9377) was restricted with NotI/ClaI to generate a 159-nt fragment containing the 52 carboxy-terminal amino acids of NS5B and the stop codon of the polyprotein (nt 9219 to 9377). This fragment was inserted into the NotI/ClaI cloning sites of pIV1002, downstream of the NS2 sequence, to yield pIV1005. Finally, pIV1005, linearized with NheI/NotI, was used for homologous recombination in E. coli with a ClaI/EcoRV restriction fragment of p90/HCV FL-Long pU (nt 708 to 9909). The resulting construct, designated pIV1056, allows the expression of the entire NS polyprotein (nt 2769 to 9374).
Protein expression in E. coli and purification.
To express NS2, NS4B, and NS5A as well as CD81 and GST in E. coli, the pGEX-4T-1 derivative bacterial expression vectors pIV1020, pIV1021, pIV1022, and pIV320 or pGEX-4T-1 were transformed into BL21 cells (Stratagene). Bacterial cultures were grown for 6 h at 20°C, and expression of GST, GST-CD81, GST-NS2, GST-NS4B, and GST-NS5A fusion proteins was induced at 20°C for 90 min with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). These conditions appear to increase the solubility of the fusion proteins. The cells from 500 ml of culture were then harvested and sonicated with a Vibra Cell apparatus (Bioblock Scientific) in 45 ml of phosphate-buffered saline (PBS) (GIBCO-Invitrogen Corporation) supplemented with a cocktail of protein inhibitors (Roche) and 1% Triton X-100. Insoluble materials were pelleted at 15,000 × g for 10 min in a Sorvall SA-600 rotor, and 500 μl of a 50% slurry of glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) was added to the clarified supernatants. The beads were allowed to bind proteins for 1 h, washed three times in PBS, and finally resuspended in 500 μl of PBS supplemented with 1% Triton X-100. All steps were performed at 4°C unless otherwise indicated.
Western blot analysis.
Protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transblotted onto Hybond-P membranes (Amersham Pharmacia Biotech), and probed with anti-NS PAbs (1:50 dilution for anti-NS2 and 1:2,500 for anti-NS3, -NS4B, and -NS5B) or MAbs against NS4A and NS5A (1:1,000 dilution) or GST IgG (1:10,000 dilution) for 1 h at room temperature. After extensive washing, a secondary rabbit-specific antibody conjugated to horseradish peroxidase (diluted to 1:5,000 or to 1:50,000 for the anti-GST PAbs; Sigma) or mouse-specific biotinylated antibody (1:1,000 dilution; Sigma) was applied to the blots for at least 1 h at room temperature. A streptavidin-biotinylated-horseradish peroxidase complex (1:1,000 dilution; Amersham Life Science) was then added for the mouse MAb-probed blots. Antibodies were diluted in 1% skimmed milk in PBS, and blots were washed in 0.1% Tween 20 in PBS. After extensive washing, the reagents for enhanced chemiluminescence (Amersham Pharmacia Biotech) were added as instructed by the manufacturer for 1 min, and signal was detected on an X-ray film (Kodak Biomax).
GST pull-down assays.
NS2, -3, -4A, -4B, -5A, and -5B proteins were in vitro transcribed from the T7 promoter present in pIV1054, pIV1051, pIV1055, pIV1024, pIV1025, and pIV1026 (derivatives of pGBKT7) and translated in the presence of [35S]methionine in TNT T7 coupled reticulocyte lysate system (Promega) as described by the manufacturer. For in vitro binding analyses, 50 μl of GST, GST-CD81, GST-NS2, GST-NS4B, or GST-NS5A proteins on glutathione-Sepharose 4B beads (50% slurry) were preincubated with bovine serum albumin (final concentration, 1 mg/ml) at room temperature for 15 min and then incubated on a rotating device with 10 μl of each in vitro-synthesized NS protein in a final volume of 200 μl in EBC buffer (150 mM NaCl-1% Triton X-100-50 mM Tris-HCl, pH 8.0). The beads were then washed three times in 600 μl of EBC buffer, pelleted at 500 × g for 30 s, and boiled in SDS-PAGE sample buffer. Labeled NS proteins, which bound to the GST-NS fusion proteins or to GST and GST-CD81, were resolved by SDS-PAGE and visualized by autoradiography.
In vitro immunoprecipitation.
The in vitro transcription-translation experiments were performed with rabbit reticulocyte lysates as described above, in the presence or absence of [35S]methionine. Ten microliters of the reaction mixture of each nonradiolabeled NS protein was preincubated at room temperature for 1 h with 10 μl of each [35S]methionine-labeled NS protein in a final volume of 200 μl in EBC buffer. Specific rabbit PAbs or mouse MAbs against the nonlabeled NS protein (1:100 dilution) were preincubated with 20 μl of protein A/G-agarose (Santa Cruz Biotechnology) in a final volume of 200 μl in EBC buffer for 1 h at room temperature. After washing, the preabsorbed antibodies were added to the NS protein reaction mixture and incubated for at least 1 h at room temperature on a rotating device. Immunoprecipitates were washed three times in 600 μl of EBC buffer. The coimmunoprecipitated radiolabeled NS proteins were analyzed directly by SDS-PAGE and autoradiography.
Protein expression and metabolic labeling in adenovirus-infected cells and immunoprecipitation.
Nearly confluent Huh-7 cell monolayers in 60-mm dishes were infected at a multiplicity of infection (MOI) of 75 with the recombinant adenovirus AdIV1043 or AdIV1053. At 65 h postinfection, cells were starved in methionine-, glutamine-, and cysteine-free DMEM (ICN Corporation) for 1 h and labeled for 4 h following the addition of 100 μCi of 35S labeling mix (NEN) per ml. Cells were washed in PBS and lysed in 300 μl of lysis buffer (Promega), and cellular debris were pelleted by centrifugation. Lysates were then precleared by the addition of 30 μl of protein A/G-agarose for 1 h at room temperature. Protein A/G-agarose-preabsorbed specific anti-NS antibodies were then incubated with the precleared cell lysates, and immunoprecipitations were performed as described above. Immunoprecipitates were analyzed by SDS-PAGE and autoradiography and/or by Western blotting.
Confocal laser scanning microscopy.
Nonconfluent Huh-7 cells grown as monolayers on plastic coverslips were infected at an MOI of 75 with the recombinant adenovirus AdIV1043 or AdIV1053. At 65 h postinfection, cells were fixed and permeabilized with 50% methanol-50% acetone at −20°C. After rehydration with PBS supplemented with 1% FCS, cells were incubated for 1 h at room temperature with two primary antibodies, a mouse MAb and a rabbit PAb, specific to two distinct NS proteins. The antibodies used in this experiment were diluted in PBS supplemented with 1% FCS as follows: a 1:100 dilution for the rabbit PAbs against NS2, NS3, NS4B, and NS5B; a 1:100 dilution for the mouse MAbs to NS4A, NS4B and NS5A; and a 1:2 dilution for the mouse MAbs to NS3 and NS5B. After extensive washing, bound primary antibodies were revealed by incubation for 1 h at room temperature in a dark chamber with Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Fluor 568-conjugated goat anti-mouse IgG (Molecular Probes), both diluted to 1:200 in PBS supplemented with 1% FCS. After washing, coverslips were mounted in Gel/Mount (Biomeda Corporation), confocal laser scanning microscopy was performed by using a Zeiss LSM 610 microscope, and images were processed with the Adobe Photoshop 5.0 LE program.
Yeast two-hybrid assays.
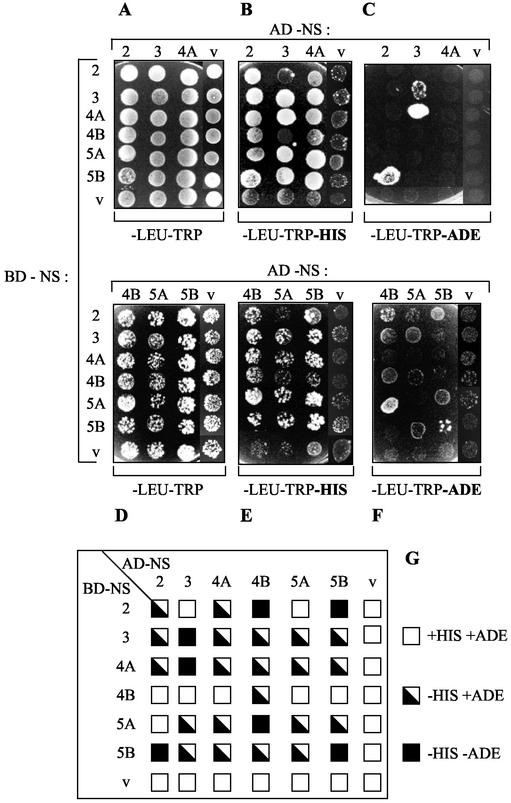
Manipulation of yeast for two-hybrid assays was performed as described in the Matchmaker GAL4 two-hybrid system 3 user manual (Clontech). In this system, the existence of an interaction between two NS proteins was indicated by the activation of the reporter genes HIS3 and ADE2, which allow growth on media lacking histidine (His) and adenine (Ade), respectively. The pGBKT7- and pGADT7-derived constructs encoding HCV NS proteins were cotransformed into AH109 (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ) cells. Cotransformants were grown for about 24 h in medium lacking leucine (Leu) and tryptophan (Trp) and then spotted onto culture plates lacking Trp and Leu to select for cotransformants and onto culture plates lacking Trp, Leu, and His or onto culture plates lacking Trp, Leu, and Ade to allow selection of interactants.
RESULTS
HCV NS proteins form complexes in vitro.
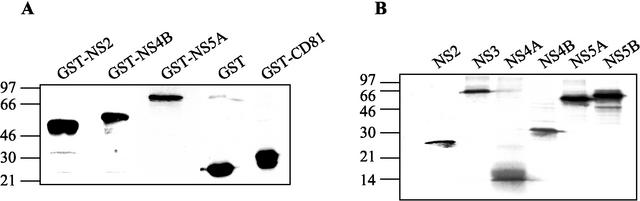
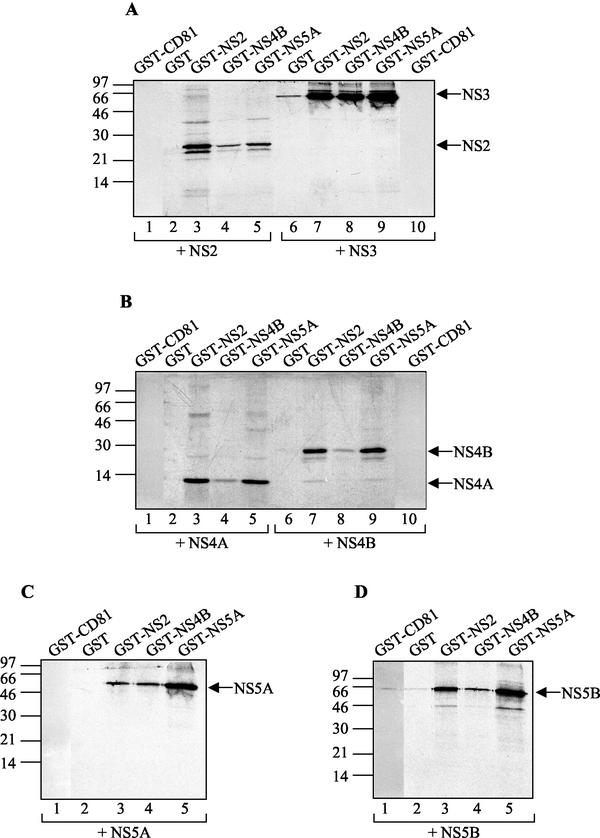
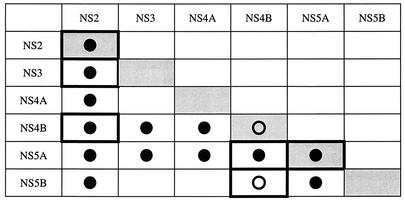
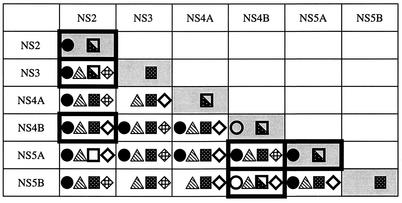
To first test the associations between HCV NS proteins in vitro, we carried out a set of analyses by using a GST pull-down assay. This assay analyzes the binding of a labeled, in vitro-translated NS protein to a purified GST-NS fusion protein bound to glutathione-Sepharose beads. Fusion proteins GST-NS2, GST-NS4B, and GST-NS5A, as well as GST and GST-CD81 (as negative controls), were expressed in E. coli and immobilized on glutathione-Sepharose beads (Fig. 1A). GST and GST-CD81 proteins were much more soluble in bacteria than the three GST-NS fusion proteins and were purified to a considerably higher extent. Thus, only 1/100 of the GST and GST-CD81 material used in the GST pull-down assays was included in the immunoblot (Fig. 1A), whereas the total amounts of the three GST-NS fusion proteins used in the same assays are shown on the blot. All six NS proteins were translated in the presence of [35S]methionine and reacted with bead-bound GST, GST-CD81, GST-NS2, GST-NS4B, and GST-NS5A. The full-length synthesis of the six in vitro-translated NS proteins was confirmed by SDS-PAGE analysis (Fig. 1B), and GST pull-down assays were carried out with approximately equimolar levels of input material (Fig. 2). Analysis of the proteins absorbed on the glutathione-Sepharose beads revealed that NS2 formed a stable complex with GST-NS2 and interacted, if weakly, with GST-NS4B and GST-NS5A but neither with GST alone nor with GST-CD81 (Fig. 2A, lanes 1 to 5). NS3 and NS5A bound specifically to all three GST fusion proteins (Fig. 2A, lanes 6 to 10, and C). NS4A, NS4B, and NS5B all interacted with GST-NS2 and GST-NS5A (Fig. 2B and D), whereas only small amounts of the last three NS proteins bound to GST-NS4B were detected (Fig. 2B, lanes 4 and 8, and D, lane 4, respectively). However, labeled NS4B was strongly retained when reacted with bead-bound GST-NS2 (Fig. 2B, lane 7) or GST-NS5A (Fig. 2B, lane 9). These observations can be explained by the existence of a weak interaction between GST-NS4B and NS4A, NS4B, NS5A, or NS5B and/or by steric hindrance due to the fusion of NS4B to GST. For NS3 and NS5B, it is likely that they also associate with GST alone (Fig. 2A, lane 6, and D, lane 2) and with GST-CD81 (Fig. 2D, lane 1), although much more weakly than with the GST-NS fusion proteins. These weak interactions are nonspecific and certainly due to the much larger amount of control proteins used in the experiment in comparison with those of the GST-NS fusion proteins. Figure 3 summarizes the results of in vitro GST pull-down assays. Several observations can be made. Indeed, we have been able to confirm the previously described interactions of NS4A with NS2 (22), NS4B, and NS5A (51), the association of NS5A with NS2, NS3, and NS5B (32, 75), and NS2/NS5B and NS3/NS4B complex formation (32). In addition, the existence of several novel NS-NS complexes was discovered: NS2 specifically interacted with NS3 and NS4B; NS4B, even in the absence of NS4A, bound directly to NS5A and to NS5B. Lastly, NS2 and NS5A proteins were shown to be able to form stable homodimers.
FIG. 1.
(A) Immunoblot of glutathione-Sepharose-immobilized GST-NS2, GST-NS4B, GST-NS5A, GST, and GST-CD81. Bead-bound bacterially expressed GST-NS fusion proteins (50 μl), as well as GST and GST-CD81 diluted to 1:100, were analyzed by SDS-PAGE followed by immunoblotting with anti-GST antibodies. (B) Autoradiogram from SDS-PAGE analysis of in vitro NS translation products. [35S]methionine-labeled NS proteins (1 μl) were subjected to SDS-PAGE and autoradiography. The positions of molecular size standards are indicated, in kilodaltons.
FIG. 2.
GST pull-down analysis of NS-NS interactions. Each panel shows an autoradiogram from a SDS-PAGE analysis of in vitro-translated NS products selected by binding to the proteins given above the lanes Glutathione-Sepharose-bound GST-CD81 and GST (as controls), GST-NS2, GST-NA4B, and GST-NS5A (50 μl) were incubated with equal amounts of [35S]methionine-labeled in vitro NS translation products, indicated below the panels. Arrows at the right of each panel indicate positions of GST-NS-bound NS proteins. Positions of molecular size standards, in kilodaltons, are shown at the left of each panel.
FIG. 3.
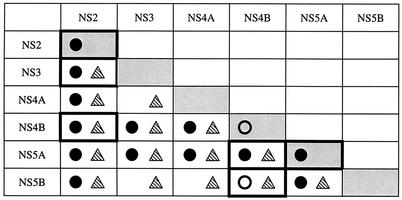
Summary of the GST pull-down assay results. The grid shows strong (•) and weak (○) interactions in the GST pull-down assay. Cells outlined with a boldface box contain new data.
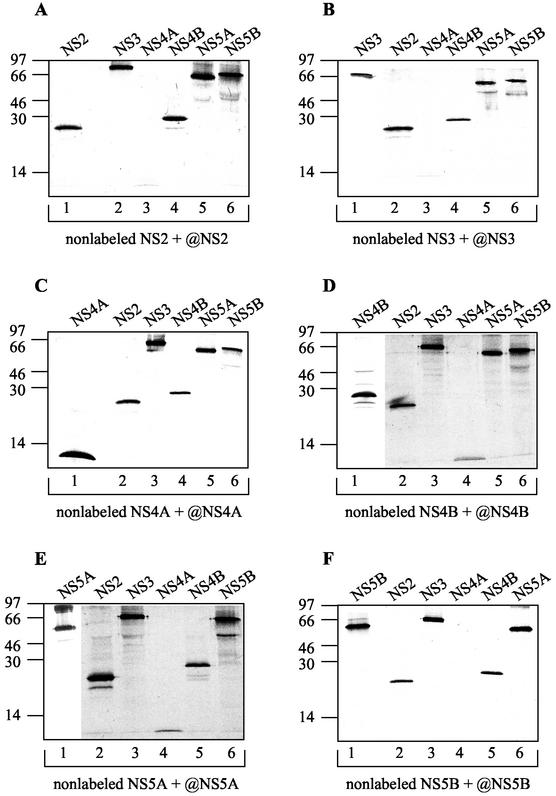
To confirm and extend the results of the GST pull-down assays, we carried out an in vitro coimmunoprecipitation study. In this study we used the six NS proteins, in vitro transcribed and translated in the presence or absence of [35S]methionine. Each translation product was efficiently immunoprecipitated by its specific antibody (polyclonal rabbit antisera to GST-NS2, NS3, NS4B, and NS5B and monoclonal mouse antibodies to NS4A and NS5A) but not by the other antibodies used in this experiment (data not shown). For coprecipitation analysis, in vitro-transcribed but nonlabeled NS2, NS3, NS4A, NS4B, NS5A, or NS5B was incubated with approximately equimolar levels of a radiolabeled NS protein in order to allow the partners to interact and then precipitated with the appropriate antibody against the nonlabeled NS protein. The coprecipitated labeled NS protein was detected following SDS-PAGE and autoradiography. As shown in Fig. 4, all of the antibodies used in this experiment coprecipitated almost all of the NS proteins to some degree. This coprecipitation of NS proteins was certainly the result of preexisting protein-protein interactions. Rabbit polyclonal antisera to NS3, for example, specifically precipitated NS3 (as a positive control) but also NS2, NS4B, NS5A, and NS5B (Fig. 4B), although the signal in the case of NS3/NS4A association was almost undetectable (Fig. 4B, lane 3). This observation is surprising, since detailed knowledge of the biochemical features of this particular complex and its crystal structure has revealed that NS4A is incorporated as an integral component into the amino-terminal β-barrel of the NS3 serine protease core (44, 96). A similar extremely weak signal was detected when NS4A was coprecipitated with antibodies to NS2 and NS5B (Fig. 4A, lane 3, and D, E, and F, lanes 4). In contrast, NS2, NS4B, NS5A, NS5B, and mostly NS3 were efficiently coprecipitated with MAbs to NS4A (Fig. 4C). The immunoprecipitation results, together with those of the GST pull-down assays (Fig. 5), thus confirm the newly observed interactions of NS2 with NS3 and NS4B as well as those of NS4B with NS5A and NS5B. However, this in vitro technique is not suited to protein homodimerization detection and does not allow monitoring of NS2 and NS5A homoassociation.
FIG. 4.
In vitro coimmunoprecipitation of NS proteins. In vitro-translated but nonlabeled NS2, NS3, NS4A, NS4B, NS5A, and NS5B were incubated with equal amounts of each [35S]methionine-labeled NS protein to allow partners to interact. Each interaction mixture was immunoprecipitated with rabbit PAbs to NS2, NS3, NS4B, and NS5B or mouse MAbs against NS4A and NS5A (shown by the “@” symbol). Positions of molecular size standards, in kilodaltons, are shown at the left of each panel.
FIG. 5.
Summary of the GST pull-down assay and in vitro coimmunoprecipitation results. The grid shows strong (•) and weak (○) interactions demonstrated in the GST pull-down assay and coimmunoprecipitation interactions (hatched triangle). Cells outlined with a boldface box contain new NS-NS interactions.
HCV NS protein interaction in vivo.
To extend the results of the in vitro binding assays, we finally carried out NS-NS interaction studies using the yeast two-hybrid system. Immunoblot analysis revealed that each NS protein was efficiently expressed in yeast cells when fused to the GAL4 BD or AD (data not shown). Auxotrophic growth of yeast cells cotransformed with a couple of appropriate NS-encoding plasmids was scored by spotting on plates lacking histidine (Fig. 6B and E) or adenine (Fig. 6C and F). In this experiments, growth on selection media is supported only when the two hybrid NS proteins interact and ensure transcription of the his and ade reporter genes. It is important to note that yeast cotransformants harboring AD-NS4B, AD-NS5A, or AD-NS5B displayed less efficient growth on each selection medium (Fig. 6). As can be seen in Fig. 6B and C, yeast cells expressing all NS protein combinations with the exception of NS2/NS5A (Fig. 6B, lane 5, and E, lane 1) displayed efficient growth on medium lacking His, whereas little or no growth was observed for the vector-cotransformed control strains, suggesting the existence of true physical interactions between the NS proteins. However, it is interesting that several interactions were directional and thus dependent on the fusion of a particular NS protein with either the BD or the AD. For example, BD-NS5A strongly interacted with AD-NS4B (Fig. 6E), while BD-NS4B failed to interact with AD-NS5A (Fig. 6E). In contrast, the NS2/NS4A, NS2/NS5B, NS3/NS4A, NS3/NS5A, NS3/NS5B, NS4A/NS5A, NS4A/NS5B, and NS5A/NS5B associations were not directional (Fig. 6G). Since all fusion proteins were expressed, the mechanism underlying directionality is unclear, but this phenomenon has been observed for numerous protein pairs in the two-hybrid system (18, 21). A further selection on medium lacking Ade was performed in order to identify the interactions of higher affinity and stability. As shown in Fig. 6C and F, NS protein pairs able to direct growth on this selection medium were NS2/NS4B, NS2/NS5B, NS3/NS3, NS3/NS4A, NS4B/NS5A, and NS5B/NS5B. Two of the corresponding interactions, namely, NS2/NS4B and NS4B/NS5B, belong to the group of novel associations discovered in our study, whereas the other three complexes were already described as conferring yeast growth on medium lacking Ade (41, 90). As expected, directionality of some interactions (NS3/NS4A and NS4B/NS5A) was also observed on this selection medium. Thus, HCV NS proteins can also physically interact in living cells. Only few of the observed interactions could be monitored as being of high affinity in the yeast two-hybrid system. The stability of the other complexes was, however, high enough to allow their detection on a less stringent medium. The lack of efficient growth on media lacking His and Ade exhibited by cotransformants containing the BD or AD vector control demonstrates the specificity of the NS-NS interactions.
FIG. 6.
Identification of NS-NS protein interactions by using the yeast two-hybrid system. (A to F) Yeast strain AH109 was cotransformed by the indicated combinations of plasmids encoding AD-NS (at the top) and BD-NS (at left). Cotransformants were spotted onto plates lacking Leu and Trp, lacking Leu, Trp, and His, or lacking Leu, Trp, and Ade, as indicated below the panels. Growth on plates lacking His or Ade is indicative of an interaction between the indicated AD fusion and BD fusion proteins. Empty vectors (v) pGADT7 (encoding GAL4 AD) and pGBKT7 (encoding GAL4 BD) (Clontech) were used as negative controls. (G) Summary of the NS-NS protein interactions in yeast. For each NS protein combination, growth on culture plates lacking His and/or Ade is represented by a square, as indicated at the right of the chart.
NS-NS interaction in the Huh-7 hepatoma cell line.
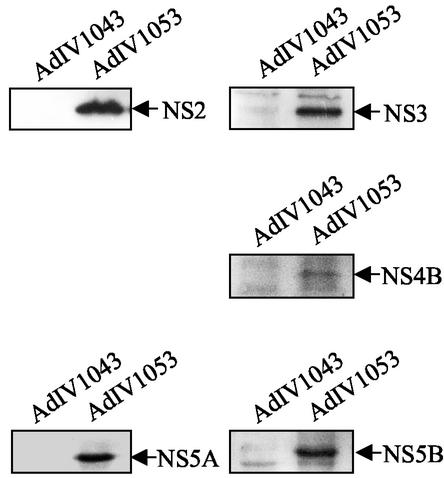
NS-NS protein interactions were finally analyzed in the context of the entire HCV NS polyprotein expressed in Huh-7 human hepatoma cells. A recombinant adenovirus (AdIV1053) allowing the synthesis and processing of the NS polyprotein after infection of Huh-7 cells was constructed. Functional NS polyprotein processing by NS2/3 and NS3 proteases was analyzed by immunoblotting with the panel of available antibodies. As shown in Fig. 7, when Huh-7 cells were infected with AdIV1053, cleavage products of correct sizes and immunoreactivities were found, with the exception of the NS4A protein, whereas no immunoreactivity was detected when the defective recombinant adenovirus AdIV1043 was used. Considering the correct size of surrounding NS4B and NS5A proteins and the importance of NS4A as a cofactor for NS3 proteolytic activity, we surmised that NS4A was also correctly processed in the case of AdIV1053.
FIG. 7.
NS protein expression in Huh-7 cells. Lysates corresponding to 8 × 105 Huh-7 cells infected with AdIV1053 or AdIV1043 (as a control) (MOI, 75) were harvested at 65 h postinfection, loaded onto a gel, separated by electrophoresis, blotted, and probed with rabbit PAbs to NS2, NS3, NS4B, and NS5B and mouse MAbs against NS5A.
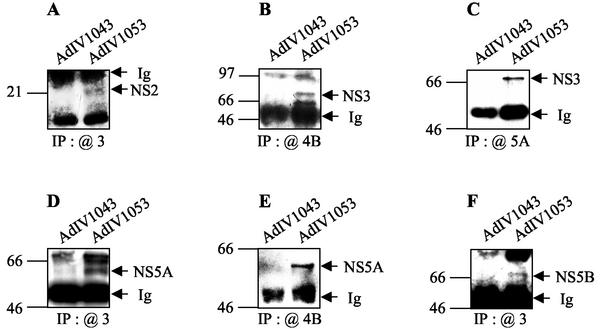
For the coprecipitation experiments, NS proteins expressed in Huh-7 cells were metabolically radiolabeled with [35S]methionine-cysteine, immunoprecipitated with the specific antibodies as described above, and analyzed by immunoblotting (Fig. 8). NS2 coimmunoprecipitated with NS3 (Fig. 8A), NS3 coimmunoprecipitated with NS4B (Fig. 8B) and NS5A (Fig. 8C), NS5A coimmunoprecipitated with NS3 (Fig. 8D) and NS4B (Fig. 8E), and, finally, NS5B coimmunoprecipitated with NS3 (Fig. 8F). In parallel, NS2 also coimmunoprecipitated with NS4A and NS5B (data not shown). In these coimmunoprecipitation experiments in Huh-7 cells expressing all six HCV NS proteins, few interacting partners could be detected. This observation, while disappointing, is not really surprising, considering the fact that in a cellular context NS proteins are tightly anchored in intracellular membranes (9, 15, 35, 36, 92). Indeed, subcellular localization could impede their binding to agarose bead-complexed antibodies in the immunoprecipitation experiments. In addition, the six NS proteins are expressed simultaneously in hepatic cells and are thus likely to interact not only between themselves but also with cellular proteins in order to form highly ordered membranous complexes. It is likely that it is more difficult in this context of cellular complexity to detect a particular association of two NS proteins when both possess multiple interaction partners. However, HCV NS protein expression in adenovirus-infected Huh-7 cells allowed us to confirm the association of NS2 with NS3 and of NS4B with NS5A discovered by other techniques, as well as the previously described complexes NS3/NS4B, NS3/NS5A, and NS3/NS5B.
FIG. 8.
Coimmunoprecipitation of NS proteins expressed in Huh-7 cells. Lysates corresponding to 8 × 105 Huh-7 cells infected with AdIV1053 or AdIV1043 (as a control) (MOI, 75) were harvested at 65 h postinfection. NS protein complexes were coimmunoprecipitated (IP) with rabbit PAbs to NS3 (A, D, and F) or NS4B (B and E) and mouse MAbs against NS5A (C). Coimmunoprecipitated NS proteins were detected by SDS-PAGE and immunoblotting with antibodies to GST-NS2 (A), NS3 (B and C), NS5A (D and E), or NS5B (F). Positions of molecular size standards, in kilodaltons, are shown at the left of each panel.
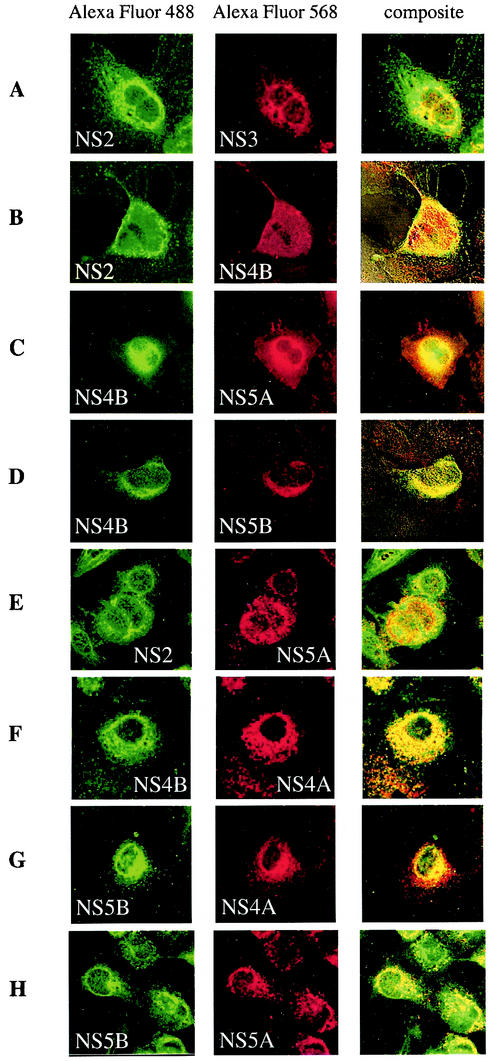
In order to confirm the coimmunoprecipitation data for this cell line and to shed light on interactions seen in the other assays that were not observed with Huh-7 cells, we examined the colocalization of individual interacting pairs in the context of expression of all NS proteins in Huh-7 cells by confocal laser scanning microscopy. In this experiment, Huh-7 cells were infected with AdIV1053 or AdIV1043 (as a control) and subsequently processed for double-labeling immunofluorescence with a large panel of rabbit PAbs and murine MAbs. No immunoreactivity was detected when the cells were infected with the defective adenovirus AdIV1043 (data not shown). The colocalization of all possible combinations of NS proteins was investigated and demonstrated. Figure 9 shows only the results obtained for the four novel interacting partners NS2/NS3 (Fig. 9A), NS2/NS4B (Fig. 9B), NS4B/NS5A (Fig. 9C) and NS4B/NS5B (Fig. 9D), as well as those of four additional NS-NS pairs, NS2/NS5A (Fig. 9E), NS4B/NS4A (Fig. 9F), NS5B/NS4A (Fig. 9G), and NS5B/NS5A (Fig. 9H), whose interaction could not be detected in Huh-7 cells. The antibodies against NS3, -4A, -4B, -5A, and -5B all revealed a reticular staining pattern, which surrounded the nucleus and extended through the cytoplasm. Similar results were previously described for tetracycline-regulated cell lines individually expressing one of these NS proteins (9, 35, 74, 92). Furthermore, these experiments indicate for the first time, to our knowledge, the subcellular localization of NS2 and show a similar reticular cytoplasmic staining pattern. As can be seen in Fig. 9, the NS proteins clearly colocalize when expressed together in Huh-7 cells. This observation confirms the coimmunoprecipitation data and suggests that interactions seen in the other assays but not in Huh-7 cells cannot be excluded in this cellular context. In addition, our study monitors, for the first time, the colocalization of all six NS proteins expressed together in mammalian cells.
FIG. 9.
Colocalization of NS proteins expressed in Huh-7 cells. Huh-7 cells (3 × 104) infected with AdIV1053 (MOI, 75) were processed for double immunolabeling at 65 h postinfection as described in Materials and Methods with rabbit PAbs against NS2 (A, B, and E), NS4B (C, D, and F), or NS5B (G and H) and with mouse MAbs against NS3 (A), NS4A (F and G), NS4B (B), NS5A (C, E, and H), or NS5B (D). Bound primary antibodies were revealed with Alexa Fluor 488- and Alexa Fluor 568-conjugated secondary antibodies, and slides were analyzed by confocal laser scanning microscopy.
DISCUSSION
In this study, we investigated the network of interactions existing among HCV NS proteins which may participate in HCV replication. While numerous associations have already been described in the literature, little information was available about the dimerization properties of NS4B and NS2, as well as about some NS protein homoassociation. Here we describe novel interactions of NS2 with itself and with NS3 and NS4B, as well as direct NS4B association with NS5A and NS5B, and NS5A homodimerization, observed by using a combination of yeast molecular genetic and in vitro and ex vivo techniques. At the same time, the formation of previously described NS complexes was monitored as an internal positive control. Thus, our study provides an exhaustive analysis of all possible interactions between HCV NS proteins, both in vitro and in and ex vivo (Fig. 10).
FIG. 10.
Compilation of GST pull-down assay results, in vitro and ex vivo coimmunoprecipitation results, and two-hybrid system results. Results are depicted as follows: • and ○, strong and weak interaction, respectively, in the GST pull-down assay;  , in vitro coimmunoprecipitation;
, in vitro coimmunoprecipitation;  ,
,  and □, interaction, weak interaction (i.e., directional interaction and/or interaction observed only once on medium lacking His), and no interaction in the two-hybrid system, respectively;
and □, interaction, weak interaction (i.e., directional interaction and/or interaction observed only once on medium lacking His), and no interaction in the two-hybrid system, respectively;  and ◊, coimmunoprecipitation and no coimmunoprecipitation in Huh-7 cells, respectively. Cells outlined with a boldface box contain new data.
and ◊, coimmunoprecipitation and no coimmunoprecipitation in Huh-7 cells, respectively. Cells outlined with a boldface box contain new data.
When considered in the context of HCV replication in living cells, NS2 homoassociation is of particular interest. Indeed, the NS2 protein extends from amino acids 810 to 1026, and autocleavage at the NS2/3 junction at amino acids 1026 to 1027 is mediated by a protease activity which is encoded by the NS2 region and the minimal NS3 protease domain flanking the cleavage site (amino acids 810 to 1206) (27, 31, 33, 64, 66). This autocleavage appears to be essential for productive replication in vivo, as validated with an HCV clone which is devoid of the NS2/3 protease activity and which fails to infect chimpanzees (47). The NS2/3 protease activity is not dependent on an active NS3 protease (27, 31, 33), yet the NS3 protease domain cannot be replaced by another NS protein (73). Since mutagenesis studies have shown that residues His952 and Cys993 are essential for the autocleavage activity (27, 31), it has been suggested that the NS2/3 protease could be a cysteine protease (26). However, the observation that the protease activity is stimulated by metal ions and inhibited by EDTA led to the suggestion that the NS2/3 protease was a metalloprotease (27, 31). Furthermore, sequence comparison with other proteases and studies with classical protease inhibitors (64, 83) have not resulted in a definitive classification. Additionally, NS2/3 autoprocessing seems to be extremely sensitive to the correct folding of the protease (27, 31, 66), and a trans-cleavage activity of an active NS2/3 protease containing a mutant cleavage site has been suggested (66). Recently, Thibeault et al. (83) reported the reconstituted autoprocessing of a purified recombinant NS2/3 protease and suggested that the NS2/3 protease might be considered as an accessory protease, unique among viral proteases in that its only known role in the viral cycle is to autoinactivate.
It is therefore of interest to consider the NS2 capacity to homodimerize in vitro and in vivo in the context of the NS2/3 protease singularity. Oligomerization has been observed for a variety of proteases, including metalloproteases and cysteine proteases. Structural models of the linkage between quaternary state and enzymatic activity are beginning to emerge for a number of these enzymes (12, 34, 39, 77, 80). Perhaps the best-understood example of an activating oligomerization event for a proteolytic enzyme is that of retroviral proteases. Indeed, retroviruses encode a protease which cleaves the structural polyprotein Gag and the enzymatic polyprotein Pol into their constituent mature proteins found in infectious virions. The protease is translated as a domain of the Gag, Gag-Pro, or Gag-Pro-Pol polyprotein, depending on the retrovirus genus (reviewed in references 79 and 87). The first cleavage during maturation is believed to be an autocatalytic event in which the protease domain is excised from the polyprotein. Retrovirus proteases are aspartate proteases with the catalytic site positioned between the two identical subunits that make up the enzymatically active protease dimer. Thus, dimerization is a requisite for enzyme function. Interestingly, it has been suggested that dimerization of the protease domains of two neighboring molecules of the polyprotein is the initial and rate-limiting step of a proteolytic cascade that results in maturation (reviewed in reference 87). In the case of HCV NS2/3 protease, we propose that following co- and posttranslational processing of the amino terminal structural proteins, the rapid intramolecular cleaving reaction between NS2 and NS3 is performed by active dimers of the NS2/3 protease. This homoassociation of neighboring polyprotein molecules could be mediated by the NS2 protein as well as by an interaction between NS2 and NS3, which is also described in this study. The hypothesis that the NS2/3 protease can act as a dimer is strongly suggested by two previous reports. Indeed, Reed et al. (66) have shown that NS2/3 constructs defective in either the NS2 or the NS3 moiety were able to trans-complement each other. The same observation was made for NS2/3 proteins carrying mutations in residue H952 or C993 and therefore unable to self-process, whose association with constructs in which cleavage was prevented through mutagenesis of the NS2/3 cleavage site results in cleavage of the active-site mutant (27, 66). The trans-cleavage activity observed in this study requires the formation of a stable protease complex. In addition, Pallaoro et al. (60) have shown that the rate of the NS2/3 autocleavage reaction in vitro is dependent on the concentration of the NS2/3 precursor protein. Such dependency is explained by assuming that the active species is a dimer. Furthermore, experiments of size exclusion chromatography revealed the existence of species whose molecular mass was compatible with that of NS2/3 dimers (60). The hypothesis of a possible NS2/3 homodimerization needs, of course, to be tested further. In addition, studies are in progress to investigate whether recombinant NS2/3 proteins are also able to self-associate in vitro and in vivo and whether this putative dimerization modulates NS2/3 protease activity.
Another possible role for NS2 homodimers could be participation in the assembly of replication complexes. Although it seems that NS2 is not directly involved in genome replication because functional HCV subgenomic RNAs replicate in cell culture even in the absence of the structural proteins and of NS2 (54), NS2 participation in the replication complex formation as an accessory protein is not excluded. This participation could contribute to the stability of the complex or to the recruitment of cellular factors. Furthermore, we have shown that NS2 is able to interact with all the HCV NS proteins while its only known role is in autocleaving at the NS2/3 junction.
Direct NS2/NS3 interaction is not surprising considering the fact that NS2 and the amino-terminal domain of NS3 together constitute the NS2/3 protease. It will be of interest to investigate whether this association in trans allows the formation of correctly folded and enzymatically functional NS2/3 protease, which will be able to act in trans at an appropriate NS2/3 cleavage site.
NS5A homoassociation is interesting considering the possible role of this protein in subverting host IFN antiviral defense mechanisms (reviewed in reference 81). NS5A seems also to be implicated in regulating translation and to interact directly or indirectly with signal transductions proteins (for a review, see reference 62). It might be justified to revisit the various activities attributed to NS5A considering its capacity to form stable homodimers which appear to be phosphorylation independent, since their detection was possible in vitro (GST pull-down assays) (Fig. 2C). Finally, NS5A homoassociation might be important for the formation of HCV replication complexes.
Three further interactions highlighted in this study all implicate the NS4B protein: NS2/NS4B, NS4B/NS5A, and NS4B/NS5B. NS4B is a relatively hydrophobic protein which contains a transmembrane domain (as does NS2) (73, 94) and colocalizes with the other NS proteins in rough-ER-derived membranes (35). No function has yet been ascribed to NS4B, in HCV or in related pesti- and flaviviruses. Interestingly, NS4B, together with NS3 and NS4A, seems to be directly involved in modulation of NS5A hyperphosphorylation (45, 59). NS4B direct association with NS5A might thus play a role in the regulation of NS5A hyperphosphorylation.
Recently, Egger et al. (15) showed that NS4B induces in cultured cells a distinct membrane alteration, called a membranous web, and colocalizes within this web together with other structural and NS proteins. The membranous web was suggested to be a structure equivalent of the HCV replication complex. Similar intracellular membrane alterations containing all NS proteins as well as HCV plus-strand RNA and newly synthesized viral RNA were also observed in cells harboring subgenomic or full-length replicons (R. Gosert, D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour, presented at the 9th International Meeting on HCV and Related Viruses, abstr. O-13, p. 28, 2002). These data clearly show that the membranous web, whose formation can be induced by NS4B alone, is the site of RNA synthesis and represents the HCV replication complex. In support of this finding, our demonstration of NS4B interaction with RDRP NS5B and with NS5A, which has been found to modulate NS5B enzymatic activity (75), strongly suggests that NS4B is a component of the HCV replication complex which is responsible for specific cytoplasmic membrane alterations but which might also be implicated in the modulation of RDRP NS5B activity. Future work will be aimed at defining whether NS4B is directly responsible for NS5B activation or inhibition and whether NS5A, in the presence of NS4B, can modulate NS5B enzymatic activity.
Since the ER-derived membranous structures appear to be the site of HCV genome replication which contain both the NS proteins and viral RNA (R. Gosert et al., 9th Int. Meet. HCV Relat. Viruses), it is of interest to investigate the mechanisms of viral RNA targeting to these cytoplasmic membranes and its association with the replication complex. Specific interaction of the NS3 helicase with the 3′-terminal sequences of HCV plus- and minus-strand RNA has already been described (2). Those authors suggest a possible role of the NS3/NS4A complex in RNA anchoring to the cytosolic membranes. Additionally, specific interaction of RDRP NS5B and of the 3′ end of viral genomic RNA has also been observed (13, 38). Since the membrane-associated NS proteins NS3 and NS5B, which unwind and direct synthesis of new viral RNA, are also capable of interacting with the other NS proteins, we are currently testing the ability of the NS3 and NS5B complexes to specifically interact with the 3′ ends of the viral positive- and negative-strand RNAs. This study should provide ultimate evidence that HCV NS proteins associate specifically to form complexes implicated in the replication of the HCV genome.
In conclusion, our results clearly demonstrate that all six HCV NS proteins interact among themselves and provide additional evidence that proteins like NS2 and NS4B with still-unknown functions in viral genome replication may also participate in the assembly of replication complexes.
ADDENDUM
While this article was in review, it was reported that NS3 and NS4B physically and functionally interact with RDRP NS5B, NS4B being a negative regulator of the NS3/NS5B complex (63).
Acknowledgments
We gratefully acknowledge Charles M. Rice for providing the p90/HCV FL-Long pU clone, ATM no. PH 13149611755; TRANSGENE S.A (Strasbourg, France) for generously supplying monoclonal antibody anti-DBP and shuttle plasmids pTG13387 and pTG6624 to generate recombinant adenoviruses; Ralf Bartenschlager, Darius Moradpour, Kohara Mishinori, and Colette Jolivet for kindly gift of the large panel of anti-NS antibodies; François Penin for stimulating discussions and critical reading of the manuscript; and Jérôme Mutterer and Mathieu Erhardt for help with confocal laser scanning microscopy. We thank Jean-Daniel Abraham for kindly providing positive control recombinant adenoviruses expressing NS3, NS5A, and NS5B; François Kien for the pIV320 construction; Marie Parnot for excellent technical assistance and sequencing; and Cathy Royer for photographic work.
We are grateful to the HCVacc Cluster, agreement no. QLK2-CT99-00356, for grants to M.D. and the Association de la Recherche pour le Cancer (ARC) and La Ligue Régionale Contre le Cancer for grants to I.I. Grants from ARC (no. 7603), FRM (no. INE20000407028), BNP Paribas Fondation (no. EPB/GP/CL011011), and the Réseau National Hépatites (agreement no. 1A133C) supported this work. The interinstitute confocal microscopy platform used in this study was cofinanced by the Région Alsace, CNRS, the Louis Pasteur University of Strasbourg and ARC.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold. Des. 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, R., and A. Dasgupta. 2001. Specific interaction of hepatitis C virus protease/helicase NS3 with the 3′-terminal sequences of viral positive- and negative-strand RNA. J. Virol. 75:1708-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager, R., and V. Lohmann. 2000. Replication of the hepatitis C virus. Baillieres Best Pract. Res. Clin. Gastroenterol. 14:241-254. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager, R., V. Lohmann, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 8.Bolten, R., D. Egger, R. Gosert, G. Schaub, L. Landmann, and K. Bienz. 1998. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent in situ hybridization. J. Virol. 72:8578-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 10.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, P., and M. Hochstrasser. 1996. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 86:961-972. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, J. C., M. F. Chang, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73:7044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 15.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77-81. [DOI] [PubMed] [Google Scholar]
- 18.Estojak, J., R. Brent, and E. A. Golemis. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Failla, C., L. Tomei, and R. De Francesco. 1995. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J. Virol. 69:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finley, R. L., Jr., and R. Brent. 1994. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc. Natl. Acad. Sci. USA 91:12980-12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flajolet, M., G. Rotondo, L. Daviet, F. Bergametti, G. Inchauspe, P. Tiollais, C. Transy, and P. Legrain. 2000. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene 242:369-379. [DOI] [PubMed] [Google Scholar]
- 23.Gale, M. J., Jr., M. J. Korth, and M. G. Katze. 1998. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin. Diagn. Virol. 10:157-162. [DOI] [PubMed] [Google Scholar]
- 24.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 25.Glue, P., R. Rouzier-Panis, C. Raffanel, R. Sabo, S. K. Gupta, M. Salfi, S. Jacobs, and R. P. Clement, et al. 2000. A dose-ranging study of pegylated interferon alfa-2b and ribavirin in chronic hepatitis C. Hepatology 32:647-653. [DOI] [PubMed] [Google Scholar]
- 26.Gorbalenya, A. E., and E. J. Snijder. 1996. Viral cysteine proteinases. Perspect. Drug Discov. Des. 6:64-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kimura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. USA 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirowatari, Y., M. Hijikata, Y. Tanji, H. Nyunoya, H. Mizushima, K. Kimura, T. Tanaka, N. Kato, and K. Shimotohno. 1993. Two proteinase activities in HCV polypeptide expressed in insect cells using baculovirus vector. Arch. Virol. 133:349-356. [DOI] [PubMed] [Google Scholar]
- 34.Hosfield, C. M., J. S. Elce, P. L. Davies, and Z. Jia. 1999. Crystal structure of calpain reveals the structural basis for Ca2+-dependent protease activity and a novel mode of enzyme activation. EMBO J. 18:6880-6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 36.Hwang, S. B., K. J. Park, Y. S. Kim, Y. C. Sung, and M. M. Lai. 1997. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology 227:439-446. [DOI] [PubMed] [Google Scholar]
- 37.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 38.Ishii, K., Y. Tanaka, C. C. Yap, H. Aizaki, Y. Matsuura, and T. Miyamura. 1999. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology 29:1227-1235. [DOI] [PubMed] [Google Scholar]
- 39.Joshua-Tor, L., H. E. Xu, S. A. Johnston, and D. C. Rees. 1995. Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6. Science 269:945-950. [DOI] [PubMed] [Google Scholar]
- 40.Kaneko, T., Y. Tanji, S. Satoh, M. Hijikata, S. Asabe, K. Kimura, and K. Shimotohno. 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem. Biophys. Res. Commun. 205:320-326. [DOI] [PubMed] [Google Scholar]
- 41.Khu, Y. L., E. Koh, S. P. Lim, Y. H. Tan, S. Brenner, S. G. Lim, W. J. Hong, and P. Y. Goh. 2001. Mutations that affect dimer formation and helicase activity of the hepatitis C virus helicase. J. Virol. 75:205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kien, F., J. D. Abraham, C. Schuster, and M. P. Kieny. 2003. Analysis of the subcellular localization of hepatitis C virus E2 glycoprotein in live cells using EGFP fusion proteins. J. Gen. Virol. 84:561-566. [DOI] [PubMed] [Google Scholar]
- 43.Kim, D. W., Y. Gwack, J. H. Han, and J. Choe. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 215:160-166. [DOI] [PubMed] [Google Scholar]
- 44.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. [DOI] [PubMed] [Google Scholar]
- 45.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 47.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 49.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin, C., J. W. Wu, K. Hsiao, and M. S. Su. 1997. The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J. Virol. 71:6465-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindsay, K. L., C. Trepo, T. Heintges, M. L. Shiffman, S. C. Gordon, J. C. Hoefs, E. R. Schiff, Z. D. Goodman, M. Laughlin, R. Yao, and J. K. Albrecht. 2001. A randomized, double-blind trial comparing pegylated interferon alfa-2b to interferon alfa-2b as initial treatment for chronic hepatitis C. Hepatology 34:395-403. [DOI] [PubMed] [Google Scholar]
- 53.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 55.Lusky, M., M. Christ, K. Rittner, A. Dieterle, D. Dreyer, B. Mourot, H. Schultz, F. Stoeckel, A. Pavirani, and M. Mehtali. 1998. In vitro and in vivo biology of recombinant adenovirus vectors with E1, E1/E2A, or E1/E4 deleted. J. Virol. 72:2022-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller, R. H., and R. H. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. V. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers. 1995. Classification and nomenclature of viruses: sixth report of the International Committee on Taxonomy of Viruses, p. 424-426. Springer Verlag, Vienna, Austria.
- 58.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 59.Neddermann, P., A. Clementi, and R. De Francesco. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J. Virol. 73:9984-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pallaoro, M., A. Lahm, G. Biasiol, M. Brunetti, C. Nardella, L. Orsatti, F. Bonelli, S. Orru, F. Narjes, and C. Steinkuhler. 2001. Characterization of the hepatitis C virus NS2/3 processing reaction by using a purified precursor protein. J. Virol. 75:9939-9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pang, P. S., E. Jankowsky, P. J. Planet, and A. M. Pyle. 2002. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 21:1168-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pawlotsky, J. M., and G. Germanidis. 1999. The non-structural 5A protein of hepatitis C virus. J. Viral Hepat. 6:343-356. [DOI] [PubMed] [Google Scholar]
- 63.Piccininni, S., A. Varaklioti, M. Nardelli, B. Dave, K. D. Raney, and J. E. McCarthy. 2002. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J. Biol. Chem. 277:45670-45679. [DOI] [PubMed] [Google Scholar]
- 64.Pieroni, L., E. Santolini, C. Fipaldini, L. Pacini, G. Migliaccio, and N. La Monica. 1997. In vitro study of the NS2-3 protease of hepatitis C virus. J. Virol. 71:6373-6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin, W., H. Luo, T. Nomura, N. Hayashi, T. Yamashita, and S. Murakami. 2002. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem. 277:2132-2137. [DOI] [PubMed] [Google Scholar]
- 66.Reed, K. E., A. Grakoui, and C. M. Rice. 1995. Hepatitis C virus-encoded NS2-3 protease: cleavage-site mutagenesis and requirements for bimolecular cleavage. J. Virol. 69:4127-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reed, K. E., and C. M. Rice. 1999. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J. Biol. Chem. 274:28011-28018. [DOI] [PubMed] [Google Scholar]
- 68.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 69.Reed, K. E., J. Xu, and C. M. Rice. 1997. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J. Virol. 71:7187-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480-484. [DOI] [PubMed] [Google Scholar]
- 71.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, et al. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santolini, E., L. Pacini, C. Fipaldini, G. Migliaccio, and N. Monica. 1995. The NS2 protein of hepatitis C virus is a transmembrane polypeptide. J. Virol. 69:7461-7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 75.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 76.Simmonds, P., D. B. Smith, F. McOmish, P. L. Yap, J. Kolberg, M. S. Urdea, and E. C. Holmes. 1994. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J. Gen. Virol. 75:1053-1061. [DOI] [PubMed] [Google Scholar]
- 77.Sommerhoff, C. P., W. Bode, G. Matschiner, A. Bergner, and H. Fritz. 2000. The human mast cell tryptase tetramer: a fascinating riddle solved by structure. Biochim. Biophys. Acta 1477:75-89. [DOI] [PubMed] [Google Scholar]
- 78.Suzich, J. A., J. K. Tamura, F. Palmer-Hill, P. Warrener, A. Grakoui, C. M. Rice, S. M. Feinstone, and M. S. Collett. 1993. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 67:6152-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 80.Tamura, T., N. Tamura, Z. Cejka, R. Hegerl, F. Lottspeich, and W. Baumeister. 1996. Tricorn protease—the core of a modular proteolytic system. Science 274:1385-1389. [DOI] [PubMed] [Google Scholar]
- 81.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 82.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thibeault, D., R. Maurice, L. Pilote, D. Lamarre, and A. Pause. 2001. In vitro characterization of a purified NS2/3 protease variant of hepatitis C virus. J. Biol. Chem. 276:46678-46684. [DOI] [PubMed] [Google Scholar]
- 84.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy. The VIIth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 86.Varaklioti, A., N. Vassilaki, U. Georgopoulou, and P. Mavromara. 2002. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 277:17713-17721. [DOI] [PubMed] [Google Scholar]
- 87.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 88.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA. 7:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wolk, B., D. Sansonno, H. G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamaga, A. K., and J. H. Ou. 2002. Membrane topology of the hepatitis C virus NS2 protein. J. Biol. Chem. 277:33228-33234. [DOI] [PubMed] [Google Scholar]
- 95.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 96.Yan, Y., Y. Li, S. Munshi, V. Sardana, J. L. Cole, M. Sardana, C. Steinkuehler, L. Tomei, R. De Francesco, L. C. Kuo, and Z. Chen. 1998. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 Å resolution structure in a hexagonal crystal form. Protein Sci. 7:837-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zeuzem, S., S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666-1672. [DOI] [PubMed] [Google Scholar]