Abstract
Porphyromonas gingivalis is a periopathogen strongly associated with the development of adult-type periodontitis. Both the virulence characteristics of periopathogens and host-related factors are believed to contribute to periodontitis. P. gingivalis lipopolysaccharide (LPS) displays a significant amount of lipid A structural heterogeneity, containing both penta- and tetra-acylated lipid A structures. However, little is known concerning how the lipid A structural content of P. gingivalis is regulated. Alterations in the lipid A content may facilitate the ability of P. gingivalis to modulate the innate host response to this bacterium. In this report, it is shown that the concentration of hemin in the growth medium significantly modulates the lipopolysaccharide lipid A structural content of P. gingivalis. Hemin is a key microenvironmental component of gingival cervicular fluid which is believed to vary depending upon the state of vascular ulceration. At low hemin concentrations, one major penta-acylated lipid A structure was found, whereas at high concentrations of hemin, multiple tetra- and penta-acylated lipid A structures were observed. Hemin concentrations, not iron acquisition, were responsible for the alterations in the lipid A structural content. The modifications of the lipid A structural content were independent of the LPS extraction procedure and occurred in a variety of laboratory strains as well as a freshly obtained clinical isolate. The known hemin binding proteins Kgp and HmuR contributed to the lipid A modulation sensing mechanism. To the best of our knowledge, this is the first report that hemin, a clinically relevant microenvironmental component for P. gingivalis, can modulate the lipid A structure found in a bacterium. Since tetra- and penta-acylated P. gingivalis lipid A structures have opposing effects on Toll-like receptor 4 activation, the alteration of the lipid A structural content may have significant effects on the host response to this bacterium.
Periodontitis is a chronic inflammatory disease of the tissue surrounding the tooth root surface. The disease is characterized by the loss of periodontal tissue and supporting alveolar bone. It is highly prevalent in the human population and is the major cause of tooth loss in the world (24). It is strongly associated with a subgingival microbial community (25) commonly referred to as periopathogenic dental plaque. Porphyromonas gingivalis, a gram-negative anaerobic bacterium, is a member of this community and displays a strong correlation with disease (25). Neither the contribution of periopathogenic plaque nor those of individual members of the periopathogenic community to the disease process are fully understood. However, it is suspected that both bacterial virulence factors, such as P. gingivalis protease secretion (17), and host factors contribute to the disease (18). The contribution of the host to the disease process is believed to result from an innate host response to periopathogenic bacteria that results in tissue damage and alveolar bone loss (7). A likely candidate for an initiator of a destructive inflammatory response is lipopolysaccharide (LPS), since it is well established that this bacterial cell wall component, as obtained from Escherichia coli, is a potent stimulator of the innate host defense system (2).
P. gingivalis LPS contains an unusual amount of lipid A heterogeneity that includes differences in the number of phosphate groups and the amount and position of lipid A fatty acids from those in LPS obtained from E. coli (1, 8, 13, 30). Clearly, the presence of multiple lipid A structures has complicated the interpretation of innate host responses elicited by P. gingivalis LPS preparations, thus hindering a more complete understanding of the contribution of P. gingivalis LPS to the pathology of periodontitis. For example, penta-acylated lipid A structures are Toll-like receptor 4 (TLR4) agonists (21), whereas tetra-acylated structures are TLR4 antagonists (5). Furthermore, little is known concerning the regulation of these different lipid A structural types. For example, for select pathogenic bacteria, environmental conditions have been shown to influence the number and types of lipid A structures found (9, 11, 12). It is believed that these bacterial pathogens regulate their lipid A structural composition in response to local host microenvironmental conditions (19).
One microenvironmental condition that may contribute to the virulence of P. gingivalis is the hemin concentration (17, 23). Hemin binds host iron and represents the major iron acquisition system for P. gingivalis (17). Although apparently contrasting results have been obtained which may depend upon the strains examined (17), it is clear that the concentration of hemin in the growth medium can regulate the expression of several virulence factors, including gingipains (17), and extracellular vesicle formation (17) as well as increasing P. gingivalis virulence in a mouse model of infection (15). Although P. gingivalis can utilize inorganic iron for growth in vitro, it is believed that iron is acquired almost exclusively through hemin uptake in vivo (17). Although not experimentally determined, it is believed that the local concentration of hemin (in the form of hemoglobin) can vary considerably depending on the state of vascular ulceration during periodontitis, as measured clinically by bleeding upon probing.
In this report, it is shown that the hemin concentration in the growth medium has a significant effect on the lipid A structural content of P. gingivalis. Although other in vitro growth conditions have been shown to alter the lipid A structural contents of other bacteria (9, 11, 12), to the best of our knowledge, this is the first report that hemin can affect the lipid A structures found in a bacterium and may represent one mechanism by which P. gingivalis is able to sense and adapt to local environments which vary in their hemin concentration.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis strains ATCC 33277, W83, and 381 and a clinical isolate were obtained from our stock collection. P. gingivalis strains A7436 and WS15 (22) were obtained from Caroline Genco. All strains were examined for purity, properly identified, and stored at −70°C. Cultures were made from frozen bacterial stocks to avoid repetitive subculturing. Bacterial culture media included both enriched Trypticase soy broth (ETSB) and Trypticase soy broth-yeast extract-hemin-vitamin K (menadione) (TYHK) and variations described in the text. The ETSB used was a modification of a previously described medium (26) and consisted of Trypticase soy broth (TSB) (30 g/950 ml), yeast extract (Difco) (1 g/950 ml), glucose (1 g/950 ml), and potassium nitrate (0.5 g/950 ml), pH 7.1 to 7.3. This basal medium was then autoclaved, and filter-sterilized supplements were added, which consisted of anhydrous sodium carbonate (0.4 g), cysteine-HCl (Sigma) (0.4 g), hemin (Sigma) stock solution (solution a) (10 ml), vitamin K (Sigma) stock solution (solution b) (0.2 ml), and distilled water (40 ml). Stock solutions were made as follows. Solution a was made by dissolving 50 mg of hemin (Sigma) in 1.0 ml of 1.0 N NaOH, adding 99 ml of distilled water, and storing the solution at 4°C. Solution b was made by dissolving 250 mg of vitamin K (Sigma) in 50 ml of 95% ethanol and storing it at 4°C. These stock solutions were replaced 2 weeks following preparation. The composition of TYHK was Trypticase soy broth (30 g/liter), yeast extract (5 g/liter) (Difco), hemin (Sigma) (0.005 g/liter), and vitamin K3 (menadione; Sigma) (0.001 g/liter), pH 7.2, and the medium was subjected to autoclaving. Bacterial growth was monitored by following the optical density at 600 nm, cells were harvested in the stationary phase of growth, and final bacterial yields were determined by wet weight after centrifugation and washing.
Purification and characterization of LPS.
P. gingivalis LPS was prepared by the Tri-Reagent procedure (30) and the phenol-water procedure (28) as previously described. Phenol-purified LPS was further treated to remove trace amounts of endotoxin protein as described by Manthey and Vogel (14), with the following modification. Following the final ethanol precipitation, LPS was lyophilized to determine the yield and was resuspended in distilled H2O to 1 mg/ml without the addition of triethanolamine. LPS obtained with Tri-Reagent was further purified by the following steps. One milligram of lyophilized LPS (the last step in the Tri-Reagent procedure) was suspended in 1 ml of cold (stored at 20°C) 0.375 M MgCl2 in 95% ethanol (EtOH) and transferred to a 1.5-ml Eppendorf tube, and after complete mixing, the suspension was centrifuged at 2,300 × g for 5 min. This step was repeated twice. The second supernatant was decanted, 1 ml of 100% EtOH (room temperature) was added, and the suspension was thoroughly mixed and subjected to centrifugation at 2,300 × g for 5 min. This process was repeated twice. The final pellet was resuspended in 0.1 ml of endotoxin-free water. Both the Tri-Reagent and phenol-purified LPS preparations were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained for protein by the enhanced colloidal gold procedure as described previously (14). The colloidal gold procedure revealed <0.1% protein contamination in either of the LPS preparations based upon the amount of LPS loaded into the gel and the intensity of the major protein band relative to that of a known bovine serum albumin standard. Gas chromatographic/mass spectrometric analysis of fatty acids present in P. gingivalis LPS1690 revealed i-3-OH C15, 3-OH C16, C16, and i3-O C17 as the major fatty acids, with trace amounts of C14:0 and C18:0. No other fatty acid peaks were detected. These data demonstrated that there was little or no phospholipid, glycolipid, or lipoprotein contamination in the P. gingivalis preparations and were consistent with the notion that the additional lipid A mass ions found clustered around m/z 1,690 were the result of an altered distribution of the fatty acids generating different lipid A structures. For example, the mass ion peak found at m/z 1,704 could be generated by containing two 3-OH C16 molecules instead of one 3-OH C16 and one i-3-OH C15 molecule. This type of lipid A heterogeneity has also been reported for LPS obtained from Leptospira interrogans (20), and further analysis will be required to elucidate each penta-acylated lipid A structure. At least three separate extractions of each P. gingivalis LPS were produced and analyzed.
MALDI-TOF analysis of lipid A.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry was performed as previously described (11), using lipid A obtained by the procedure described by Caroff et al. (3).
RESULTS
Hemin concentration alters the lipid A structural content of P. gingivalis.
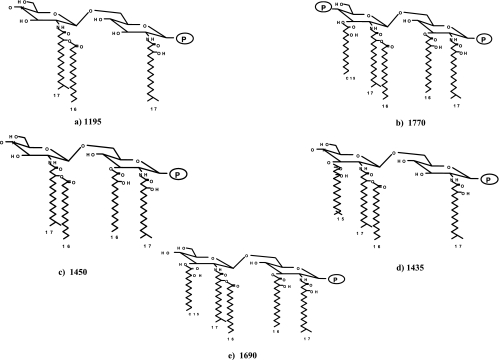
Several different structural types of lipid A have been reported for P. gingivalis LPS (1, 8, 13, 16, 30), and Fig. 1 depicts those lipid A structures that have been characterized structurally (13, 16). The major lipid A structures consist of di- and monophosphoryl penta-acylated lipid A and monophosphoryl tetra-acylated lipid A forms (see the legend to Fig. 1 for structural assignments to lipid A mass ions identified by MALDI-TOF). Furthermore, additional lipid A structural heterogeneity is found in that the mono- and diphosphoryl penta-acylated structures as well as the monophosphoryl tetra-acylated structure contain additional lipid A mass ions that differ by 14 atomic mass units from m/z 1,770, 1,690, and 1,449, respectively. This type of heterogeneity was originally observed by Kumada et al. (13) and was suspected to represent fatty acid heterogeneity but was not defined. Subsequently, it has been found that while the lipid A biosynthetic genes lpxA and lpxD in some bacterial species demonstrate strict acyl-chain-length specificity (29), in other bacteria these genes can transfer fatty acids with differing chain lengths to the nascent lipid A (25a). We suspect that P. gingivalis LpxA and LpxD can transfer fatty acids with different chain lengths to the nascent lipid A and that this can account for the “clusters” of lipid A structural forms observed in the MALDI-TOF spectrum (data not presented). For example, centered around the penta-acylated monophosphoryl structure at m/z 1,690 is a mass ion peak at m/z 1,704 which could be generated by containing two 3-OH C16 molecules instead of one 3-OH C16 and one i-3-OH C15 molecule, and conversely, the mass ion peak found at m/z 1,676 could contain two i-3-OH C15 molecules instead of one 3-OH C16 and one i-3-OH C15 molecule. This type of lipid A heterogeneity has also been reported for LPS obtained from Leptospira interrogans (20), and further analysis will be required to determine if relaxed fatty acid acyl-chain-length specificity can account for this type of lipid A heterogeneity.
FIG. 1.
Structures of previously characterized P. gingivalis lipid A mass ions. P. gingivalis LPS contains several different lipid A structures that differ in their numbers of phosphate groups and fatty acids. In this figure, the MALDI-TOF lipid A peaks that have been characterized with respect to their structure are presented. (A) Structure determined by Ogawa (16); (B to E) structures elucidated by Kumada et al. (13).
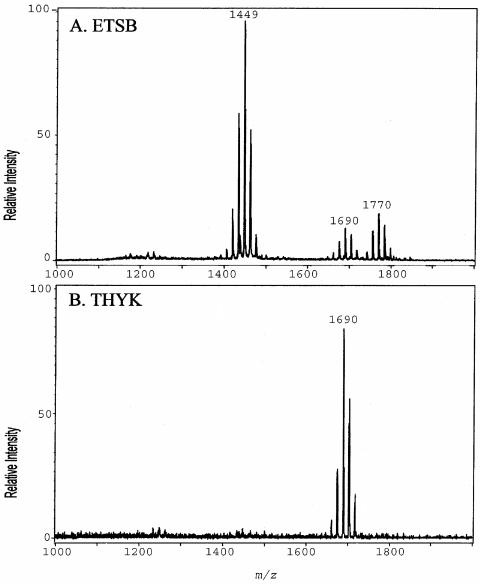
We have reported that the culture medium can affect the number and types of lipid A structures found in P. gingivalis LPS, significantly affecting the lipid A content of the bacterium (8). It was found that growth of strain 33277 to early stationary phase in TYHK medium yielded an LPS preparation that contained a single penta-acylated lipid A cluster (21). In contrast, growth of the same P. gingivalis 33277 strain in ETSB medium yielded a mix of lipid A structures that included both tetra- and penta-acylated forms (8). These results are depicted in Fig. 2. For these experiments, the Tri-Reagent LPS extraction procedure (30) was employed since it was found that this isolation procedure obtains the most representative sample of the different lipid A structural types (8). Examination of the lipid A content in the logarithmic or late stationary phase of growth revealed that the culture medium composition, not the phase of growth, affected the lipid A content.
FIG. 2.
Effect of in vitro culture medium on P. gingivalis lipid A structural content. (A) P. gingivalis strain 33277 incubated in ETSB medium; (B) the same strain incubated in TYHK medium (see the text for details). P. gingivalis LPS was obtained by the Tri-Reagent procedure (30), and lipid A was cleaved and separated from the LPS as described by Caroff et al. (3). MALDI-TOF analysis was performed as previously described (11). All values given are average masses rounded to the nearest whole numbers for singly charged deprotonated molecules. Note that the in vitro culture medium significantly affected the lipid A structural content.
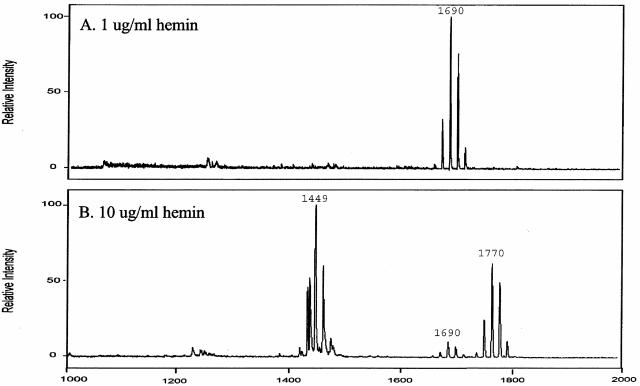
For this report, the medium component or components responsible for the effect on the lipid A content were examined. ETSB contains glucose, cysteine-HCl, sodium carbonate, and filter-sterilized hemin, while TYHK does not contain these components and employs autoclaved hemin. Each of these components was examined individually and in combination by selectively adding them to TYHK medium and determining the resultant lipid A content of strain 33277 after growth. No effect on the growth rate or final yield of bacteria was observed with any of these medium additions. It was found that employing filter-sterilized hemin (5 μg/ml) in TYHK medium was sufficient to elicit the lipid A content observed when P. gingivalis was grown in ETSB. None of the other ETSB medium component additions to TYHK significantly altered the lipid A content (data not shown). It was reasoned that although the hemin concentration was 5 μg/ml for both ETSB and TYHK, autoclaving the hemin in TYHK medium may have reduced the available hemin concentration. Therefore, the lipid A content of P. gingivalis was examined after growth in TYHK medium containing either 1 or 10 μg/ml hemin (Fig. 3). With 1 μg/ml hemin, a single major monophosphoryl penta-acylated lipid A cluster (centered at m/z 1,690) was observed, while the lipid A content of bacteria incubated with 10 μg/ml hemin showed a significantly reduced amount of this lipid A structure and a significant increase in both monophosphoryl tetra-acylated lipid A structures (m/z 1,435 and 1,449) and a diphosphoryl penta-acylated lipid A cluster (centered at m/z 1,770), consistent with the lipid A content observed after growth in ETSB medium.
FIG. 3.
Effect of hemin concentration on lipid A content of P. gingivalis strain 33277. P. gingivalis strain 33277 was incubated in TYHK medium with either 1 (A) or 10 (B) μg/ml hemin. P. gingivalis LPS was obtained by the Tri-Reagent procedure (28), and lipid A was cleaved and separated from the LPS as described by Caroff et al. (3). MALDI-TOF analysis was performed as previously described (11). All values given are average masses rounded to the nearest whole numbers for singly charged deprotonated molecules. Note that at 1 μg/ml hemin, the lipid A content was similar to that for growth in TYHK medium, whereas at 10 μg/ml hemin, the lipid A content resembled that found in ETSB medium (see Fig. 2 for comparison).
Phenol-isolated LPS displays similar hemin-mediated effects on lipid A content.
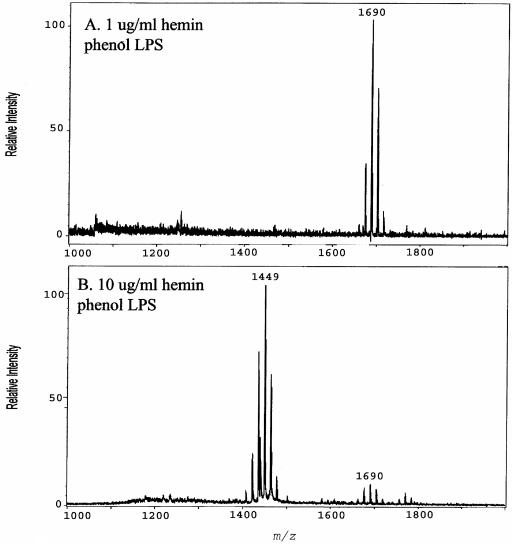
Next, the effect of the LPS extraction procedure on the lipid A content of P. gingivalis grown in TYHK medium containing either a high (10 μg/ml) or low (1 μg/ml) hemin concentration was examined, since we have previously reported that the MgCl2-EtOH LPS extraction procedure can influence the lipid A contents of LPS preparations obtained from P. gingivalis grown in TYHK medium (8). Therefore, the effect of the commonly employed phenol-water procedure (28) on the lipid A content was examined (Fig. 4). At the high hemin concentration, there was a loss of the penta-acylated lipid A cluster centered around m/z 1,690 and a corresponding increase in tetra-acylated lipid A structures at m/z 1,435 and 1,449 (Fig. 4), similar to those observed with Tri-Reagent-isolated LPS (Fig. 3). However, in contrast to the results obtained with Tri-Reagent-isolated LPS, at the high hemin concentration, phenol-isolated LPS displayed a significant reduction in the diphosphorylated penta-acylated lipid A cluster centered around m/z 1,770 (compare Fig. 3, bottom panel, and 4B). These results demonstrate that phenol isolation either does not extract this lipid A structure as efficiently as the Tri-Reagent procedure or selectively degrades the lipid A structure at m/z 1,770 during isolation. Nevertheless, the effect of a high hemin concentration yielding a significant decrease in penta-acylated structures and a corresponding increase in tetra-acylated lipid A structures was observed with both isolation procedures. All subsequent experiments were performed with the Tri-Reagent extraction procedure.
FIG. 4.
Effect of phenol-water LPS isolation procedure on lipid A content of P. gingivalis strain 33277. P. gingivalis strain 33277 was incubated in TYHK medium with either 1 (A) or 10 (B) μg/ml hemin. P. gingivalis LPS was obtained by the phenol-water isolation procedure (30), and lipid A was cleaved and separated from the LPS as described by Caroff et al. (3). MALDI-TOF analysis was performed as previously described (11). All values given are average masses rounded to the nearest whole numbers for singly charged deprotonated molecules. Note that the phenol-water isolation procedure failed to effectively extract lipid A structures centered at m/z 1,770 (see Fig. 3, bottom panel, for comparison).
Hemin, not an increase in iron acquisition, is responsible for the alteration in lipid A content.
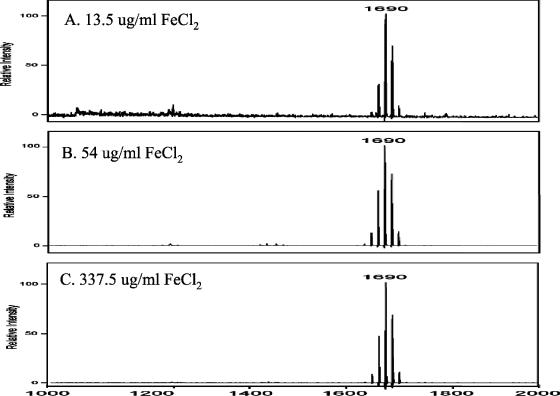
Hemin uptake by P. gingivalis represents an important iron acquisition system in vivo (17); however, P. gingivalis is able to utilize inorganic iron for growth in vitro (22). Therefore, the possibility that the effect of increased hemin on the lipid A content of P. gingivalis was mediated by increased iron acquisition was examined. In these experiments, P. gingivalis was grown in TYHK medium containing various concentrations of FeCl2 instead of hemin. Each culture was monitored for growth, and no difference in the generation time or final yield of bacteria was observed at the different FeCl2 concentrations. As shown in Fig. 5, when P. gingivalis was incubated with 13.5 μg/ml FeCl2 (a concentration found in most media employed to grow P. gingivalis in the absence of hemin) or with FeCl2 at a 4- or 25-fold greater concentration, no effect on the lipid A content was observed. In each of these cultures, one major lipid A cluster corresponding to monophosphoryl penta-acylated lipid A was observed. Therefore, it is the concentration of hemin, not an increase in complexed iron, that is responsible for the alteration in the lipid A content.
FIG. 5.
Effects of various FeCl2 concentrations on lipid A content of P. gingivalis strain 33277. P. gingivalis strain 33277 was incubated in TYHK medium containing no hemin and either 13.5 (A), 54 (B), or 337.5 (C) μg/ml FeCl2. P. gingivalis LPS was obtained by the Tri-Reagent procedure (30), and lipid A was cleaved and separated from the LPS as described by Caroff et al. (3). MALDI-TOF analysis was performed as previously described (11). All values given are average masses rounded to the nearest whole numbers for singly charged deprotonated molecules. Note that the FeCl2 concentration did not significantly alter the lipid A content.
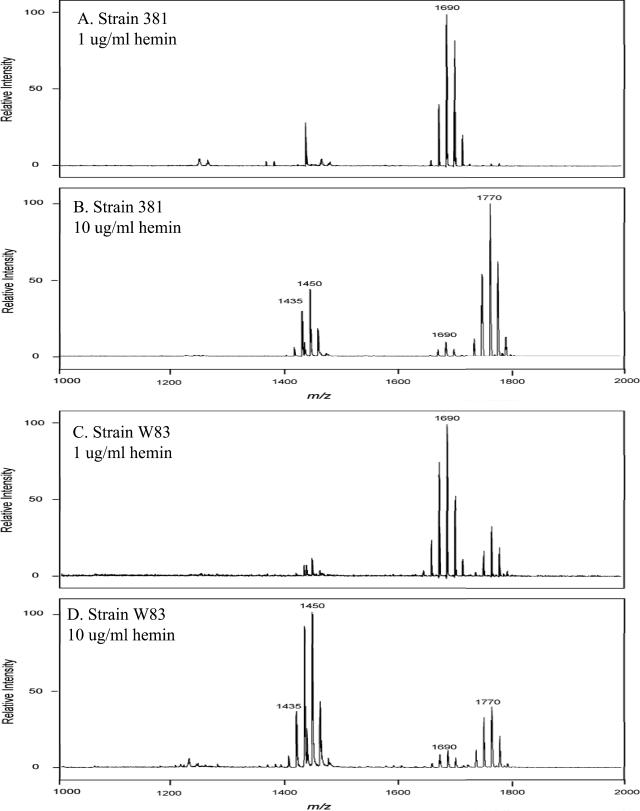
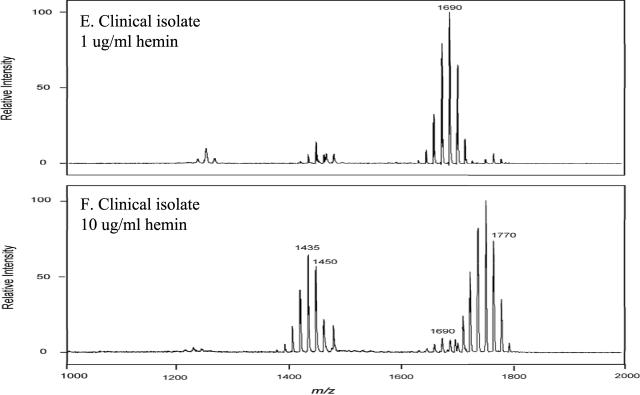
Hemin concentration alters the lipid A content in multiple strains of P. gingivalis.
Next, the effects of hemin concentration on the lipid A contents of different strains of P. gingivalis were examined. In these experiments, two commonly employed laboratory strains (W83 and 381) as well as a low-passage clinical isolate were incubated in TYHK medium containing either 1 or 10 μg/ml hemin and LPS extracted by the Tri-Reagent procedure. As shown in Fig. 6, each strain demonstrated the same effect in that at the high hemin concentration (10 μg/ml), there was a significant reduction in the monophosphoryl penta-acylated lipid A cluster centered around m/z 1,690 and a corresponding increase in tetra-acylated lipid A structures (m/z 1,435 and 1,449) as well as diphosphorylated penta-acylated lipid A (m/z 1,770). The relative amounts of these lipid A structures varied among the strains examined. However, the observation that an increased hemin concentration significantly alters the lipid A content, with a consistent loss of monophosphoryl penta-acylated lipid A structures and an increase in tetra-acylated as well as diphosphorylated penta-acylated lipid A structures, was established for multiple strains.
FIG. 6.
Effect of hemin concentration on lipid A contents of different P. gingivalis strains. P. gingivalis laboratory strains 381 (A and B) and W83 (C and D) and a freshly obtained P. gingivalis clinical isolate (E and F) were incubated in TYHK medium with either 1 (A, C, and E) or 10 (B, D, and F) μg/ml hemin. P. gingivalis LPS was obtained by the Tri-Reagent procedure (30), and lipid A was cleaved and separated from the LPS as described by Caroff et al. (3). MALDI-TOF analysis was performed as previously described (11). All values given are average masses rounded to the nearest whole numbers for singly charged deprotonated molecules. Note that the concentration of hemin affected the lipid A contents of several different strains in a similar fashion.
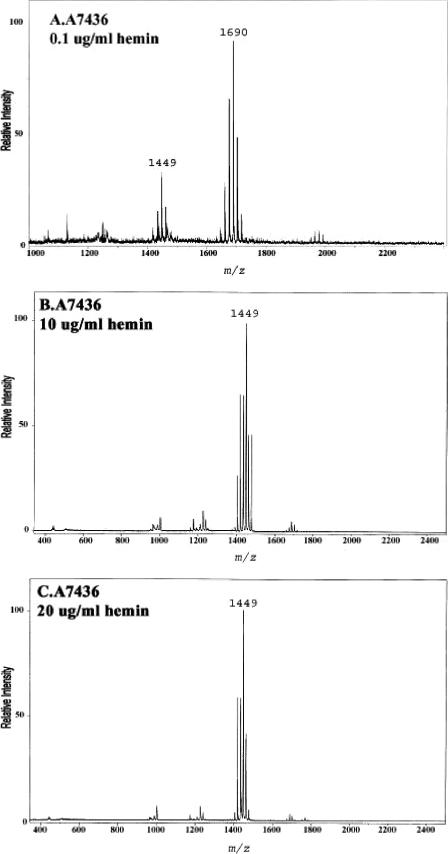
Previously identified hemin acquisition proteins participate in alteration of the lipid A content.
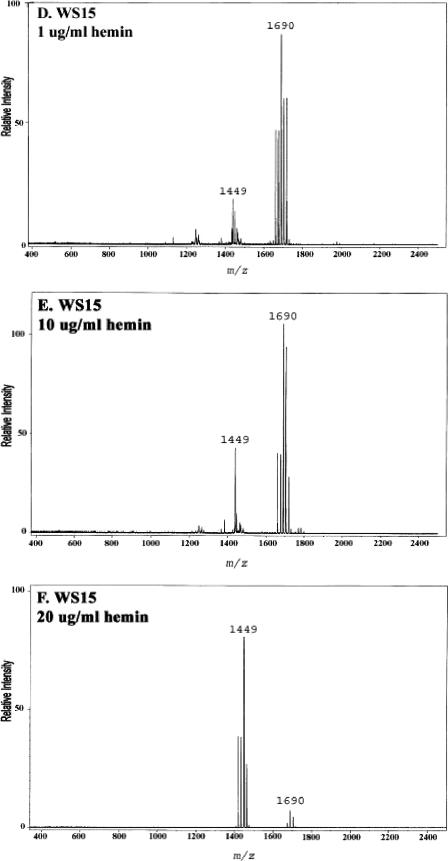
The data presented above demonstrated that P. gingivalis can detect the amount of hemin in the growth medium and respond by altering its lipid A content. Next, the contributions of P. gingivalis proteins known to participate in hemin acquisition to the changes observed in the lipid A content were examined. In these experiments, the lipid A contents of a mutant of P. gingivalis which has impaired hemin uptake and of its isogenic parent were compared after growth at various hemin concentrations (Fig. 7). The mutant strain, designated WS15, and its isogenic parent strain, designated A7436, have been described previously (22). Briefly, strain WS15 demonstrates impaired hemin binding due to mutations in two hemin uptake proteins, one of which is a well-characterized protease designated Kgp, and the other of which, designated HmuR, is involved in hemin transport across the periplasmic space. Strains A7436 and WS15 were incubated in TYHK medium containing various concentrations of hemin, and their lipid A contents were determined after growth to early stationary phase as described above. Both strains grew equally well at each hemin concentration, as demonstrated by similar growth curves and similar final bacterial yields. As shown in Fig. 7, however, there was a significant difference in the lipid A content of each strain at the different hemin concentrations. The parental strain, A7436, displayed one major lipid A cluster at m/z 1,690 corresponding to the monophosphoryl penta-acylated lipid A structures when incubated with 0.1 μg/ml hemin and tetra-acylated lipid A structures when incubated at higher hemin concentrations. Interestingly, this strain displayed some variability in the amounts of penta- and tetra-acylated lipid A structures when incubated with 1 μg/ml hemin, indicating that this concentration of hemin may be close to the level which facilitates the change in the lipid A content. In contrast to the parental strain, the mutant strain, WS15, contained a single penta-acylated lipid A cluster centered around m/z 1,690 when incubated with 1 and 10 μg/ml hemin and tetra-acylated lipid A structures when incubated with 20 μg/ml hemin. These data demonstrate that when hemin uptake is impaired, as in strain WS15, more hemin is required to elicit a change in the lipid A content. The data suggest that previously characterized hemin acquisition and detection proteins, such as Kgp and HmuR, contribute to the ability of P. gingivalis to “sense” the hemin concentration and alter the lipid A structural content.
FIG.7.
Effect of hemin concentration on lipid A content of a P. gingivalis hemin acquisition mutant. P. gingivalis strain A7436 (A, B, and C) and its isogenic Kgp/HmuR double mutant strain WS15 (D, E, and F) were incubated in TYHK medium with various concentrations of hemin, as indicated in each panel. P. gingivalis LPS was obtained by the Tri-Reagent procedure (30), and lipid A was cleaved and separated from the LPS as described by Caroff et al. (3). MALDI-TOF analysis was performed as previously described (11). All values given are average masses rounded to the nearest whole numbers for singly charged deprotonated molecules. Note that strain WS15 required a higher concentration of hemin to alter its lipid A content.
DISCUSSION
The identification of hemin as a medium component that can significantly alter the lipid A structural content of P. gingivalis has important implications for innate host responses to LPS. Specifically, it has recently been shown that the major penta-acylated lipid A structural forms found when P. gingivalis is grown at low hemin concentrations are TLR4 agonists and facilitate E-selectin activation on human endothelial cells (21). In contrast, the tetra-acylated structural forms found at high hemin concentrations have been shown to be TLR4 antagonists (5, 21). Therefore, increased hemin concentrations facilitate the production of lipid A structures that are TLR4 antagonists. The ability to significantly alter P. gingivalis LPS interactions with TLR4 represents a form of immunomodulation (19) that may facilitate survival of the bacterium in different host environments. With respect to the development of periodontitis, the concentration of hemin may change significantly during stages of vascular ulceration, as measured by bleeding upon probing. Consistent with hemin having an effect on the lipid A content, as described here, two previous studies have shown that the hemin concentration can affect the potency (4) and composition (6) of P. gingivalis LPS. These studies were performed before MALDI-TOF analysis of lipid A structures was available, and it is highly likely that the changes in lipid A content described here contributed to the effects described in these publications.
The observation that the in vitro hemin concentration can significantly modulate the lipid A structural composition may help to explain some of the unusual and apparently conflicting results obtained with P. gingivalis LPS preparations obtained from different laboratories (8). Firstly, the structure of lipid A of P. gingivalis has been controversial, although this now may be explained by the observation that P. gingivalis may contain multiple lipid A structures and that growth conditions can influence which structures are present. Furthermore, we have previously shown that the MgCl2-EtOH LPS extraction procedure enriches for tetra-acylated lipid A structures (8), and in this work it was shown that the commonly employed phenol-water isolation procedure also does not yield an accurate representation of what is present in the bacterium. These factors have almost certainly contributed to some of the confusion regarding P. gingivalis lipid A structure and host responses.
The mechanisms responsible for the alteration of the P. gingivalis lipid A structural content in response to the hemin concentration are not currently understood. As shown in this report, hemin concentration sensing involves the known hemin acquisition proteins Kgp and/or HmuR but may also involve as yet unidentified hemin binding or transport proteins. Consistent with this, a recent report presented evidence that hemin uptake may be mediated by proteins other than Kgp and HmuR (22). After the concentration of hemin is detected, the structural changes in the lipid A content suggest that at least two different phenomena occur. Firstly, at high hemin concentrations, deacylase activity (27) may be induced, which would account for the loss of the major penta-acylated lipid A structures at m/z 1,690 and the formation of the major tetra-acylated lipid A structures found at m/z 1,449. A recent report (10) described the distribution of PagL deacylase homologs in gram-negative bacteria, and although a PagL homologue was not found in P. gingivalis, other potential analogues or unrelated deacylase enzymes may be present. Secondly, the appearance of the penta-acylated diphosphorylated lipid A structure at m/z 1,770 suggests that an increased hemin concentration may induce more LPS biosynthesis. This lipid A structure represents the most complete lipid A synthesized, and its appearance in the LPS when P. gingivalis is grown at high hemin concentrations suggests that new lipid A is being synthesized. The effects of hemin concentration on lipid A modification and synthesis are currently under investigation.
Acknowledgments
This work was supported by NIH grants DE-12768 and DE-09161.
Editor: J. B. Bliska
REFERENCES
- 1.Bainbridge, B. W., S. R. Coats, and R. P. Darveau. 2002. Porphyromonas gingivalis lipopolysaccharide displays functionally diverse interactions with the innate host defense system. Ann. Periodontol. 7:1-9. [DOI] [PubMed] [Google Scholar]
- 2.Beutler, B. 2000. Tlr4: central component of the sole mammalian LPS sensor. Curr. Opin. Immunol. 12:20-26. [DOI] [PubMed] [Google Scholar]
- 3.Caroff, M., A. Tacken, and L. Szabó. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175:273-282. [DOI] [PubMed] [Google Scholar]
- 4.Champagne, C. M., S. C. Holt, T. E. Van Dyke, B. J. Gordon, and L. Shapira. 1996. Lipopolysaccharide isolated from Porphyromonas gingivalis grown in hemin-limited chemostat conditions has a reduced capacity for human neutrophil priming. Oral Microbiol. Immunol. 11:319-325. [DOI] [PubMed] [Google Scholar]
- 5.Coats, S. R., T. T. Pham, B. W. Bainbridge, R. A. Reife, and R. P. Darveau. 2005. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J. Immunol. 175:4490-4498. [DOI] [PubMed] [Google Scholar]
- 6.Cutler, C. W., P. I. Eke, C. A. Genco, T. E. Van Dyke, and R. R. Arnold. 1996. Hemin-induced modifications of the antigenicity and hemin-binding capacity of Porphyromonas gingivalis lipopolysaccharide. Infect. Immun. 64:2282-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darveau, R. P. 2000. Oral innate host defense responses: interactions with microbial communities and their role in the development of disease, p. 169-218. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bacterial ecology: the molecular basis. Horizon Scientific Press, Wymondham, Norfolk, United Kingdom.
- 8.Darveau, R. P., T. T. Pham, K. Lemley, R. A. Reife, B. W. Bainbridge, S. R. Coats, W. N. Howald, S. S. Way, and A. M. Hajjar. 2004. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both Toll-like receptors 2 and 4. Infect. Immun. 72:5041-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 10.Geurtsen, J., L. Steeghs, J. T. Hove, P. van der Ley, and J. Tommassen. 2005. Dissemination of lipid A deacylases (pagL) among gram-negative bacteria: identification of active-site histidine and serine residues. J. Biol. Chem. 280:8248-8259. [DOI] [PubMed] [Google Scholar]
- 11.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 12.Kawahara, K., H. Tsukano, H. Watanabe, B. Lindner, and M. Matsuura. 2002. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 70:4092-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumada, H., Y. Haishima, T. Umemoto, and K.-I. Tanamoto. 1995. Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J. Bacteriol. 177:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manthey, C. L., and S. N. Vogel. 1994. Elimination of trace endotoxin protein from rough chemotype LPS. J. Endotoxin Res. 1:84-91. [Google Scholar]
- 15.Marsh, P. D., A. S. McDermid, A. S. McKee, and A. Baskerville. 1994. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis W50. Microbiology 140:861-865. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa, T. 1993. Chemical structure of lipid A from Porphyromonas (Bacteroides) gingivalis lipopolysaccharide. FEBS Lett. 332:197-201. [DOI] [PubMed] [Google Scholar]
- 17.Olczak, T., W. Simpson, X. Liu, and C. A. Genco. 2005. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol. Rev. 29:119-144. [DOI] [PubMed] [Google Scholar]
- 18.Page, R. C., S. Offenbacher, H. E. Schroeder, G. J. Seymour, and K. S. Kornman. 1997. Advances in the pathogenesis of periodontitis: summary of developments, clinical implications and future directions. Periodontol. 2000 14:216-248. [DOI] [PubMed] [Google Scholar]
- 19.Portnoy, D. A. 2005. Manipulation of innate immunity by bacterial pathogens. Curr. Opin. Immunol. 17:1-4. [DOI] [PubMed] [Google Scholar]
- 20.Que-Gewirth, N. L., A. A. Ribeiro, S. R. Kalb, R. J. Cotter, D. M. Bulach, B. Adler, I. S. Girons, C. Werts, and C. R. Raetz. 2004. A methylated phosphate group and four amide-linked acyl chains in Leptospira interrogans lipid A. The membrane anchor of an unusual lipopolysaccharide that activates TLR2. J. Biol. Chem. 279:25420-25429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reife, R. A., S. R. Coats, M. Al-Qutub, D. R. Dixon, P. A. Braham, R. J. Billharz, W. N. Howald, and R. P. Darveau. 2006. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell. Microbiol. 8:857-868. [DOI] [PubMed] [Google Scholar]
- 22.Simpson, W., T. Olczak, and C. A. Genco. 2004. Lysine-specific gingipain K and heme/hemoglobin receptor HmuR are involved in heme utilization in Porphyromonas gingivalis. Acta Biochim. Pol. 51:253-262. [PubMed] [Google Scholar]
- 23.Smalley, J. W., A. J. Birss, A. S. McKee, and P. D. Marsh. 1996. Haemin binding as a factor in the virulence of Porphyromonas gingivalis. FEMS Microbiol. Lett. 141:65-70. [DOI] [PubMed] [Google Scholar]
- 24.Socransky, S. S., and A. D. Haffajee. 1992. The bacterial etiology of destructive periodontal disease: current concepts. J. Periodontol. 63:322-331. [DOI] [PubMed] [Google Scholar]
- 25.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 25a.Sweet, C. R., A. Preston, E. Toland, S. M. Ramirez, R. J. Cotter, D. J. Maskell, and C. R. Raetz. 2002. Relaxed acyl chain specificity of Bordetella UDP-N-acetylglucosamine acyltransferases. J. Biol. Chem. 277:18281-18290. [DOI] [PubMed] [Google Scholar]
- 26.Syed, S. A. 1980. Characteristics of Bacteroides asaccharolyticus from dental plaques of beagle dogs. J. Clin. Microbiol. 11:522-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trent, M. S., W. Pabich, C. R. Raetz, and S. I. Miller. 2001. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 276:9083-9092. [DOI] [PubMed] [Google Scholar]
- 28.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure, p. 83-91. In R. Whistler (ed.), Methods in carbohydrate chemistry, vol. 5. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 29.Wyckoff, T. J., S. Lin, R. J. Cotter, G. D. Dotson, and C. R. Raetz. 1998. Hydrocarbon rulers in UDP-N-acetylglucosamine acyltransferases. J. Biol. Chem. 273:32369-32372. [DOI] [PubMed] [Google Scholar]
- 30.Yi, E. C., and M. Hackett. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651-656. [DOI] [PubMed] [Google Scholar]