Abstract
The affinity of the origin-binding domain (OBD) of simian virus 40 large T antigen for its cognate origin was measured at equilibrium using a DNA binding assay based on fluorescence anisotropy. At a near-physiological concentration of salt, the affinities of the OBD for site II and the core origin were 31 and 50 nM, respectively. Binding to any of the four 5′-GAGGC-3′ binding sites in site II was only slightly weaker, between 57 and 150 nM. Although the OBD was shown previously to assemble as a dimer on two binding sites spaced by 7 bp, we found that increasing the distance between both binding sites by 1 to 3 bp had little effect on affinity. Similar results were obtained for full-length T antigen in absence of nucleotide. Addition of ADP-Mg, which promotes hexamerization of T antigen, greatly increased the affinity of full-length T antigen for the core origin and for nonspecific DNA. The implications of these findings for the assembly of T antigen at the origin and its transition to a non-specific DNA helicase are discussed.
Small DNA tumor viruses such as simian virus 40 (SV40), polyomavirus, or papillomavirus have become important model systems for the study of molecular events involved in DNA replication. These viruses encode an initiator protein, large T antigen in the case of SV40 and polyomavirus and E1 in the case of papillomavirus, that catalyzes viral DNA replication in conjunction with host replication factors (reviewed in references 3, 6, and 12). These initiator proteins are structurally and functionally related (7, 20). They posses sequence-specific DNA binding and helicase activities that are used, respectively, to recognize the origin and unwind the DNA ahead of the replication fork. These two essential activities map to different domains of these proteins. For SV40 large T antigen and papillomavirus E1, a minimal sequence-specific DNA binding domain has been identified in the middle of the protein, N-terminal to the ATPase/helicase domain (5, 15, 23, 17, 27). The minimal origin-binding domain (OBD) of SV40 large T antigen, located between amino acids 131 and 260 (Fig. 1A), has been studied extensively, and its solution structure has been solved by nuclear magnetic resonance (19). It is structurally similar to the minimal DNA-binding domain (DBD) of the bovine papillomavirus E1 protein despite the fact that both domains share only 6% identity at the primary amino acid sequence level (10). The T-antigen OBD and E1 DBD are monomeric proteins in solution but bind DNA preferentially as dimers to pairs of inverted binding sites in electrophoretic mobility shift assays (EMSA) (5, 15, 19). The fact that EMSA are not performed at true equilibrium and are not suitable to measure weak interactions prompted us to reexamine the binding of the SV40 T antigen OBD to its origin using an assay based on fluorescence anisotropy (13, 18). In this assay, binding of the OBD to a duplex oligonucleotide labeled with fluorescein results in a change in anisotropy that reaches maximum when all of the DNA molecules are bound. This type of assay is performed in solution at equilibrium and is well suited for affinity measurements. Specifically we were interested in measuring the affinity of the T-antigen OBD for pairs of inverted binding sites and for single sites to determine the contribution of dimerization to affinity, similarly to what we have done for the papillomavirus E1 DBD (26).
FIG. 1.
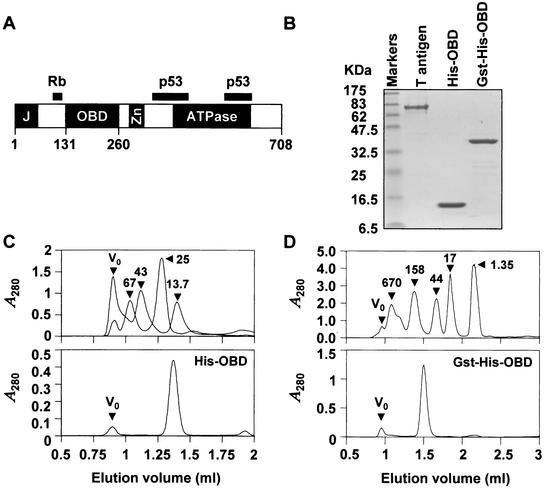
Purified proteins used in this study. (A) Schematic diagram of the 708-amino-acid SV40 large T antigen. The position of the OBD, located between amino acids 131 and 260, is indicated relative that of other functional domains including the J domain (J), the ATPase domain and the zinc finger domain (Zn). Regions of the protein involved in binding to the cell cycle regulatory protein Rb and p53 are indicated as black bars. (B) Sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis of purified proteins stained with Coomassie blue. Aliquots (3 μg) of full-length large T antigen, of polyhistidine-tagged OBD (His-OBD), and of the corresponding GST fusion protein (GST-His-OBD) were analyzed. Concentrations of the His-OBD and Gst-His-OBD proteins were estimated by absorbance at 280 nm in 6 M guanidine hydrochloride using the following molar extinction coefficients: His-OBD = 9,320 M−1cm−1; GST-His-OBD = 50,480 M−1cm−1. The concentration of full-length T antigen was determined by Bradford analysis. (C and D) Gel filtration profiles of His-OBD (C) and Gst-His-OBD (D) proteins. His-OBD (75 μg) and GST-His-OBD (150 μg) were chromatographed on a Superdex 75 PC 3.2/3.0 and a Superdex 200 PC 3.2/3.0 gel filtration column, respectively, using the SMART system (Pharmacia) in a solution containing 25 mM Tris (pH 8.0), 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol. Molecular mass standards in (C) were blue dextran (2,000 kDa), albumin (67.0 kDa), ovalbumin (43.0 kDa), chymotrypsinogen A (25.0 kDa), and RNase A (13.7 kDa). These standards were chromatographed in two groups to increase peak resolution; both elution profiles are superimposed in the figure. Molecular mass standards in panel D were thyroglobulin (670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa). The positions of each standard and of the void volume (V0) are indicated by black triangles. The molecular masses of the His-OBD and of GST-His-OBD proteins calculated from their retention times relative to those of the standards are 15.8 and 92 kDa, respectively. These values are consistent with the His-OBD and of GST-His-OBD proteins being primarily monomeric and dimeric, respectively (molecular mass of a monomer calculated from the primary amino acid sequence is 16.0 kDa for His-OBD and 42 kDa for Gst-His-OBD).
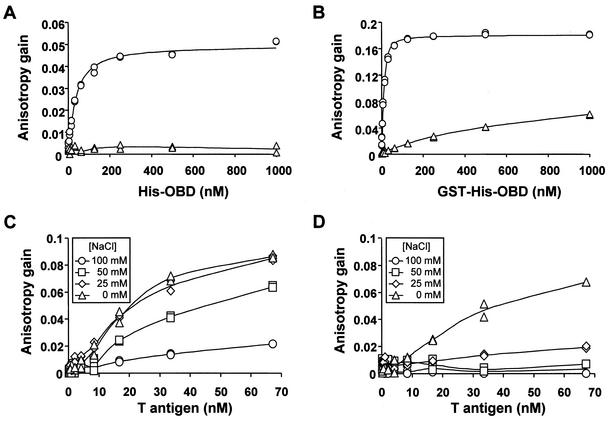
The T antigen OBD (amino acids 131 to 260) was purified from bacteria as a double fusion with glutathione S-transferase (GST) and a hexahistidine tag (Fig. 1B). The GST moiety was subsequently removed by proteolytic cleavage with thrombin, essentially as we reported previously for the papillomavirus E1 DBD (26). The resulting polyhistidine-tagged T antigen OBD (Fig. 1B) was found by analytical gel filtration to be primarily monomeric (Fig. 1C) in solution whereas the GST-His-OBD fusion protein was found to be mostly dimeric (Fig. 1D). We then measured the binding of the His-OBD to a 24-bp, fluorescein-labeled duplex oligonucleotide containing a single T-antigen binding site (TBS) (5′-GAGGC-3′) (Fig. 2A). As a control for specificity we measured binding to a probe in which the pentanucleotide binding site was mutated to 5′-AGAAT-3′ (Fig. 2A). Each DNA duplex was generated by annealing the oligonucleotide that was labeled at its 5′ end with fluorescein via a six-carbon linker (purchased from Genset Corp.) to a complementary unlabeled oligonucleotide, as described previously (26). As can be seen in Fig. 3A titration of the T-antigen OBD in presence of the TBS-containing probe resulted in a dose-dependent change in anisotropy. This gain in anisotropy was due to sequence-specific binding of the OBD to its target site since little change was observed with the control probe. To calculate dissociation constants (Kd), the anisotropy of the probe either in the free state (Af) or at a saturating protein concentration (Ab), as well as the observed anisotropy of each sample (A) and the fractional change in fluorescein quantum yield upon binding (q), were measured and used to determine the fraction of oligonucleotide bound at each protein concentration as described by the following equation: fraction of bound probe = (A − Af)/[(Ab − A)q + (A − Af)]. The fractional change in fluorescein quantum yield was determined as follows: q = (I‖ + 2I⊥)sample/(I‖ + 2I⊥)free, where I‖ and I⊥ are the parallel and perpendicular fluorescence intensities respective to the orientation of the excitation polarizer. Finally the fraction of bound probe was plotted versus the concentration of free protein and the resulting isotherms were fitted by nonlinear least-squares regression with the program GraFit 3.09b (Erithicus Software Ltd.) to the equation describing the following tight binding equilibrium, D + E↔DE (D = duplex DNA; E = T-antigen OBD; DE = DNA-OBD complex): fraction bound (FB) = [(DT + ET + Kd) − {(DT + ET + Kd)2 − (4DTET)}1/2]/(2DT) where total concentrations of T-antigen OBD and fluorescent probe are defined by ET and DT. Duplicates of each binding reaction were prepared in a 96-well plate format and three readings registered on a Victor 2 Multilabel HTS Counter (Wallac) using a 480/535 nm excitation/emission filter set. All sample measurements were corrected for blank fluorescence (buffer only). From the binding isotherms in Fig. 3, dissociation constants of 31 and >1,000 nM were obtained for the binding of the OBD to the TBS probe and the control probe, respectively, and dissociation constants of 1.5 and >1,000 nM were obtained for the binding of GST-His-OBD to the TBS probe and the control probe, respectively.
FIG. 2.
DNA duplexes used in this study. (A) Fluorescent probes and analogous competitor oligonucleotides. The sequences of the fluorescein (F)-labeled strand of the probe containing a single T antigen-binding site (TBS) and of the control probe are indicated. Similarly, the top strand of competitor oligonucleotides containing one, two, or no TBS is indicated. TBS are boxed in black. (B) Oligonucleotides derived form the SV40 origin. The sequence of the 64-bp double-stranded core origin is indicated. The AT-rich and early palindrome regions are boxed. The positions of site II and of the four pentanucleotide TBS are indicated. The bottom of the figure describes the sequence of the top strand of competitor oligonucleotides derived from the 31-bp site II or from the 64-bp core origin. Functional (not mutated) TBS are boxed in black and their numbers are specified in the nomenclature of each oligonucleotide. Site II oligonucleotides in which the spacer region between TBS 1 and 3 has been increased by 1, 2, or 3 bp are referred to as +1, +2, and +3, respectively.
FIG. 3.
DNA-binding activity of the large T-antigen OBD and of full-length large T antigen detected by fluorescence anisotropy. Binding isotherms were generated with purified His-OBD (A) or GST-His-OBD (B) at room temperature in binding buffer (50 mM Tris [pH 7.6], 0.005% NP-40, 1 mM EDTA, 1 mM dithiothreitol) containing 100 mM NaCl and a 15 nM concentration of either a one-TBS-containing probe (circles) or a control probe lacking any TBS (triangles). Binding isotherms generated with large T antigen (C and D) were obtained at room temperature in binding buffer containing the indicated concentration of NaCl and either a one-TBS-containing probe (C) or a control probe lacking any TBS (D).
The fluorescein moiety of the probe can potentially interfere with the binding of the OBD to its target sites. To rule out this possibility and obtain more accurate affinity values, we used competition experiments to measure the affinity of the OBD for unlabeled duplex oligonucleotides containing one or no TBS. The competitor duplex DNAs were titrated in binding reactions performed with the TBS-containing probe and affinity values (Ki) were calculated from the 50% inhibitory concentration (IC50). Each IC50 value was obtained from duplicate inhibition curves fitted by a nonlinear least-square regression using the SAS program package (software release 6.12; SAS Institute Inc., Cary, N.C.). Ki values were calculated using the following equation: FB0 = ([Kd(1 + IC50/Ki) + DT + ET] − {[Kd(1 + IC50/Ki) + DT + ET]2 − (4DTET)}1/2)/DT where FB0 = fraction bound in absence of inhibitor and Ki = dissociation constant of the competitor. This equation is derived from the equilibrium EI↔I + E + D↔DE. By this method, the Ki of the His-OBD was found to be 28 nM for the duplex containing a single TBS and >924 nM for the one devoid of TBS (Table 1). These Ki values are in close agreement to the Kd values determined from direct binding to fluorescent probes, suggesting that the fluorescein moiety does not substantially affect binding.
TABLE 1.
Ki for His- and GST-His-OBD and IC50 for T antigena
| Competitor |
Ki (nM) for:
|
IC50 (nM) for T antigen
|
||
|---|---|---|---|---|
| His-OBD | GST-His-OBD | −ADP-Mg | +ADP-Mg | |
| 0 TBS | >924 | >60 | >3,875 | 237 |
| 1 TBS | 28 | 4 | 53 | 50 |
| 2 TBS | 48 | 0.9 | 60 | 112 |
| Site II 1234 | 31 | 0.5 | 14 | 13 |
| Site II mut | >1,550 | >33 | 3,758 | 75 |
| Site II 1 | 107 | 7 | 47 | 38 |
| Site II 2 | 150 | 11 | 211 | 45 |
| Site II 3 | 57 | 4 | 60 | 21 |
| Site II 4 | 123 | 8 | 159 | 27 |
| Site II 13 | 39 | 0.7 | 18 | 28 |
| Site II 24 | 62 | 1 | 56 | 35 |
| Site II 13+1 | 50 | ND | 33 | ND |
| Site II 13+2 | 58 | ND | 27 | ND |
| Site II 13+3 | 80 | ND | 29 | ND |
| Core | 50 | 0.9 | 19 | 1 |
| Core mut | >376 | >24 | >3,875 | 15 |
Ki values and IC50 were calculated as indicated in the text from inhibition curves obtained with the indicated competitor duplex DNA in binding reactions performed with the one TBS-containing probe. Ki values for the His-OBD and GST-His-OBD proteins were obtained from binding reactions performed in buffer containing 100 mM NaCl. IC50 for T antigen were obtained from binding reactions containing 50 mM NaCl. The concentrations of proteins and probe were, respectively, 100 and 15 nM for His-OBD, 100 and 15 nM for GST-His-OBD, and 34 and 15 nM for full-length T antigen. When indicated, ADP and Mg were used at concentrations of 4 and 7 mM, respectively (ADP-Mg). Values presented are the average of two independent measurements that typically varied by less than 10% from the mean value. ND, not determined.
The OBD was shown by EMSA to bind as a dimer to two inverted pentanucleotides binding sites separated by 7 bp (15, 16). To assess the contribution of dimerization to affinity, we measured the binding of the OBD to this arrangement of sites. In competition experiments, the affinity of the OBD for two TBS (Ki =48 nM) was found to be comparable to that for a single site (Ki = 28 nM) suggesting that dimerization of the OBD has little effect on affinity. To extend this finding we performed competition experiments with a 31-bp duplex oligonucleotide corresponding to site II of the SV40 origin. Site II was shown previously to be necessary and sufficient for the binding of the OBD to the origin (16). This sequence contains four TBS arranged in two pairs of inverted sites (sites 1 and 3 and sites 2 and 4, Fig. 2B), which could each potentially bind an OBD dimer. Mutant versions of site II in which three or two of the four binding sites were inactivated by mutations were also used as competitors (Fig. 2B). A sequence in which all four sites were mutated served as a specificity control. The affinity of the OBD for site II (Ki = 31 nM) was found to be comparable to that for the longer 64-bp core origin (Fig. 2) (Ki = 50 nM) but only two- to fivefold higher than its affinity for any of the single pentanucleotide, which varied between 57 and 150 nM. Of the four TBS, site 3 was the highest-affinity binding site. Since all four pentanucleotides have the same sequence, we surmised that residues flanking each site are responsible for their slightly different affinity. Binding to pairs of pentanucleotides, either to a combination of sites 1 and 3 (Ki = 39 nM) or 2 and 4 (Ki = 62 nM) was only two- to threefold stronger than to the corresponding individual sites. These results suggest that the inhibitory activity of each oligonucleotide is related primarily to the number of TBS it contains, irrespective of how these sites are arranged relative to each other. In further support of this notion, we found that the affinity of the OBD for the combination of sites 1 and 3 decreased by twofold or less when the 7-bp spacer region between both sites was lengthened by 1 to 3 bp (Fig. 2B and Table 1). Collectively these results reinforce the notion that dimerization of the OBD on pairs of binding sites contributes little to affinity.
We were surprised by the findings presented above since we expected that dimerization of the T-antigen OBD would increase substantially its affinity for DNA, as we have observed for the related DBD of the papillomavirus E1 helicase (26). As a control experiment to verify that dimerization of the T-antigen OBD would indeed increase its affinity for DNA, we performed binding experiments with the GST-His-OBD fusion protein. In this protein context the OBD is artificially dimeric (Fig. 1D) because GST itself is a dimer. In experiments measuring the direct binding of GST-His-OBD to fluorescent probes (Fig. 3B), or in competition experiments (Table 1), this protein was found to bind DNA in a sequence-specific manner with an approximately 15-fold-higher affinity for a single TBS than the monomeric OBD. Furthermore, its affinity for two TBS, or for site II and the core origin, was approximately 50-fold higher than that of the monomeric OBD. As expected, the GST-His-OBD fusion proteins showed a substantially higher affinity for pairs of pentanucleotides relative to the corresponding single sites. For example, its affinity for the combination of sites 2 and 4 (Ki = 1.0 nM) was approximately 10-fold higher that its affinity for site 2 (Ki = 11 nM) or site 4 (Ki = 8 nM). This is in contrast to the monomeric His-OBD, which showed only a two- to threefold-higher affinity for sites 2 and 4 (Ki = 62 nM) than for any of these two individual sites (site 2 Ki = 150 nM, site 4 Ki = 123 nM). Collectively, these results indicate that artificial dimerization of the OBD by fusion with GST does increase its affinity for two TBS. From these results one might expect that dimerization of the monomeric OBD, if it were strong, should increase substantially its affinity for two TBS relative to one. Since this was not observed, we surmise that the OBD does not dimerize upon binding to DNA, or dimerizes only weakly, but rather binds primarily in an independent manner to each TBS under our assay conditions. On the basis of these results, we suggest that the OBD dimer observed in EMSA (15) was formed predominantly by the independent binding of two OBD molecules to two adjacent inverted pentanucleotides. Their subsequent cross-linking would have generated an artificial dimer whose higher affinity for site II would have allowed it to resist the harsh conditions of EMSA. In support of this suggestion we verified that artificial dimerization of the OBD, in our case by fusion with GST, does increase its affinity for pairs of binding sites. The fact that cross-linking was essential to observe binding of the ODB to pairs of binding site in EMSA and that binding to a single site could not be observed even with cross-linking (15) further supports our suggestion.
For comparison with the OBD, we performed binding studies with full-length T antigen immunopurified as described previously (24) from baculovirus-infected insect cells (Fig. 1B). The experiments with the isolated OBD were performed in buffer containing 100 mM NaCl so we first attempted to detect binding of full-length T antigen to fluorescent probes under the same salt condition. As can be seen in Fig. 3C and D, T antigen was able to bind to the TBS-containing probe in a sequence-specific manner but with a lower affinity than that of the OBD. In an attempt to increase binding of T antigen to the probe, we performed binding isotherms at lower salt concentrations, either 25 or 50 mM NaCl, or in absence of NaCl. As expected, binding to the TBS-containing probe (Fig. 3C) and to the control probe (Fig. 3D) was increased at lower salt concentrations. We chose to perform all subsequent experiments at 50 mM NaCl since at this concentration binding of T antigen to the TBS-containing probe was increased while binding to the control-probe remained low. Under these conditions, binding of T antigen was sufficiently weak that saturation of the TBS-containing probe could not be reached at the highest protein concentration tested. This prevented us from obtaining a Kd and consequently from calculating Ki values in subsequent competition experiments. Nevertheless, it was still possible to compare the affinities of T antigen for different duplex DNA carrying one, two, or no TBS, or for site II and the core origin by comparing the IC50 of each competitor (Table 1). IC50 values were determined both in absence or presence of ADP-Mg in the binding reactions. ADP was used instead of ATP because ADP can promote assembly of hexamers and double hexamers, but unlike ATP, it does not support DNA unwinding which would complicate interpretation of the results (8, 21). Several observations were made. First in the absence of ADP-Mg, T antigen bound with a comparable affinity to a competitor 24-bp duplex carrying two TBS (IC50=60 nM) as to one carrying a single TBS (IC50=53 nM). Similar results were also obtained with the 31-bp site II oligonucleotide. In this case binding to pentanucleotide pairs 1 and 3 or 2 and 4 was approximately 2.5 to 3.5 times stronger than to the corresponding individual sites in absence of ADP-Mg. Furthermore the affinity of T antigen for binding sites 1 and 3 decreased by less than twofold when the spacing between both sites was increased by 1 to 3 bp (Table 1). These results are consistent with the notion that full-length T antigen binds primarily in an independent manner to all four binding sites within the 31-bp site II oligonucleotide, similarly to what we observed for the isolated OBD.
When the experiments described above were repeated in presence of ADP-Mg, two main observations were made. First, we found that ADP-Mg increased the affinity of full-length T antigen for the longer core origin by approximately 20-fold but had little effect on the affinity for the 31-bp site II oligonucleotide. Specifically the IC50 of the core origin changed from 19 to 1 nM upon addition of ADP-Mg. Second, ADP-Mg dramatically increased nonspecific binding of T antigen to DNA, as observed for the three control probes lacking any TBS (0 TBS, site II mut, and core mut). ADP-Mg increased nonspecific binding to the mutant site II and the mutant core origin by 50- and >250-fold, respectively. By doing so, ADP-Mg greatly reduced the ability of T antigen to discriminate between wild type and mutant site II or the core origin. For example, the difference in affinity between the wild type and mutant core origin that was >200-fold in absence of nucleotide was reduced to 15-fold in presence of ADP-Mg. A possible mechanism by which ADP-Mg could increase the binding of T antigen to the longer core origin and to nonspecific DNA would be by promoting the oligomerization of the protein and/or the interaction of the C-terminal helicase/ATPase domain with DNA. Consistent with this proposal is the observation that ATP increases the size of the T-antigen footprint on the origin (2, 9). Note that although our results suggest that T antigen binds to its recognition sites primarily in an independent manner in absence of ADP, they do not address whether the increased binding to the core origin and to nonspecific DNA induced by ADP is due to cooperative binding. Although this is a very likely possibility, additional experiments are required to address this question.
Concluding remarks.
In this study we found that the T-antigen OBD binds primarily in an independent manner to the four TBS in the SV40 origin. This is in contrast to the related papillomavirus E1 DBD, whose affinity for its cognate origin or for pairs of inverted binding sites is approximately 10-fold higher than that for a single site and is dependent on dimerization of the protein and on having the correct spacing between both binding sites (26). Interestingly, we note that the affinity of the T-antigen OBD for a single TBS (Ki between 57 and 150 nM) is comparable to that of a dimer of the E1 DBD for two E1 binding sites (Ki= 80 nM for the HPV11 E1 DBD at 100 mM NaCl [26]). Therefore, it may be that dimerization is necessary for papillomavirus E1 to form a stable protein-DNA complex at the origin whereas it is dispensable for T antigen.
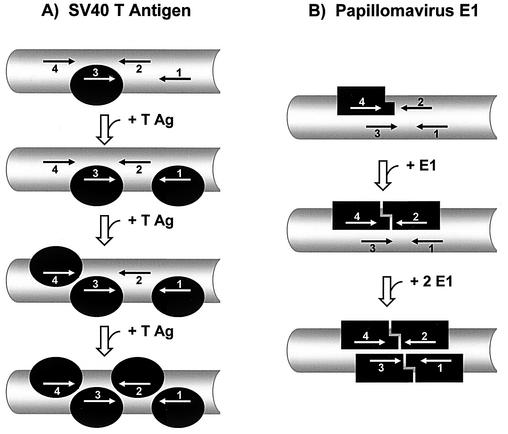
The affinities measured in this study are consistent with a two-step model of the assembly of T antigen at the origin. In the first step, the T-antigen OBD or the full-length protein in absence of ADP-Mg would bind to all four pentanucleotides in site II, predominantly in an independent manner. It is likely that binding to all four sites is necessary for proper assembly since double hexamers formed on only two of the four TBS, either a combination of sites 1 and 3 or 2 and 4, do not catalyze unwinding (14, 25). Formation of a tetramer of T antigen at the origin would make the T-antigen system similar to that of the related papillomavirus E1 helicase, which was shown to assemble at the origin via a tetrameric intermediate (11). An important difference however would be that formation of the T-antigen tetramer would proceed by the sequential and independent binding of four monomers to the origin, in sharp contrast to the E1 tetramer which is assembled from two independent dimers (11) (Fig. 4).
FIG. 4.
Model for the assembly of SV40 T antigen at the origin and comparison with the assembly of the related papillomavirus E1. (A) Assembly of the T-antigen OBD on site II. The DNA is represented in gray. The four pentanucleotides TBS are indicated by arrows and numbered according to Fig. 2. The figure shows the stepwise, independent binding of four OBD (colored in black) onto the four TBS in site II. Assembly is shown to proceed stepwise starting with binding of the OBD to the highest affinity site (site 3) and ending with binding to the weakest site (site 2). (B) Assembly of the papillomavirus E1 DBD at its cognate origin. The four E1 binding sites found in many papillomavirus origins (4) are indicated by arrows as two overlapping pairs of inverted repeats. Assembly is shown to begin by the binding of a monomer of the E1 DBD (black box) to the highest-affinity binding site, site 4 for HPV11 (26). The affinity of a monomer for the origin is weak (Ki = 850 nM for the HPV11 E1 DBD at 100 mM NaCl [26]), but its binding is increased approximately 10-fold upon dimerization (for a dimer of the HPV11 E1 DBD Ki = 80 nM at 100 mM NaCl [26]). Assembly is shown to proceed via the cooperative binding of two E1 DBD on sites 2 and 4 to form an initial stable dimer. This is followed by the assembly of a second dimer on the weaker affinity sites 1 and 3 (26). A recent crystal structure of the tetrameric E1 DBD-DNA complex revealed that both dimers do not interact with each other significantly (11). The low affinity and specificity of E1 for its target site are such that, in vivo, dimerization of full-length E1 probably occurs only if the protein is recruited to the origin by E2, a transcription/replication factor with high affinity for the origin.
In the second step following formation of this tetrameric intermediate, ADP-Mg would promote the assembly of T antigen into a higher-affinity complex on the core origin (IC50 = 1 nM). Our results indicate that assembly of this high-affinity complex requires sequences outside of site II, in agreement with a previous report indicating that T antigen binds more tightly to the 64-bp core origin than to site II (15). Importantly, we found that ADP-Mg reduces the DNA binding specificity of T antigen. Specifically, the difference in affinity between the wild type and the mutant core origin, which was greater than 200-fold in absence of nucleotide, was reduced to only 15-fold in the presence of ADP-Mg. This ADP-stimulated step likely corresponds to the formation of hexamers and double-hexamers on the origin whose higher-affinity for nonspecific DNA is consistent with their role as a nonspecific DNA helicase. It will be interesting in the future to investigate the effect of phosphorylation of T antigen, in particular of threonine 124, on the affinity of T antigen for its origin since phosphorylation of this residue promotes double hexamer formation and unwinding activity (1, 22, 28).
Acknowledgments
We thank Peter Bullock, Craig Fenwick, and Steve Mason for critical reading of the manuscript.
REFERENCES
- 1.Barbaro, B. A., K. R. Sreekumar, D. R. Winters, A. E. Prack, and P. A. Bullock. 2000. Phosphorylation of simian virus 40 T antigen on Thr 124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J. Virol. 74:8601-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borowiec, J. A., and J. Hurwitz. 1988. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 7:3149-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 4.Chen, G., and A. Stenlund. 2001. The E1 initiator recognizes multiple overlapping sites in the papillomavirus origin of DNA replication. J. Virol. 75:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, G., and A. Stenlund. 1998. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J. Virol. 72:2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, L. T., and T. R. Broker. 1994. Papillomavirus DNA replication. Intervirology 37:150-158. [DOI] [PubMed] [Google Scholar]
- 7.Clertant, P., and I. Seif. 1984. A common function for polyoma virus large-T and papillomavirus E1 proteins? Nature 311:276-279. [DOI] [PubMed] [Google Scholar]
- 8.Dean, F. B., M. Dodson, H. Echols, and J. Hurwitz. 1987. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc. Natl. Acad. Sci. (USA). 84:8981-8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deb, S. P., and P. Tegtmeyer. 1987. ATP enhances the binding of simian virus 40 large T antigen to the origin of replication. J. Virol. 61:3649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enemark, E. J., G. Chen, D. E. Vaughn, A. Stenlund, and L. Joshua-Tor. 2000. Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol. Cell 6:149-158. [PubMed] [Google Scholar]
- 11.Enemark, E. J., A. Stenlund, and L. Joshua-Tor. 2002. Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex. EMBO J. 21:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fanning, E., and R. Knippers. 1992. Structure and function of simian virus 40 large T antigen. Annu. Rev. Biochem. 61:55-85. [DOI] [PubMed] [Google Scholar]
- 13.Heyduk, T., Y. Ma, H. Tang, and R. H. Ebright. 1996. Fluorescence anisotropy: rapid, quantitative assay for protein-DNA and protein-protein interaction. Methods Enzymol. 274:492-503. [DOI] [PubMed] [Google Scholar]
- 14.Joo, W. S., H. Y. Kim, J. D. Purviance, K. R. Sreekumar, and P. A. Bullock, P. A. 1998. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol. Cell. Biol. 18:2677-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joo, W. S., X. Luo, D. Denis, H. Y. Kim, G. J. Rainey, C. Jones, K. R. Sreekumar, and P. A. Bullock. 1997. Purification of the simian virus 40 (SV40) T antigen DNA-binding domain and characterization of its interactions with the SV40 origin. J. Virol. 71:3972-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, H. Y., B. A. Barbaro, W. S. Joo, A. E. Prack, K. R. Sreekumar, and P. A. Bullock. 1999. Sequence requirements for the assembly of simian virus 40 T antigen and the T-antigen origin binding domain on the viral core origin of replication. J. Virol. 73:7543-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leng, X., J. H. Ludes-Meyer, and V. G. Wilson. 1997. Isolation of an amino-terminal region of bovine papillomavirus type 1 E1 protein that retains origin binding and E2 interaction capacity. J. Virol. 71:848-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lundbald, J. R., M. Laurence, and R. H. Goodman. 1996. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 10:607-612. [DOI] [PubMed] [Google Scholar]
- 19.Luo, X., D. G. Sanford, P. A. Bullock, and W. W. Bachovchin. 1996. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat. Struct. Biol. 3:1034-1039. [DOI] [PubMed] [Google Scholar]
- 20.Mansky, K. C., A. Batiza, and P. F. Lambert. 1997. Bovine papillomavirus type 1 E1 and simian virus 40 large T-antigen share regions of sequence similarity required for multiple functions. J. Virol. 71:7600-7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mastrangelo, I. A., P. V. Hough, J. S. Wall, M. Dodson, F. B. Dean, and J. Hurwitz. 1989. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338:658-662. [DOI] [PubMed] [Google Scholar]
- 22.McVey, D., B. Woelker, and P. Tegtmeyer. 1996. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J. Virol. 70:3887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarafi, T. R., and A. A. McBride. 1995. Domains of the BPV-1 E1 replication protein required for origin-specific DNA binding and interaction with the E2 transactivator. Virology 211:385-396. [DOI] [PubMed] [Google Scholar]
- 24.Simanis, V., and D. P. Lane. 1985. An immunoaffinity purification procedure for SV40 large T antigen. Virology 144:88-100. [DOI] [PubMed] [Google Scholar]
- 25.Sreekumar, K. R., A. E. Prack, D. R. Winters, B. A. Barbaro, and P. A. Bullock, P. A. 2000. The simian virus 40 core origin contains two separate sequence modules that support T-antigen double-hexamer assembly. J. Virol. 74:8589-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Titolo, S., K. Brault, J. Majewski, P. W. White, and J. Archambault. 2003. Characterization of the minimal DNA binding domain of the human papillomavirus E1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein. J. Virol. 77:5178-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Titolo, S., A. Pelletier, A. M. Pulichino, K. Brault, E. Wardrop, P. W. White, M. G. Cordingley, and J. Archambault. 2000. Identification of domains of the human papillomavirus type 11 E1 helicase involved in oligomerization and binding to the viral origin. J. Virol. 74:7349-73461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. 1999. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 73:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]