Abstract
Viral RNA-dependent RNA polymerases exhibit great sequence diversity. Only six core amino acids are conserved across all polymerases of positive-strand RNA viruses of eukaryotes. While exploring the function of one of these completely conserved residues, asparagine 297 in the prototypic poliovirus polymerase 3Dpol, we identified three viable mutants with noncanonical amino acids at this conserved position. Although asparagine 297 could be replaced by glycine or alanine in these mutants, the viruses exhibited Mn2+-dependent RNA replication and viral growth. All known RNA polymerases and replicative polymerases of bacterial, eukaryotic, and viral organisms are thought to be magnesium dependent in vivo, and therefore these mutant polioviruses may represent the first viruses with a requirement for an alternative polymerase cation. These results demonstrate the extreme functional flexibility of viral RNA-dependent RNA polymerases. Furthermore, the finding that strictly conserved residues in the nucleotide binding pocket of the polymerase can be altered in a manner that supports virus production suggests that drugs targeting this region of the enzyme will still be susceptible to the problem of drug-resistant escape mutants.
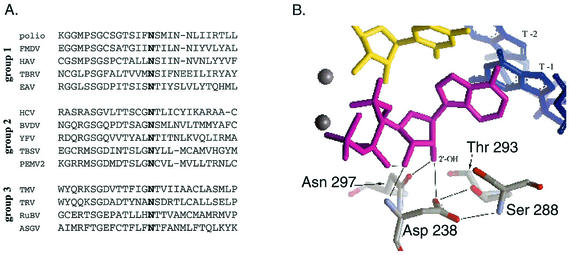
RNA viruses exhibit extreme genetic diversity and an extraordinary ability to evolve in new environmental conditions. The genetic diversity of RNA viruses is most evident when the protein-coding sequences of related family members (for example, the positive-strand RNA viruses of eukaryotes) are analyzed. The RNA-dependent RNA polymerase protein-coding sequence is the only protein-coding sequence with clear sequence homology across this class of viruses (1, 18, 19). Even within the RNA-dependent RNA polymerase protein-coding sequence (∼300 to 500 amino acids in the core catalytic domain), there is an enormous variety of sequences, with only six residues being conserved across all species of positive-strand RNA viruses of eukaryotes (18). With the poliovirus polymerase 3Dpol sequence as a reference, those six completely conserved amino acids are lysine 159, glycine 289, aspartic acid 233, aspartic acid 238, asparagine 297 (Fig. 1A), and the two aspartic acids (positions 328 and 329) of the canonical GDD motif (the SDD motif in coronaviruses) (12). The functions of these residues have not been fully elucidated. Lysine 159 has been proposed to interact with and stabilize the triphosphate moiety of the incoming nucleoside triphosphate (NTP). Glycine 289 is part of the NTP binding pocket. Aspartic acids 233 and 328 are involved in the coordination of the two magnesium cations (Mg2+) essential for catalyzing the incorporation of the incoming NTP (5a, 10). Aspartic acid 238 most likely binds and positions the 2′ and 3′ hydroxyls (OH) of the incoming NTP, linking sugar selection to catalysis (10). Asparagine 297 appears to assist in NTP binding (likely at the 2′OH) and may facilitate discrimination between NTPs and dNTPs (10) (Fig. 1B).
FIG. 1.
Conserved asparagine 297. (A) The asparagine at position 297 of the poliovirus RNA-dependent RNA polymerase (3Dpol) is absolutely conserved among all eukaryotic positive-strand RNA viruses, in all three supergroups. The viruses indicated are from reference 18. (B) Asparagine 297 is postulated to play an important role in nucleotide selection in the NTP binding site. By hydrogen bonding with the 2′ hydroxyl of incoming NTPs, the asparagine positively selects for NTPs and discriminates against incorrect dNTPs (10). The NTP is in magenta, the primer strand is in yellow, the template is in blue, magnesium ions (Mg2+) are grey spheres, and 3Dpol residues are in grey, with oxygens in red and nitrogens in blue.
As these residues are the only six amino acids conserved across all species of positive-strand RNA viruses of eukaryotes, they are each expected to be critical for the function of the polymerase. Thus, it was not surprising to find that mutations engineered into these positions of the poliovirus polymerase 3Dpol have generally resulted in loss of infectivity (10, 15, 16, 22). Recently, one exception to this prediction was found at position 297: changing the asparagine to an aspartic acid resulted in a minimally viable, highly temperature-sensitive virus that produced minute plaques after 6 days at 32°C (10).
In the present study, we further explored the genetic flexibility of the polymerase and demonstrated that asparagine 297 is not essential for efficient growth. The virus appears to tolerate multiple changes even at this extremely conserved locus, as a glycine 297 mutant was viable and two double mutants involving an alanine at position 297 were also viable. These mutants provide new insights into the function of conserved asparagine 297 in the polymerase NTP binding pocket. Interestingly, two of the viruses containing mutant polymerases were dependent on Mn2+ for RNA replication and growth. Thus, it appears that the mutations in these noncanonical RNA polymerases resulted in altered cation utilization of the enzyme. This is the first demonstration of a replicative polymerase with an alternative cation requirement.
MATERIALS AND METHODS
Mutation identification.
Candidate mutant viruses were isolated from individual plaques and used to infect HeLa cells at a multiplicity of infection (MOI) of 0.1. Infected cells were incubated in medium in the presence of 1 mM Mn2+ at 37°C for 8 h, and total RNA was then harvest from the cells by using RNeasy (Qiagen). Oligo(dT)-primed cDNA of each candidate mutant was made by using Superscript II. The 3Dpol-coding sequence was PCR amplified from cDNA, the entire length of the 3Dpol-coding region was sequenced by using BigDye terminator cycle sequencing and an ABI 310 DNA sequencer, and the data were analyzed with DNASTAR SeqManII.
Plasmids and molecular biology.
Desired mutations were introduced into the poliovirus polymerase-coding region of a poliovirus cDNA clone by oligonucleotide-directed mutagenesis with PCR. Primers 3DPST1 and 3D297G.R were used to amplify PCR fragment G5 from a pMoBPKN template (10). Primers 3D297G.F and 3DNHE1 were used to amplify PCR fragment G3. The G5 and G3 products were then used as the combined template in a hybrid PCR to produce fragment 297Gh, which was digested with PstI and NheI and cloned into PstI-NheI-digested subcloning plasmid pLIT-3CD-BPKN. All PCRs were done with PfuTurbo under the manufacturer's recommended conditions. The sequences of all resulting plasmids were confirmed by DNA sequencing. All of the plasmids generated contain an Ampr selectable marker. The BglII-EcoRI fragment of pLIT-3CD-297G was then cloned into a BglII-EcoRI-digested pMoRA backbone. A similar cloning strategy was used to generate p297A/286L. Primers 3DPST1 and 3DMnD1.R were used to amplify PCR fragment D5 from a pMoBPKN template. Primers 3DMnD1.F and 3DNHE1 were used to amplify PCR fragment D3. Fragments D3 and D5 were used as a template for a hybrid PCR, and the resulting product was subcloned into pLIT-3CD-BPKN and then inserted into the pMoRA backbone to generate p297A/286L. The cloning strategy for making p297A/239G required an additional level of complexity. Primers 3DPST1 and 3D236G.R were used to amplify PCR fragment 13.N from a pMoBPKN template. Primers 3D239G.F and 3D297A.R were used to amplify PCR fragment 13.M. Primers 3D297A.F and 3DNHE1 were used to amplify PCR fragment 13.C. The three fragments 13.N, 13.M, and 13.C were used as a combined template for a hybrid PCR, and the resulting product MnD13 h was cloned basically as described above to generate p297A/286L.
Subcloning plasmid pLIT-3CD-BPKN was made by cloning the BglII-EcoRI fragment of pMoRA-BPKN (10) into a BglII-EcoRI-digested pLIT28S backbone. pLIT28S was made by taking the plasmid pLITMUS28 (New England Biolabs, Beverly, Mass.) and removing the PstI-AflII region of the polylinker.
Replicon clones were made by cloning the BglII-ApaI fragments of pLIT-3CD-297G, pLIT-3CD-286L/297A, and pLIT-3CD-239G/297A into BglII-ApaI-digested pRLucRA (pRLuc-rib+polyAlong) (9, 13) to produce the respective plasmids pRLucRA-297G, pRLucRA-286L/297A, and pRLucRA-239G/297A.
3Dpol expression vectors were constructed by cloning the PstI-NheI fragments of pLIT-3CD-297G, pLIT-3CD-286L/297A, and pLIT-3CD-239G/297A into PstI-NheI-digested pET26-Ub-3D-BPKN-I92T (10) to produce the respective plasmids pET-3D-286L/297A (pET-3D-MnD1), pET-3D-297G, and pET-3D-239G/297A (pET-3D-MnD13). The pET-3Dpol expression plasmids each contain a Kanr selectable marker.
Oligonucleotides.
The sequences of the oligonucleotides used are as follows: 3DPST1, GATAACAGGTTCTGCAGT; 3D297G.R, TAATCATTGATCCAAAAATTGAGGTACCTGAG; 3D297G.F, CCTCAATTTTTGGATCAATGATTAACAACTTGAT; 3DNHE1, TTCCTGATTGGGCTAGC; 3DMnD1.F, CAAGGGCGGTTTGCCATCTGGCTGCTCAGGTACCTCAATTTTTGCTTCAATGATTAACAACTTGAT; 3DMnD1.R, TAATCATTGAAGCAAAAATTGAGGTACCTGAGCAGCCAGATGGCAAACCGCCCTTGACACAGTATG; 3D239G.F, AGGGTATGATGGATCTCTCAGCCCTGCTTG; 3D239G.R, GGCTGAGAGATCCATCATACCCTGTGTAGT; 3D297A.F, CCTCAATTTTTGCTTCAATGATTAACAACTTGAT; and 3D297A.R, TAATCATTGAAGCAAAAATTGAGGTACCTGAG.
Cells and transfections.
HeLa S3 cells (American Type Culture Collection stock plus 10 to 30 passages) were propagated in Dulbecco modified Eagle medium (DMEM)-F12 (Gibco/Life Technologies) supplemented with 10% fetal calf serum (Gibco/Life Technologies), with the cultures always kept between 20 and 80% confluence. For infectious-center assays (7, 8), viral RNA was produced by in vitro transcription of linearized plasmids with T7 RNA polymerase as described previously (8). Ten micrograms of each viral RNA transcript was electroporated into 1.2 × 106 HeLa cells in 400 μl in a 0.2-cm cuvette with the electroporation settings 600 μF, 24 Ω, and 130 V on a BTX electroporator, giving an average pulse length of 5 ms. Electroporated cells were plated on 2 × 105 HeLa cells in a six-well dish in a total volume of 1.0 ml.
The original Mo297A infectious-center assay was done by electroporating cells as described above and then plating the cells on 3 × 106 HeLa cells (prepared 1 day in advance) in a 10-cm-diameter dish in a total volume of 3.0 ml. Cells were allowed to adsorb to the plate for 1 to 2 h at 37°C, and then the medium with phosphate-buffered saline was aspirated and the cells were overlaid with 20 ml of a mixture of 1× DMEM-F12-10% fetal calf serum and 1% agar as described previously (10), with or without 0.5 mM Mn2+. Infectious-center assay mixtures were then incubated at 37°C, and plaques were identified and picked at day 5.
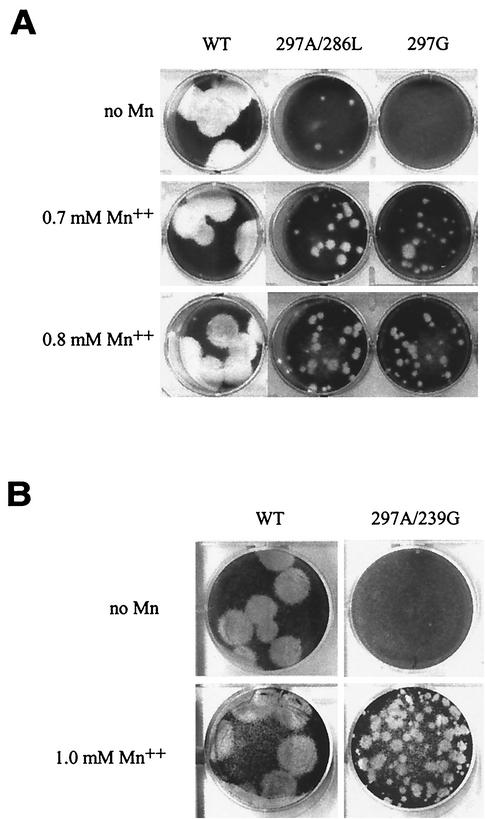
Some cytotoxicity was observed when plaque assay mixtures were supplemented with Mn2+. However, this did not affect the size or quality of wild-type or 297A/286L virus plaques (see Fig. 4), indicating that the Mn2+ cytotoxicity was not relevant to the observed Mn2+-dependent plaque assay growth of the 297G and 297A/239G mutants.
FIG. 4.
Plaque assays were supplemented with Mn2+ to tested for a possible Mn2+-dependent growth phenotype of the mutant viruses. (A) Three-day plaque assays of wild-type (WT), 297A/286L, and 297G viruses were done in the absence of manganese or in the presence of 0.7 or 0.8 mM Mn2+. Significant increases in plaque size and the number of plaques were seen for 297A/286L and 297G. No change was seen for wild-type virus. (B) Higher concentrations of Mn2+ were necessary for optimal plaque formation by 297A/239G. Shown is a 3-day plaque assay comparison of the wild type and 297A/239G in the absence or presence of 1.0 mM Mn2+. One minute 297A/239G plaque is visible in the absence of Mn2+, while ∼75 larger plaques are visible in the presence of 1.0 mM Mn2+.
Plaque assays were done as described previously (8, 9), with Mn2+ added to the medium as necessary. MnCl2 stocks were stored at −80°C.
Replicon transfections and firefly luciferase assays were performed with pFLucRA (formally called pRLucRA)-derived FLuc RNA or RNA from clones pFLuc-3DMnD13286L/297A, pFLucRA-3Dpol297G, and pFLucRA-3Dpol239G/297A as previously described (9, 10), using electroporation. The dual luciferase assay was done using the conditions recommended by the manufacturer (Promega, Madison, Wis.).
Measurement of viral RNA synthesis.
HeLa cells were infected as described above with an MOI of 5 for revertant 297A/239G and wild-type polioviruses. After 15 min of adsorption of the virus at 37°C, the cells were washed with phosphate-buffered saline and DMEM-F12 containing actinomycin D (5 μg/ml) was added. [3H]uridine (20 μCi/ml) (purchased from Du Pont NEN Research Products, Boston, Mass.) was added at 1 h postinfection. Cells were collected described at various time points as described above. Acid-insoluble materials were collected onto Millipore glass microfiber filters, and radioactivity was determined by scintillation counting (5).
Biochemical characterization of 3D alleles.
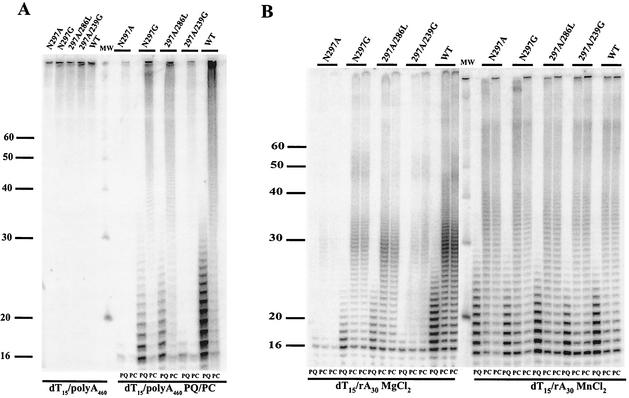
3Dpol expression and purification were done as previously described. The polymerase activities in Tables 1 and Table 2 were measured by using reagents and methods previously described (3, 10, 11). dT15/polyA460 extension reactions contained 1 mM dT15, 0.15 mM polyA460, 0.4 mCi of [α-32P]UTP per ml, and 500 mM UTP in 50 mM HEPES (pH 7.5)-5 mM MgCl2-10 mM β-mercaptoethanol-60 μM ZnCl2 (final concentrations). Reactions were initiated by the addition of 3Dpol to a final concentration of 0.5 μM and allowed to proceed for 5 min at 30°C. Reactions were stopped by the addition of an equal volume of 0.5 M EDTA. For pulse-chase and pulse-quench experiments, 3Dpol was added to the above-described reaction mixture in the absence of unlabeled UTP and left for 3 min. Initially labeled products were either quenched or chased into long products by the addition of unlabeled UTP and heparin-3000 to final concentrations of 500 and 10 mM, respectively. At a single time point 5 min after the addition of unlabeled UTP-heparin, the reaction was stopped as described above. The reaction products were resolved on a 15% denaturing polyacrylamide gel as previously described (10). The template switching assay (see Fig. 7) was done similarly to that previously described (4) except that the final concentration of rA30 was 1 mM and the reaction mixtures contained either MgCl2 or MnCl2 to final concentration of 5 mM. At 3 and 7 min after the addition of unlabeled UTP-heparin, the reactions were quenched as described above.
TABLE 1.
Rates of AMP incorporation into sym/sub-U by wild-type 3Dpol and 3Dpol derivatives
| Enzyme | AMP incorporation rate (s−1)a with:
|
Virus viability | |
|---|---|---|---|
| Mg2+ | Mn2+ | ||
| Wild type | 45 ± 4 | 11 ± 2 | +++++ |
| N297A | 3.3 ± 0.3 | 1.4 ± 0.1 | − |
| D238A | 0.044 ± 0.002 | 0.24 ± 0.02 | − |
| N297G | 14 ± 2 | 3.0 ± 0.2 | ++ |
| N297A/M286L | 20 ± 4 | 4.5 ± 0.5 | +++ |
| N297A/A239G | 6.0 ± 0.6 | 3.3 ± 0.3 | + |
Results are means and standard deviations.
TABLE 2.
Extension activities of wild-type 3Dpol and 3Dpol derivatives with dT15/rA30 as a substrate
| Enzyme | Phenotype
|
||
|---|---|---|---|
| In vitro activity (pmol of UMP/min/mg of protein) with:
|
Virus viability | ||
| Mg2+ | Mn2+ | ||
| Wild type | 42 | 225 | +++++ |
| N297A | 13 | 127 | − |
| N297G | 43 | 220 | ++ |
| N297A/M286L | 20 | 166 | +++ |
| N297A/A239G | 43 | 287 | + |
FIG. 7.
Polymerase initiation and elongation in vitro. (A) Pulse-chase and pulse quench on dT15/polyA460. Elongation reactions were initiated by the addition of 3Dpol to a final concentration of 0.5 μM and allowed to proceed for 5 min at 30°C in the presence of 0.2 μM [α-32P]UTP and 500 μM unlabeled UTP (left side). Full-length products were observed with all five polymerases tested. For pulse-chase and pulse quench experiments (right side), 3Dpol was added to the reaction mixture in the absence of unlabeled UTP for three minutes. Initially labeled products were either quenched (PQ) or chased into long products (PC) by the addition of unlabeled UTP and heparin-3000 to final concentrations of 500 and 10 μM, respectively. A single time point was taken 5 min after the addition of unlabeled UTP-heparin. All reaction products were resolved on a 15% denaturing polyacrylamide gel. (B) Pulse-chase and pulse quench on dT15/rA30. Reactions contained either MgCL2 (left side) or MnCl2 (right side) to final concentration of 5 mM. A spectrum of initiation differences was observed with Mg2+, while all polymerases were competent for initiation with Mn2+.
RESULTS
Isolation and characterization of viable viruses with alterations in highly conserved polymerase amino acids.
Jablonski and Morrow have previously shown that mutations of the aspartic acid residues of the conserved GDD sequence motif of poliovirus RNA polymerase result in enzymes with altered metal ion requirements (16). In a previous study (10), we analyzed the biological phenotypes of polioviruses containing a series of mutations in the nucleotide binding pocket. Several of these mutations were changes at position 297 in 3Dpol. All mutations tested resulted in nonviable viruses at 37°C, including mutant Mo-3DpolN297A (10). In addition, we have observed that the RNA-dependent RNA polymerase activity in vitro can be stimulated by Mn2+ (3, 4). Thus, we examined whether this nonviable mutant could be rescued by supplementing the medium with 0.5 mM manganese (Mn2+). Transfection with mutant Mo-3DpolN297A RNA, which encodes a polymerase with very low activity in vitro (10), yielded few (∼20) plaques in the presence of Mn2+ at 5 days after transfection. These plaques were isolated for further study. Surprisingly, these virus isolates grow, albeit poorly, even in the absence of Mn2+, suggesting that the RNA transfection in the presence of Mn2+ facilitated the emergence of viable revertant viruses. Based on plaque size in the absence of Mn2+, these viruses can be classified into three groups: those with small, very small, and minute plaques.
Given that a simultaneous double or triple point mutation would be necessary to revert alanine 297 (codon GCT) back to asparagine (codon AAT or AAC), we expected that these emerging viable viruses were single point mutation pseudorevertants rather than true revertants. To test this possibility, two small-plaque viruses were sequenced. Both viruses contained a single point mutation converting the methionine at position 286 of 3Dpol to a leucine (M286L), and they retained the N297A mutation (this mutant is referred to as 297A/286L). Two very-small-plaque viruses were sequenced, and these isolates contained a single point mutation that changed the mutant alanine at position 297 of 3Dpol to a glycine (A297G); no other mutations were observed in these clones (this mutant is referred to as 297G). Two minute-plaque viruses were then sequenced, and these contained a single point mutation changing the alanine at position 239 of 3Dpol to a glycine (A239G) and retained the N297A mutation (this mutant is referred to as 297A/239G).
Strikingly, all three of these mutant viruses contained amino acids which are different from the highly conserved asparagine at position 297. As described above, asparagine 297 of 3Dpol is completely conserved among positive-strand RNA viruses of eukaryotes and is situated in an apparently critical position in the nucleotide binding pocket. Asparagine 297 appears to participate in NTP binding and may assist in the crucial function of discriminating between NTPs and dNTPs. As this amino acid residue is central to the function of the polymerase but its role in RNA replication is somewhat unclear, we further explored the biological phenotypes of these novel polymerase mutants.
In vivo characterization of 297 pseudorevertants.
The 3Dpol mutations were introduced into poliovirus plasmid clones by site-directed mutagenesis to produce three mutant plasmid clones, pMo-297A/286L, pMo-297G, and pMo-297A/239G, from which well-defined viral stocks were produced. Viruses obtained from molecular clones recapitulated the plaque phenotypes observed in the original revertant viruses, having small, very small, or minute plaques (Fig. 2A).
FIG. 2.
Viable 3Dpol297 mutants. (A) Viable polioviruses with mutations at 3Dpol position 297. Three-day plaque assays are shown. Approximately 50 minute plaques (and 1 small plaque) are visible in the 297A/239G well. (B) Virus production from high-MOI (10 PFU/cell) infections with wild-type, 297A/286L, 297G, and 297A/239G viruses. Wild-type virus reached maximum titers of 109 PFU/ml by 6 h postinfection. 297A/286L, 297G, and 297A/239G viruses reached maximum titers of 3 × 108, 2 × 108, and 5 × 106 PFU/ml, respectively, by 12 h postinfection. Results from a representative experiment are shown; titer variations between experiments were generally within a threefold range.
All three viruses had severely defective growth kinetics under one-step growth conditions (Fig. 2B). Wild-type poliovirus reached maximum virus production of 1.5 × 109 PFU/ml by 6 h postinfection, but no virus production by the three mutants was detected at that time point. Limited virus production of 297A/286L, 297G, and 297A/239G was detected at 8 h (data not shown), and maximum titers were observed at 12 h postinfection (Fig. 2B). Pseudorevertants 297A/286L and 297G grew to relatively high titers of 1 × 108 to 3 × 108 PFU/ml under these conditions, whereas mutant 297A/239G was severely defective for virus production, generating only 3 × 106 PFU/ml, a 500-fold decrease from wild-type levels.
RNA replication by 297 mutant polymerases.
We used a poliovirus replicon (FLuc) to evaluate the levels of viral RNA replication generated by the pseudorevertants in cell culture. FLuc is a poliovirus replicon that consists of a full-length poliovirus genome with the capsid genes replaced by a firefly luciferase reporter gene (2, 9, 13, 14) (Fig. 3B). Transfection of HeLa cells with replicon RNA results in the production of a polyprotein containing luciferase that is processed by the viral 2A protease to liberate active luciferase, and the replicon translates and replicates comparably to wild-type poliovirus (2, 13). The mutations in 3Dpol observed in 297A/286L, 297G, and 297A/239G were introduced into pFLuc luciferase replicon plasmids to generated pFLuc297A/286L, pFLuc297G, and pFLuc297A/239G, respectively.
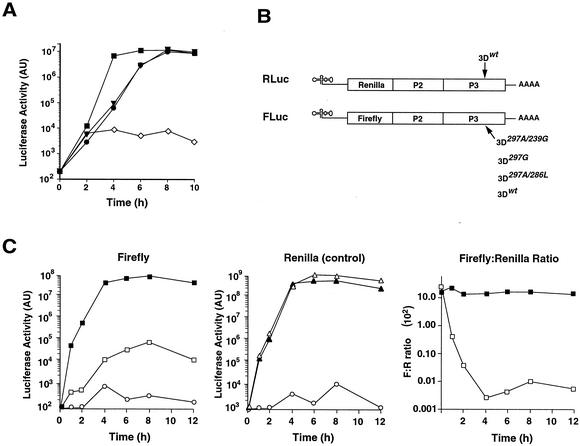
FIG. 3.
Effect of mutations as explored with a replicon system. (A) A poliovirus replicon (FLuc) was used to evaluate RNA replication by the mutant polymerases in cell culture. An FLuc replicon with a wild-type polymerase is shown as a positive control (▪), and FLuc238A (⋄), which has no detectable RNA replication activity in cell culture (10), is shown as a negative control. ▾, FLuc297A/286L; •, FLuc297G. AU, arbitrary units. (B) FLuc is a poliovirus replicon that consists of a full-length poliovirus genome with the capsid genes replaced by a firefly luciferase reporter gene (14). RLuc is a poliovirus replicon that consists of a full-length wild-type poliovirus genome with the capsid genes replaced by a renilla luciferase reporter gene. The two-replicon system was used for panel C. (C) Replicon activity after transfection. FLuc replication is shown on the left as firefly luciferase activity. □, FLuc297A/239G; ▪, FLucwt; ○, mock. RLuc replication is shown in the middle panel as renilla luciferase activity. RLucwt was transfected concurrently with each of the FLuc constructs as an internal control as follows: ▵, RLucwt transfected with FLuc297A/239G; ▴, RLucwt transfected with FLucwt; ○, mock. The right panel shows the ratio of firefly to renilla luciferase activity. ▪, FLucwt/RLucwt; □, FLuc297A/239G/RLucwt.
FLuc238A was previously reported to be a polymerase mutant that translates but does not replicate (10), and it is used here as a translation-alone control (∼6 × 103 relative light units [RLU]) (Fig. 3A). Wild-type FLuc rapidly replicated to levels 1,000-fold higher than those of the negative control FLuc238A by 4 h posttransfection (7 × 106 RLU), leveling off at ∼1.1 × 107 RLU by 6 h posttransfection. FLuc297A/286L replicated to levels similar to those of wild-type FLuc but with 2-h-slower kinetics. FLuc297G replicated to levels equal to those of wild-type FLuc but with a 2-h time lag, comparable to the replication kinetics of FLuc297A/286L (Fig. 3A).
To measure the crippled replication of 297A/239G, a more robust dual-replicon system was employed. As an internal control we used a second replicon (RLuc) that is identical to FLuc but carries renilla luciferase as the reporter gene and utilizes a different enzymatic substrate (Fig. 3B). Thus, RLuc (renilla replicon wild type) was cotransfected in each experimental condition as an internal control. FLuc297A/239G presented a severe defect in replication compared with wild-type FLuc (Fig. 3C). Wild-type FLuc rapidly replicated to produce up to 108 luciferase counts, while FLuc297A/239G produced only 3 × 104 luciferase counts by 8 h posttransfection, showing that RNA replication is severely defective in FLuc297A/239G. The data were also graphed as ratios of renilla/firefly luciferase activity. The ratio between firefly and renilla luciferases indicates that FLuc297A/239G replicates to levels approximately 1,000- to 10,000-fold lower than those of the wild-type replicon (Fig. 3C, right panel). These data correlated well with the plaque assay phenotypes observed, as FLuc297A/239G had a significantly smaller plaque phenotype than the wild type and produced only 0.2% of the wild-type titer (Fig. 2).
Luciferase assays could not be done on the FLuc replicon mutants in the presence of Mn2+ because it inhibited the luciferase activity (data not shown).
Manganese-dependent growth.
As these mutant viruses were originally observed in an infectious-center assay in the presence of 0.5 mM Mn2+, we assessed the growth phenotypes of 297A/286L, 297G, and 297A/239G in the presence of supplemental Mn2+. Optimal plaque formation was seen in the range of 0.5 to 1.0 mM Mn2+ (data not shown). The presence of Mn2+ had no obvious effect on wild-type poliovirus plaque formation (Fig. 4A). In contrast, 297A/286L made plaques approximately 40% larger in the presence of 0.7 to 0.8 mM Mn2+, and its plaquing efficiency was twofold greater in Mn2+. 297G had a more pronounced phenotype, making easily visible plaques at day 3 in the presence of Mn2+ but no or pinpoint plaques in the absence of Mn2+ (Fig. 4A). The most extreme phenotype was the growth of 297A/239G, which required 1.0 mM Mn2+ for maximum plaque formation (Fig. 4B).
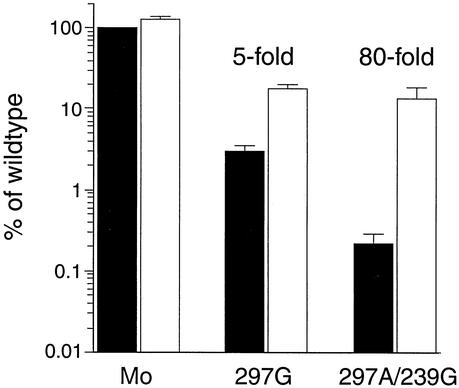
Given these unusual Mn2+-dependent phenotypes observed by plaque assay, we tested the effect of different levels of supplemental Mn2+ on the replication of these mutant viruses under one-step growth conditions. Low levels of Mn2+ (0.5 μM) had no effect on any of the mutant viruses or wild-type virus (data not shown). However, 0.5 mM Mn2+ had a dramatic effect on 297A/239G growth: virus production increased 80-fold (Fig. 5). With 297G, a fivefold increase in virus production was observed in the presence of 0.5 mM Mn2+ (Fig. 5). In the case of the 297A/286L mutant, Mn2+-dependent growth was not observed under these conditions (data not shown).
FIG. 5.
Mn2+-dependent virus growth. Virus production in high-MOI (10 PFU/cell) infections with wild-type, 297G, and 297A/239G viruses in the absence (black bars) or presence (white bars) of 0.5 mM Mn2+ is shown. All data are normalized to wild-type virus production in the absence of Mn2+ (100%). Error bars depict standard errors of the mean from four or five experiments. A fivefold stimulation of 297G virus production was observed in the presence of 0.5 mM Mn2+, and a striking 80-fold stimulation of Mo-3Dpol 239G/297A virus production was seen in the presence of 0.5 mM Mn2+.
Manganese-dependent RNA synthesis.
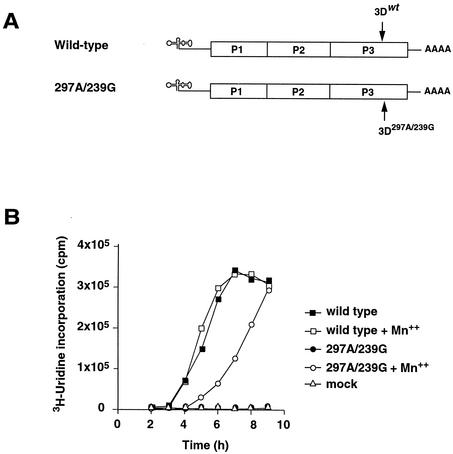
Because mutant 297A/239G had a major alteration of a conserved residue in the 3D polymerase and exhibited Mn2+-dependent plaque formation and virus production, we analyzed the ability of the 297A/239G virus to produce viral RNA in the absence and the presence of Mn2+. HeLa cells were infected with wild-type or 297A/239G virus, and infected cells were incubated at 37°C in the presence of [3H]uridine. Incorporation of [3H]uridine into replicating viral RNA was determined by acid precipitation at several time points. In the absence of Mn2+, mutant 297A/239G was unable to incorporate [3H]uridine above background levels (about 1,000 cpm) (Fig. 6). In contrast, the addition of 1.0 mM Mn2+ greatly stimulated RNA synthesis, such that by 9 h postinfection 297A/239G reached wild-type levels of viral RNA accumulation (Fig. 6). This result strongly suggests that 297A/239G possesses an Mn-dependent polymerase and that the presence of Mn2+ is required for RNA synthesis and the generation of progeny virus.
FIG. 6.
Mn2+-dependent RNA replication. (A) Wild-type and 297A/239G viruses. (B) HeLa cells were infected with wild-type or 297A/239G virus, and infected cells were incubated at 37°C in the presence of 3[H]uridine with or without supplemental 1.0 mM Mn2+. Incorporation of 3[H]uridine into replicating viral RNA was determined by acid precipitation at several time points.
Biochemical analysis of the poliovirus mutant polymerases. (i) Single-nucleotide incorporation.
In order to determine whether the phenotypes observed biologically correlated with the ability of the mutants to catalyze phosphoryl transfer, we performed an experiment using “sym/sub,” a primer-template of defined sequence that permits evaluation of up to four consecutive cycles of nucleotide incorporation (3, 9, 10). In the presence of Mg2+, there was a direct correlation between the efficiency of single-nucleotide incorporation and virus viability for all three pseudorevertant 3Dpol polymerases (Table 1). All three pseudorevertant 3Dpol polymerases had higher nucleotide incorporation rates than the original mutant 297A under single-turnover conditions in the presence of Mg2+, although the incorporation rates were significantly lower than those for wild-type polymerase (Table 1); these data may suggest that there are differences in the affinity of each derivative for nucleotide.
In the presence of Mn2+, the binding constant of 3Dpol for the correct nucleotide is reduced substantially and the rate-limiting step is phosphoryl transfer (J. J. Arnold and C. E. Cameron, unpublished data). Therefore, under these conditions any differences in nucleotide binding should disappear and the capacity of these enzymes to perform catalysis should be apparent. Note that for wild-type 3Dpol, Mn2+ reduced the observed rate of nucleotide incorporation fourfold (Table 1). This may be due to the fact that this cation is larger than Mg2+ and would increase the distance between the 3′OH of the primer and the α-phosphate of the incoming nucleotide. 297A/286L, 297G, and 297A/239G had incorporation rates 25 to 40% of that of wild-type polymerase in the presence of Mn2+. The viable mutant polymerases had more rapid NTP incorporation in the presence of Mn2+ than compared to the original nonviable mutant 297A (Table 1). This difference may suggest that the overall organization of the catalytic site is changed by the mutations in the nucleotide binding pocket.
Of note is that the mutant polymerases showed no obvious differences in nucleoside misincorporation rates compared to wild-type polymerase on a sym/sub-U template in the presence of ATP, using Mg2+ or Mn2+ (data not shown).
(ii) Processive synthesis.
The assays described above evaluated only incorporation of a single nucleotide. It was possible that the changes observed were only part of the polymerase defect and that the ability of the enzymes to perform processive synthesis was also impaired. As a first step towards evaluating this possibility, we assessed the primer elongation capacities of the polymerase mutants on a longer template (4). Homopolymeric RNA template rA30, was used with a dT15 primer. In this assay, 297A/286L, 297G, and 297A/239G all exhibited higher activity than 297A in the presence of Mg2+ (Table 2). Additionally, 297A/286L, 297G, and 297A/239G exhibited higher activity than 297A in the presence of Mn2+ (Table 2).
These data could result from a change in the number of primers initiated or a change in the processivity of the enzyme. In order to evaluate these possibilities, we performed pulse-quench and pulse-chase analyses of product formation on an rA460 template in the presence of Mg2+ (Fig. 7A). For pulse-chase experiments, 3Dpol was added to a reaction mixture containing 0.2 μM [α-32P]UTP and dT15/rA460 primer-template, in the absence of unlabeled UTP, for a 3-min pulse. Products labeled during the pulse period were chased into longer products (PC lanes in Fig. 7A) by the addition of unlabeled 500 μM UTP and heparin. In pulse-quench experiments, quenching was done immediately after the 3-min pulse by the addition of 0.5 M EDTA (PQ lanes in Fig. 7A).
If the various mutant polymerases differed in their extension activity due to poor processivity, we would anticipate that a pulse-chase experiment would result in the formation of products whose average length would be shorter than that for the wild-type polymerase. If the mutant polymerases instead differed in the extension activity due to an inability to initiate synthesis, we would anticipate that the total amount of radioactivity incorporated into products during the pulse (as observed in the pulse-quench experiments) would be less than the amount in the wild type.
We observed that the polymerases exhibited clear differences in their ability to initiate synthesis, as fewer products were observed in reactions primed with 297A and 297D than in reactions primed with 297A/286L, 297G, or wild type, and all of the products formed during the pulse were competent for extension into full-length product (Fig. 7A). Interestingly, 297A/239G, which is marginally viable in vivo in the absence of supplemental Mn2+, was severely defective for initiation of RNA synthesis in vitro. This spectrum of initiation activity was not observed with the sym/sub template, possibly due to the higher concentration of NTP employed in this experiment.
To further examine the biochemical phenotypes of these enzymes, we performed a second pulse-chase, pulse-quench experiment. In this case, rA30 was used instead of rA460 in order to assess the template switching ability of these polymerases, as poliovirus exhibits frequent RNA recombination in vivo (17), and the formation of long products by 3Dpol in the presence of an rA30 template in vitro requires template switching (4). On this rA30 template the polymerases again exhibited clear differences in their ability to initiate synthesis in the presence of Mg2+, with 297A/286L and 297G performing much better than the nonviable mutant polymerase 297A (Fig. 7B). The polymerases did not exhibit any obvious differences in template switching capacity, as long products were formed by all of the polymerases in proportion to the amount of radiolabeled nucleotide incorporated during the pulse (Fig. 7B). These experiments were also performed in the presence of Mn2+, and all of the polymerases exhibited an increased rate of initiation with Mn2+ (Fig. 7B).
DISCUSSION
In this report we demonstrate that an absolutely conserved residue within the nucleotide binding pocket of the poliovirus polymerase can be substituted for a different amino acid, yielding replication-competent virus. Unexpectedly, two of the resulting viruses exhibit a dependence on Mn2+ for growth. These results attest to the extreme evolutionary flexibility of the viral polymerase, in terms of both structure and cation usage, since the introduction of two point mutations is sufficient to replace the function of an amino acid residue that is completely conserved across the entire class of positive-strand RNA viruses of eukaryotes. This observation is particularly unexpected given that only six conserved amino acid positions exist in this class of polymerases (18).
Mutants 297G and 297A/286L.
From a structure standpoint, the 297G mutation was the most striking of the three mutations observed. Asparagine 297 is thought to interact with the incoming nucleotide through its amide group. How is it then possible that glycine could replace asparagine? Molecular modeling suggests that a glycine at position 297 leaves a sufficiently large pocket for an additional water molecule (Fig. 8B). Therefore, glycine may substitute for asparagine 297 by allowing a water molecule to become the hydrogen bonding partner for the NTP 2′OH. Notably, mutant 297A/286L may utilize the same strategy of accommodating an H2O molecule as a surrogate hydrogen bonding partner. Although the N297A mutation does not leave sufficient space for an H2O and the mutant is nonviable, a compensatory M286L mutation may enlarge the hole created by N297A, providing sufficient volume for an H2O to enter and replace the H bond to the NTP lost by the N297A mutation (Fig. 8C).
FIG. 8.
Molecular modeling of the 3Dpol NTP binding pocket. (A) van der Waal's representation of the amino acids present in the NTP binding pocket of 3Dpol. Amino acid side chains are labeled. The incoming nucleotide (ATP) is also labeled. (B) The same view as in panel A; however, the N297 side chain has been removed (i.e., N297A- or N297G-containing derivatives), resulting in an expansion of the NTP binding pocket. (C) 286L/297A derivative. (D) 239G/297A derivative. (E) 239G/297A derivative, rotated by 90° counterclockwise about the vertical axis of the page. In this view the gap produced by the A239G substitution is observable. The N297A substitution is occluded by the D238 side chain. In addition, replacement of the alanine at position 239 by glycine might result in additional rotational freedom for the adjacent side chain (D238).
The 297G virus was viable under normal growth conditions, but it was partially Mn2+ dependent, producing fivefold more virus and having a higher plaquing efficiency in the presence of Mn2+. Mn2+ generally improves NTP Kd values (D. Gohara and C. E. Cameron, unpublished data) and may improve processivity or initiation rates of the 3Dpol297G mutant polymerase due to the ability of Mn2+ to overcome contortional problems in the NTP binding pocket and make NTP positioning requirements less stringent. UTP concentrations are relatively low in vivo, and the affinity of wild-type 3Dpol for UTP is also low (3). Small perturbations in the NTP binding pocket that affect UTP binding would therefore be expected to have a severe effect on initiation, as poliovirus RNA genome replication begins with the incorporation of two UTP nucleotides to generate VPgpU(pU) primers and then the subsequent incorporation of sequential UTP nucleotides to initiate negative-strand synthesis (6, 20, 21, 23). This may explain the fivefold viral titer increase of 297G in the presence of Mn2+. Alternatively, this phenotype may result from more rapid elongation by the polymerase, due to an in vivo mixture of Mg2+ and Mn2+ that is not easy to reproduce in vitro. Also, the normal plaque phenotype of 297G is variable (compare Fig. 2 with Fig. 4), which is possibly a reflection of the sensitivity of 3Dpol297G to environmental conditions. Although the glycine may provide sufficient space for an H2O molecule, the extra flexibility that a glycine allows may destabilize neighboring residues and make the polymerase more sensitive to environmental conditions. For example, 3Dpol297G may have a basal requirement for trace Mn2+ or other cations that are variable in the medium and serum used.
Biochemical analysis of 3Dpol297G and 3Dpol297A/286L revealed that both of these polymerases have a good rate of NTP incorporation, five- to sevenfold higher than that of the nonviable 297A single mutant. In vitro NTP incorporation by 3Dpol297G and 3Dpol297A/286L was 50% of that seen with wild-type polymerase (Table 1), which correlates with the RNA replication observed in vivo (Fig. 3A). Notably, the 3Dpol297G, 3Dpol297A/286L, and wild-type polymerases also showed a spectrum of initiation activity on long templates that correlated with viability (Fig. 7).
Mutant 297A/239G.
Mutant 297A/239G had the most extreme growth defect of the three viable mutants isolated. Importantly, this mutant provides the first genetic evidence for cooperation between the active-site N297 and residues around D238. Luciferase replicon experiments demonstrated that 297A/239G had a severe RNA replication defect (Fig. 3C). Virus replication experiments in the presence of [3H]uridine demonstrated that RNA replication by 297A/239G is profoundly Mn2+ dependent (Fig. 6). Polymerase 3Dpol297A had minimal activity in Mg2+, i.e., an incorporation rate only twofold better than that of nonviable 297A and 13% of wild-type activity with Mg2+ (Table 1) (10). In contrast to mutants 3Dpol297G and 297A/286L, which appear to compensate for the lost asparagine at position 297 by recruiting an H2O molecule to interact with the 2′OH of the incoming NTP, 297A/239G may solve the same problem by strengthening the interaction of D238 with the 2′OH of the incoming NTP. The replacement of alanine at position 239 with a glycine may provide sufficient flexibility in the polypeptide backbone to let neighboring aspartic acid 238 accommodate into an optimal position for interacting with the 2′OH (Fig. 8D and E). However, since D238 also appears to have important interactions with the 3′OH of the incoming NTP (10), the glycine mutation at position 239 may hinder that 3′OH interaction, resulting in a functional defective polymerase as observed in vivo and in vitro.
Manganese dependence.
297A/239G exhibited a striking 80-fold Mn2+-dependent growth phenotype in vivo due to its unusual polymerase mutations. [3H]uridine incorporation experiments revealed that measurable RNA synthesis by 3Dpol297A/239G depended on the presence of supplemental Mn2+. 3Dpol297A/239G may depend primarily on Mn2+ for catalysis in vivo. Alternatively, 3Dpol297A/239G may depend primarily on magnesium in vivo for high-fidelity RNA synthesis but require Mn2+ as a supplemental metal ion for certain critical functions, for example, initiation of RNA synthesis. It is possible that an improved Kd for UTP in the presence of Mn2+ may allow efficient synthesis of VPgpU(pU) primers in vivo. Alternatively, Mn2+ could enhance polymerase function through an indirect effect on the folding of the poliovirus polymerase or on the structure of the incoming NTPs. As mentioned above, it is also possible that a mixture of Mg2+ and Mn2+ is employed by the mutant RdRp.
There are two plausible scenarios for why viable mutants were recovered after transfection of Mo-3Dpol297A RNA in the presence of Mn2+. (i) Recovery may have been dependent on the presence of mutated T7 polymerase-derived in vitro transcripts capable of significant plaque formation only after transfection in Mn2+ (e.g., 297A/239G). (ii) Alternatively, Mn2+ is an RNA virus mutagen (S. Crotty and R. Andino, unpublished data), the presence of Mn2+ could encourage the synthesis of mutated Mo-3Dpol297A derivatives during the low-level Mo-3Dpol297A replication that occurs posttransfection, and several of those new mutants were then viable for plaque formation. Jablonski and Morrow reported that they were able to recover infectious poliovirus in the presence of iron (FeSO4) after transfection of a plasmid containing a mutant poliovirus genome possessing a GDN sequence instead of the canonical GDD motif (16). However, they were unable to isolate viral genomes of the viable virus or sequence that virus, and so the polymerase sequence of the virus that emerged could not be determined and RNA replication of the virus could not be characterized (16).
Our results with 297A/239G clearly demonstrate that a viable virus can have a requirement for an alternative cation. This may be of particular relevance to the RNA-dependent RNA polymerases of plant viruses, as plants generally contain significant intracellular stores of Mn2+. We speculate that certain plant viruses may have polymerases with a requirement for Mn2+. Additionally we wonder if putative plant RNA-dependent RNA polymerases involved in RNA interference could possibly utilize Mn2+.
In summary, here we show that the poliovirus polymerase tolerates multiple changes even at the extremely conserved asparagine 297 position. Thus, the polymerase NTP binding pocket appears to be more genetically flexible than previously anticipated. Importantly, two of the viruses containing mutant polymerases were heavily dependent on Mn2+ for RNA replication and growth. The mutations in these unusual RNA polymerases resulted in an alteration in the cation utilization of the enzyme. Finally, the observation that a virus can acquire mutations at a completely evolutionarily conserved residue and maintain viability should serve as a cautionary note about the probable efficacy of polymerase-targeted antiviral therapeutic compounds.
REFERENCES
- 1.Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296:1270-1273. [DOI] [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, J. J., and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275:5329-5336. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, J. J., and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 274:2706-2716. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, H. D., N. Sonenberg, and D. Baltimore. 1985. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol. Cell. Biol. 5:2913-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Cameron, C. E., D. W. Gohara, and J. J. Arnold. 2003. Poliovirus RNA-dependent RNA polymerase (3Dpol): structure, function, and mechanism, p. 255-267. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 6.Crawford, N. M., and D. Baltimore. 1983. Genome-linked protein VPg of poliovirus is present as free VPg and VPg-pUpU in poliovirus-infected cells. Proc. Natl. Acad. Sci. USA 80:7452-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crotty, S., B. L. Lohman, F. X. Lu, S. Tang, C. J. Miller, and R. Andino. 1999. Mucosal immunization of cynomolgus macaques with two serotypes of live poliovirus vectors expressing simian immunodeficiency virus antigens: stimulation of humoral, mucosal, and cellular immunity. J. Virol. 73:9485-9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 10.Gohara, D. W., S. Crotty, J. J. Arnold, J. D. Yoder, R. Andino, and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 275:25523-25532. [DOI] [PubMed] [Google Scholar]
- 11.Gohara, D. W., C. S. Ha, S. Kumar, B. Ghosh, J. J. Arnold, T. J. Wisniewski, and C. E. Cameron. 1999. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17:128-138. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 13.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jablonski, S. A., M. Luo, and C. D. Morrow. 1991. Enzymatic activity of poliovirus RNA polymerase mutants with single amino acid changes in the conserved YGDD amino acid motif. J. Virol. 65:4565-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jablonski, S. A., and C. D. Morrow. 1995. Mutation of the aspartic acid residues of the GDD sequence motif of poliovirus RNA-dependent RNA polymerase results in enzymes with altered metal ion requirements for activity. J. Virol. 69:1532-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell. 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 19.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 20.Lee, Y. F., A. Nomoto, B. M. Detjen, and E. Wimmer. 1977. A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. USA 74:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 22.Richards, O. C., S. Baker, and E. Ehrenfeld. 1996. Mutation of lysine residues in the nucleotide binding segments of the poliovirus RNA-dependent RNA polymerase. J. Virol. 70:8564-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takegami, T., R. J. Kuhn, C. W. Anderson, and E. Wimmer. 1983. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc. Natl. Acad. Sci. USA 80:7447-7451. [DOI] [PMC free article] [PubMed] [Google Scholar]