Abstract
The obligatory intracellular bacterium Chlamydophila psittaci is the causative agent of psittacosis in birds and humans. The capability of this zoonotic pathogen to develop a persistent phase is likely to play a role in chronicity of infections, as well as in failure of antibiotic therapy and immunoprophylaxis. To elucidate three different in vitro models for transition of C. psittaci to persistence (iron depletion, penicillin G treatment, and gamma interferon [IFN-γ] exposure), a set of 27 genes was examined by mRNA expression analysis using quantitative real-time PCR. While the phenotypical characteristics were the same as in other chlamydiae, i.e., aberrant morphology of reticulate bodies, loss of cultivability, and rescue of infectivity upon removal of inducers, the transcriptional response of C. psittaci to persistence-inducing factors included several new and distinctive features. Consistent downregulation of membrane proteins, chlamydial sigma factors, cell division protein, and reticulate body-elementary body differentiation proteins from 24 h postinfection onward proved to be a general feature of C. psittaci persistence. However, other genes displayed considerable variations in response patterns from one model to another, which suggests that there is no persistence model per se. In contrast to results for Chlamydia trachomatis, late shutdown of essential genes in C. psittaci was more comprehensive with IFN-γ-induced persistence, which is probably due to the absence of a functional tryptophan synthesis operon.
Chlamydophila psittaci (formerly avian Chlamydia psittaci) is the causative agent of psittacosis, a systemic disease in psittacine birds which can be of acute, protracted, chronic, or subclinical manifestation and which represents the most important animal chlamydiosis of zoonotic character (2, 20). Avian strains are pathogenic to humans, the symptoms being mainly nonspecific and influenza-like, but severe pneumonia, endocarditis, and encephalitis are not uncommon (34). Besides, recent surveys showed that C. psittaci can also be found in nonavian domestic animals and wildlife (17, 38, 40), although its role in these hosts remains to be clarified.
Generally speaking, chlamydial infections can generate a variety of clinical diseases, ranging from acute self-limiting infection, as in cases of human urogenital disease, neonatal conjunctivitis, and pneumonia, to chronic inflammation, as in cases of trachoma, pelvic inflammatory disease, reactive arthritis, chronic obstructive pulmonary disease, and cardiovascular disease. On the other hand, the infection often takes a mild or subclinical course. This remarkable variety of clinical manifestations should, at least in part, be a consequence of the distinctive features of the causative agents. Chlamydiae are a group of obligately intracellular bacteria distinguished by their unique biphasic developmental cycle. In the course of a replication cycle, infectious but metabolically inactive elementary bodies (EBs) evolve into noninfectious but metabolically active reticulate bodies (RBs). The latter reside in a vacuole-like inclusion of the host cell and undergo binary fission before transforming back into elementary bodies to start a fresh cycle.
However, the classical hypothesis of a lytic biphasic cycle seems to reflect only optimized growth conditions and needs to be extended by persistence as a third state (16). Chlamydial persistence is known as a state of infection during which the pathogen remains viable but noncultivable, while the host immune system is incapable of eliminating it (4, 9). Morphologically, this reversible state is characterized by aberrant bodies, i.e., enlarged pleomorphic RBs, and reduced inclusion size.
In the literature, eight different in vitro models have been used to study the behavior of chlamydiae and the host cell response in the course of persistent infection. These include antibiotic treatment (23), amino acid depletion (10), iron depletion (31), and gamma interferon (IFN-γ) exposure (3). The publication of complete genome sequences of Chlamydia trachomatis and Chlamydophila pneumoniae (32, 37) has opened up the possibility of systematically investigating the transcriptional response during persistence in these human pathogens. Recent studies have focused on genes involved in regulation of the developmental cycle (14, 26), energy metabolism (13), and cell membrane structures (15).
Despite some general similarities in mRNA expression profiles, such as downregulation of genes that are expressed late in the productive developmental cycle, downregulation of genes that encode proteins involved in cell division, and a decrease in the ratio of MOMP to HSP60 (ompA-to-groEL ratio), significant differences among the various in vitro persistence models have been noted (16). The failure to derive a common pattern of transcriptional response from the studies published so far is a consequence of the different molecular mechanisms of persistence induction underlying each model, and it also reflects the variations in chlamydial species and strains, host cell lines, and other experimental conditions.
Although earlier data from morphological, ultrastructural, and cell biological studies showed that C. psittaci was also capable of persistence (7, 23, 25, 33), changes in the transcriptome have not been investigated. Systematic studies are somewhat hampered, as the complete genome of C. psittaci has yet to be sequenced. However, molecular data are required to improve our understanding of persistence as not only a chlamydial phenomenon but also a potentially important factor in C. psittaci infections and in the pathogenesis of the ensuing human and animal diseases.
In the present study, 27 genes of C. psittaci were examined by mRNA expression analysis in three different in vitro models of persistence. Immunofluorescence and electron microscopy were used to visualize specific morphological alterations, and mRNA expression rates were measured by quantitative real-time PCR.
MATERIALS AND METHODS
Host cells and bacterial strain.
HEp-2 cells (ATCC-CCL23) were cultured at 37°C in 5% CO2 in Eagle's minimal essential medium containing Earle's salts (Cambrex Biosciences, Verviers, Belgium) and supplemented with 5% fetal bovine serum (Cambrex Biosciences) and 2 mM glutamine (Sigma-Aldrich, Munich, Germany). Strain DC15 of C. psittaci, which was isolated in the lab of the authors from an aborted calf fetus, was grown in HEp-2 cell monolayers without the addition of cycloheximide. The species identity of the strain was established by sequencing the 16S rRNA and ompA genes. Chlamydial EBs were harvested by scraping off cultured cells with a cell scraper at 48 h postinfection (p.i.). Cells containing mature EBs were centrifuged at 1,500 × g for 10 min. Cell pellets were resuspended in medium A (phosphate-buffered saline [pH 7.2] containing 2% [vol/vol] fetal bovine serum, 0.2 M sucrose, and 0.002% [wt/vol] phenol red), aliquoted, titrated, and stored at −80°C until use. Prior to the experiments, the HEp-2 cell line and the chlamydial strain were examined for the absence of mycoplasma contamination by PCR using primers MW28 (5′-CCAGACTCCTACGGGAGGCA-3′) and MW29 (5′-TGCGAGCATACTACTCAGGC-3′), which are specific for the class Mollicutes (18).
Infection and induction of persistence.
Confluent HEp-2 monolayers (106 cells) grown in T25 cell culture flasks (10 Cellstar; Greiner Bio-One, Frickenhausen, Germany) were infected with strain DC15 at a multiplicity of infection (MOI) of 1 and centrifuged at 3,400 × g at 37°C for 1 h. Infected cells were further incubated for an additional hour at 37°C and 5% CO2. The removal of unbound EBs by aspiration of the supernatant was followed by the addition of fresh Eagle's minimal essential medium containing the respective persistence inducer (persistence models) or lacking it (acute infection as the control). The final concentrations of the inducers were 150 μM deferoxamine methanesulfonate (DAM) (Sigma-Aldrich) in the iron depletion model and 200 U/ml of penicillin G (Jenapharm, Jena, Germany) in the penicillin G model. In the IFN-γ exposure model, HEp-2 monolayers were preincubated 24 h before infection with 240 U/ml of recombinant human IFN-γ (PeproTech, London, United Kingdom). After medium exchange at 2 h p.i., another 240 U/ml of IFN-γ was added. Infected HEp-2 monolayers were incubated at 37°C and 5% CO2 for the indicated time periods. For long-term observation, infected and treated HEp-2 monolayers were maintained for up to 96 h, with medium refreshment and addition of fresh persistence inducer at 48 h p.i.
Immunofluorescence staining.
Infected HEp-2 monolayers grown on coverslips were fixed with methanol at 48 h p.i. Samples were incubated with an anti-lipopolysaccharide Chlamydiaceae-specific fluorescein isothiocyanate-labeled antibody using an IMAGEN Chlamydia kit (DakoCytomation, Glostrup, Denmark). Inclusion-containing cells were visualized using a BX-51 M fluorescence microscope (Olympus, Ballerup, Denmark) at 400- and 1,000-fold magnification.
Electron microscopy.
The cell culture supernatant was replaced with 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.2) at 48 h p.i. After 2 h of fixation at 4°C, cells were removed from the cell culture dish using a cell scraper, collected in an Eppendorf tube, and centrifuged for 5 min at 1,500 × g. The cell pellet was embedded in 2% agarose and sectioned to 1-mm3 cubes. Cubes were postfixed in 2% osmium tetroxide and embedded in Araldite Cy212. Semithin sections were prepared to select areas of high cell density for ultramicrotomy. Ultrathin sections (85 nm) were stained with uranyl acetate and lead citrate and examined with a transmission electron microscope (Tecnai 12; FEI, Eindhoven, The Netherlands).
Infectivity assay.
Infectivity in the course of induced persistence was determined by harvesting chlamydial organisms from infected monolayers by sonication with an amplitude of 80% for 8 s with 10 pulses (Branson 450D sonifier; Branson, Danbury, CT) at 48 h p.i. and reinoculating them onto fresh, untreated HEp-2 monolayers. The following concentrations of persistence inducers were used: 0 to 250 μM of DAM for the iron depletion model, 0 to 400 U/ml of penicillin G for the penicillin model, and 0 to 480 U/ml of IFN-γ for the IFN-γ exposure model. Forty-eight hours after reinfection, infected monolayers were fixed with methanol and stained as described above. The numbers of inclusions were counted in 20 random fields and calculated as inclusion-forming units (IFU) per milliliter.
Reactivation assay.
HEp-2 monolayers grown on coverslips were infected with a MOI of 1, and persistence was induced as described above. Twenty-four hours or 48 h p.i., the medium was replaced with complete growth medium, i.e., medium containing no penicillin (penicillin G model) or containing 50 μM FeCl3 (iron depletion model) or a 10-fold amount of tryptophan (102 mg/liter; IFN-γ exposure model). Chlamydial organisms were harvested by sonication 24 h later and inoculated onto fresh HEp-2 cell monolayers using complete standard medium. Forty-eight hours later, monolayers were fixed with methanol, samples were stained as described above, and the infectious titer was estimated by counting 20 random fields per coverslip.
RNA extraction and reverse transcription.
Two independent rounds of infection experiments were performed, where 1 × 106 HEp-2 cells were infected at an MOI of 1 and exposed to the persistence models described above. At 12, 24, 36, and 48 h p.i., total RNA was isolated using an RNeasy Mini Kit (QIAGEN, Hilden, Germany) including DNase I treatment, according to the instructions of the manufacturer. The quality and quantity of total RNA were determined using agarose gel electrophoresis and UV spectrophotometry. In each case, 5 μg of total RNA was reverse transcribed into cDNA using random hexamer primers and a SuperScript III First-Strand Synthesis system for reverse transcription-PCR (Invitrogen, Karlsruhe, Germany) following the manufacturer's instructions.
Design of gene-specific primers.
Since the complete genome sequence of C. psittaci is not yet available, the genes of interest were searched using the Chlamydophila abortus genome as a matrix (http://www.sanger.ac.uk/Projects/C_abortus/). Additionally, unpublished sequences of the type III secretion system operon (provided by Matthias Horn, Vienna, Austria) and primers of the pmp genes (provided by David Longbottom, Edinburgh, United Kingdom) of C. psittaci were included. Primers for C. psittaci were defined using Primer Express (Applied Biosystems) and tested by conventional PCR using chromosomal DNA of this species as template. Amplified products were isolated from gel bands using a QIAquick gel extraction kit (QIAGEN, Hilden, Germany), further processed using a BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems), and sequenced with an ABI Prism 310 genetic analyzer (Applied Biosystems, Darmstadt, Germany). For verification and identification, the sequencing data were subjected to BLAST analysis at http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi.
qrt-PCR.
Measurement of mRNA expression levels using quantitative real-time PCR (qrt-PCR) was performed by using a Mx3000P thermocycler system (Stratagene, La Jolla, CA) with the following temperature-time profile: initial denaturation at 95°C for 10 min, 40 cycles of 95°C for 30 s and 59°C for 1 min, and subsequent melting curve analysis of the amplification product. Each reaction vessel contained a reaction mix of 10 ng cDNA template, 1 μl of each primer (final concentration, 200 nM), 12.5 μl of qPCR Mastermix Plus for SYBR Green I (Eurogentec, Seraing, Belgium), and 8.5 μl of deionized water. All qrt-PCR assays from two independent series of infections were performed at least in triplicate for each target gene with samples collected from each persistence model at 12, 24, 36, and 48 h p.i. The relative gene expression ratio of a target gene was calculated as described by Pfaffl (30) on the basis of real-time amplification efficiency and cycle threshold deviation of a given sample versus the control (acute infection) in comparison to reference gene 16S rRNA or gyrA. Genes showing relative expression ratios higher than 2 or lower than 0.5 with both internal control genes compared to acute infection were considered to be up- or downregulated, respectively, at the given time point.
Statistical analysis.
To compare two independent samples (i.e., mRNA expression ratios of 27 genes from two independent infection experiments, each based on 16S rRNA or gyrA) at the same time point, Student's t test was used. A P value of <0.05 was considered to show a statistically significant difference at the 95% confidence level. Analysis of variance was performed to test multiple data (i.e., all values per gene from independent experiments) along the time axis for significantly different results.
RESULTS
Infectivity and morphological characteristics of C. psittaci cultures in persistent infection.
Three different in vitro cell culture models of persistent infection were studied. Figure 1 illustrates that the addition of either DAM, penicillin G, or IFN-γ caused transition of chlamydial cells to the persistent state. The infectivity of the C. psittaci strain was drastically reduced at 48 h p.i. in all three models. The following concentrations of persistence-inducing substances were considered optimal and used in all subsequent trials: 150 μM DAM (infectivity reduced to 15.6%), 200 U/ml penicillin G (17.0%), or 240 U/ml IFN-γ (14.9%).
FIG. 1.
Decrease of infectivity of C. psittaci after exposure of infected HEp-2 cell cultures to different amounts of persistence-inducing reagents. The number of inclusion-forming units was determined from methanol-fixed immunostained cell cultures at 48 h p.i. (A) Iron depletion by addition of DAM. (B) Penicillin G treatment. (C) Exposure to IFN-γ.
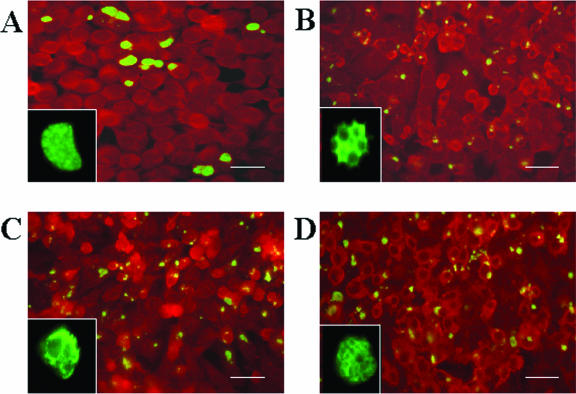
Chlamydial bodies exhibiting abnormal morphology were observed from 24 h p.i. onward for all three persistence models. Immunofluorescent images revealed aberrant forms, i.e., the inclusions appeared to be smaller and RBs larger than those seen with acute, untreated C. psittaci infection (Fig. 2). At higher magnification, images of aberrant inclusions revealed characteristic inhomogeneities resulting from the enlarged bodies, which distinguished them from normal inclusions.
FIG. 2.
Immunohistological staining of chlamydial cells in acute and persistent infection. Infected HEp-2 monolayers were incubated at 37°C and 5% CO2 for 48 h, fixed with methanol, stained as described in Materials and Methods, and visualized using fluorescence microscopy at a magnification of ×400. (A) HEp-2 monolayers infected with C. psittaci DC15 without persistence inducer. (B) Persistent infection with iron depletion (150 μM DAM). (C) Persistent infection after treatment with 200 U/ml penicillin G. (D) Persistent infection upon exposure to IFN-γ (240 U/ml). To illustrate inclusion morphology, magnifications of typical chlamydial inclusions are shown in the lower left corner of each image. Please note that the natural size of the inclusion shown in panel A is four to five times those of the persistent inclusions shown in panels B to D. Bars, 20 μm.
In electron microscopy, large inclusions containing numerous EBs, numerous intermediate condensing bodies, and low numbers of RBs were observed for untreated controls at 48 h p.i. The addition of either DAM, penicillin, or IFN-γ led to smaller inclusions, enlarged RBs, and few or no EBs (Fig. 3). The morphology of the inclusions differed among the persistence model systems. The iron depletion model generated RBs about twice the size of those seen with acute infection. RBs were irregularly shaped. Budding of small bodies under the wavy outer membrane was observed. Numerous intrainclusional membrane vesicles were observed. Treatment with penicillin G caused inclusions containing few, closely packed RBs. Within the same inclusions, RBs varied from normal size to 10 times enlarged and had a highly variable shape. Especially large RBs had an electron-lucent cytoplasm with a loose network of filaments and multiple electron-dense foci. Multifocal deposition of electron-dense material was observed multifocally on the outer membrane of RBs. Images of the IFN-γ exposure model exhibited inclusions containing 2- to 4-times-enlarged RBs of variable shape in the periphery and with amorphous granular material in the center. RBs had a clumped cytoplasm with electron-lucent foci.
FIG. 3.
Transmission electron microscopy of chlamydial inclusions in acute and persistent infection at 48 h p.i. (A) HEp-2 monolayers infected with C. psittaci DC15 without persistence inducer. A large inclusion, with numerous EBs, numerous intermediate condensing bodies, and few RBs is shown. (B) Persistent infection at iron depletion (150 μM DAM). A small inclusion containing only a few RBs is shown. (C) Persistent infection after treatment with 200 U/ml penicillin G. An inclusion densely packed with RBs of variable size and shape is shown. Two RBs are severely enlarged. Note the electron-dense depositions at the outer membranes of RBs. (D) Persistent infection upon exposure to IFN-γ (240 U/ml). A small inclusion with large RBs at the periphery and amorphous material in the center is shown. RBs are highly variable in size, shape, and appearance. Bars, 2 μm.
Rescue of infectious chlamydiae from persistent cultures.
To check the reversibility of the persistent state in each model system, the capability of persisting chlamydial bodies to recover and infect fresh and nontreated host cells was examined. The results of the reactivation assay in Fig. 4 illustrate that C. psittaci cells were rescued from persistence and returned to be infectious. Compared to acute infection, recovery rates at 24 h p.i. were 85.4% for the iron depletion model, 68.1% for antibiotic treatment, and 62.3% for IFN-γ exposure.
FIG. 4.
Reactivation of persistent chlamydial organisms. The numbers of recoverable infectious chlamydial cells (in IFU) from acute infection (Acute inf.) and persistence trials based on iron depletion by addition of DAM, penicillin G (PenG) treatment, or exposure to IFN-γ (filled columns) are compared to those from the reactivation assay, which included removal of persistence inducers at 24 h p.i. (open columns), and 48 h p.i. (shaded columns) and subsequent culture in complete growth medium for 24 h.
Differential gene expression in the persistent state in three cell culture models.
HEp-2 cell monolayers were infected with C. psittaci DC15 (MOI, 1) and treated with DAM, penicillin G, or IFN-γ to induce persistence in two independent experimental series. Acute nontreated infections served as controls. Total RNA was isolated at 12, 24, 36, and 48 h p.i. and reverse transcribed into cDNA. The study focused on 27 chlamydial genes, which encode membrane proteins and stress response proteins as well as proteins involved in transcriptional regulation, signal transduction, and RB-EB differentiation (see Table S1 in the supplemental material). Their transcript abundance levels were analyzed by qrt-PCR. The 16S rRNA and gyrA genes served as internal standards for comparisons of gene expression between persistence-induced and nontreated chlamydial cultures.
Relative mRNA expression levels in the course of development of persistence in all three cell culture models are summarized in Table 1 (see Table S2 in the supplemental material for data normalized for gyrA). From 24 h p.i. onward, qrt-PCR measurements revealed downregulation of genes encoding membrane proteins (omcA, omcB, pomp91A, pomp91B), chlamydial sigma factors (rpoD [sig66], rpsD [sig28], rpoN [sig54]), cell division protein (ftsW), and RB-EB differentiation proteins (ctcB, ctcC) in all three models.
TABLE 1.
Relative mRNA expression levels of C. psittaci genes at different time points in persistence modelsa
| Gene | Relative expression ratio at:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 h p.i.
|
24 h p.i.
|
36 h p.i.
|
48 h p.i.
|
|||||||||
| DAM | PenG | IFN-γ | DAM | PenG | IFN-γ | DAM | PenG | IFN-γ | DAM | PenG | IFN-γ | |
| groEL1 | 1.29 ± 0.35 | 0.84 ± 0.37 | 0.85 ± 0.28 | 0.80 ± 0.06 | 2.32 ± 0.29 | 0.26 ± 0.12*b | 0.38 ± 0.12* | 1.10 ± 0.17 | 0.15 ± 0.06* | 0.79 ± 0.22 | 1.64 ± 0.32 | 0.11 ± 0.07*b |
| grpE | 1.46 ± 0.33 | 1.37 ± 0.39 | 0.78 ± 0.10 | 1.00 ± 0.35 | 2.72 ± 0.52 | 0.17 ± 0.11*b | 0.84 ± 0.33 | 1.85 ± 0.08 | 0.18 ± 0.12* | 0.41 ± 0.12* | 1.70 ± 0.53 | 0.04 ± 0.03* |
| dnaK (hsp70) | 0.91 ± 0.10 | 0.86 ± 0.16 | 0.51 ± 0.09 | 0.17 ± 0.07* | 1.13 ± 0.23 | 0.22 ± 0.13* | 0.20 ± 0.05* | 0.69 ± 0.12 | 0.16 ± 0.07* | 0.28 ± 0.09* | 0.33 ± 0.13* | 0.06 ± 0.03* |
| groES | 1.37 ± 0.27 | 2.26 ± 0.21 | 0.63 ± 0.14 | 0.51 ± 0.27 | 0.20 ± 0.11*b | 0.36 ± 0.18 | 0.88 ± 0.28 | 2.97 ± 0.29b | 0.26 ± 0.13* | 0.80 ± 0.15 | 1.60 ± 0.38 | 0.09 ± 0.05* |
| ompA | 1.84 ± 0.50 | 1.33 ± 0.63 | 0.72 ± 0.15 | 0.51 ± 0.19 | 1.09 ± 0.45 | 0.72 ± 0.09 | 0.32 ± 0.19* | 1.61 ± 0.72 | 0.19 ± 0.14* | 0.34 ± 0.17 | 2.45 ± 0.33 | 0.15 ± 0.05*b |
| omcB | 1.52 ± 0.27 | 1.15 ± 0.49 | 2.68 ± 0.62 | 0.04 ± 0.04* | 0.22 ± 0.03* | 0.20 ± 0.16* | 0.04 ± 0.03* | 0.13 ± 0.06* | 0.05 ± 0.03* | 0.11 ± 0.04* | 0.15 ± 0.08* | 0.01 ± 0.02* |
| pomp91A | 2.46 ± 0.33b | 1.69 ± 0.15 | 0.57 ± 0.20 | 0.17 ± 0.03* | 0.34 ± 0.15* | 0.08 ± 0.03* | 0.25 ± 0.08* | 0.88 ± 0.13 | 0.06 ± 0.04* | 0.28 ± 0.12* | 0.32 ± 0.09* | 0.08 ± 0.04* |
| pomp91B | 0.93 ± 0.31 | 2.62 ± 0.46 | 9.19 ± 1.35 | 0.14 ± 0.06* | 0.39 ± 0.25 | 0.17 ± 0.05* | 0.28 ± 0.12* | 0.44 ± 0.05* | 0.24 ± 0.07* | 0.17 ± 0.10* | 0.11 ± 0.05* | 0.05 ± 0.04* |
| omcA | 3.27 ± 0.57b | 3.68 ± 0.73 | 6.18 ± 0.84 | 0.04 ± 0.01* | 0.16 ± 0.13* | 0.20 ± 0.18* | 0.07 ± 0.06* | 0.26 ± 0.19* | 0.07 ± 0.03* | 0.08 ± 0.05* | 0.11 ± 0.05* | 0.01 ± 0.002* |
| euo | 1.32 ± 0.23 | 2.49 ± 0.30 | 2.60 ± 0.35b | 1.37 ± 0.16 | 1.97 ± 0.13 | 0.98 ± 0.31 | 1.15 ± 0.13 | 2.75 ± 0.14 | 1.38 ± 0.31 | 0.22 ± 0.14* | 0.35 ± 0.11* | 0.13 ± 0.07* |
| cpaF | 1.52 ± 0.23 | 2.47 ± 0.29 | 1.15 ± 0.52 | 0.59 ± 0.29 | 0.99 ± 0.18 | 0.19 ± 0.14*b | 0.38 ± 0.14* | 1.78 ± 0.17 | 0.35 ± 0.06*b | 0.32 ± 0.18 | 0.98 ± 0.35 | 0.22 ± 0.02*b |
| efp2 | 1.21 ± 0.39 | 1.27 ± 0.34 | 0.61 ± 0.18 | 0.94 ± 0.47 | 1.02 ± 0.50 | 0.95 ± 0.20 | 3.71 ± 0.22b | 2.49 ± 0.39b | 1.08 ± 0.20 | 0.09 ± 0.06* | 0.26 ± 0.14* | 0.22 ± 0.10*b |
| hctA | 1.71 ± 0.99 | 1.56 ± 0.22 | 11.81 ± 0.85 | 0.05 ± 0.03* | 0.20 ± 0.13* | 0.06 ± 0.08* | 0.18 ± 0.09* | 1.44 ± 0.17 | 1.17 ± 0.16 | 0.12 ± 0.03* | 0.33 ± 0.12* | 0.01 ± 0.002* |
| CADD (CT610) | 1.12 ± 0.52 | 0.62 ± 0.15 | 2.10 ± 0.32 | 0.95 ± 0.25 | 1.07 ± 0.43 | 2.39 ± 0.32 | 0.77 ± 0.35 | 0.94 ± 0.12 | 2.83 ± 0.53b | 0.41 ± 0.11* | 0.36 ± 0.18* | 0.35 ± 0.09* |
| ftsW | 1.22 ± 0.20 | 0.68 ± 0.19 | 0.52 ± 0.18 | 0.31 ± 0.15* | 0.39 ± 0.10* | 0.19 ± 0.18* | 0.23 ± 0.10* | 0.40 ± 0.07* | 0.26 ± 0.04* | 0.22 ± 0.03* | 0.38 ± 0.08* | 0.05 ± 0.015* |
| ctcC | 1.29 ± 0.38 | 1.21 ± 0.41 | 0.77 ± 0.13 | 0.17 ± 0.15* | 0.10 ± 0.03* | 0.11 ± 0.02* | 0.07 ± 0.05* | 0.35 ± 0.13* | 0.06 ± 0.05* | 0.12 ± 0.06* | 0.23 ± 0.09* | 0.09 ± 0.03* |
| ctcB | 1.84 ± 0.33 | 1.32 ± 0.26 | 3.47 ± 0.83 | 0.26 ± 0.14* | 0.14 ± 0.03* | 0.18 ± 0.09* | 0.19 ± 0.03* | 0.41 ± 0.07* | 0.24 ± 0.13* | 0.29 ± 0.06* | 0.23 ± 0.09* | 0.09 ± 0.01* |
| rpoD (sigA) | 1.68 ± 0.33 | 1.05 ± 0.20 | 0.98 ± 0.27 | 0.45 ± 0.19 | 0.64 ± 0.07 | 0.61 ± 0.25 | 0.44 ± 0.10 | 1.15 ± 0.46 | 0.94 ± 0.12 | 0.39 ± 0.03* | 0.47 ± 0.23 | 0.11 ± 0.04* |
| rpsD (sig28) | 1.17 ± 0.13 | 1.28 ± 0.49 | 0.77 ± 0.25 | 0.06 ± 0.04* | 0.18 ± 0.05* | 0.15 ± 0.16* | 0.69 ± 0.07 | 0.82 ± 0.48 | 0.13 ± 0.08* | 0.23 ± 0.06* | 0.25 ± 0.07* | 0.03 ± 0.01* |
| rpoN (sig54) | 2.06 ± 0.32 | 1.34 ± 0.24 | 4.66 ± 0.40 | 0.25 ± 0.12* | 0.76 ± 0.18 | 1.46 ± 0.20 | 0.30 ± 0.18* | 1.05 ± 0.13 | 1.24 ± 0.35 | 0.38 ± 0.06* | 0.29 ± 0.06* | 0.16 ± 0.04* |
| dcrA | 1.25 ± 0.59 | 1.32 ± 0.32 | 0.87 ± 0.12 | 0.58 ± 0.12 | 1.01 ± 0.11 | 0.22 ± 0.09* | 1.12 ± 0.16 | 1.76 ± 0.14 | 1.08 ± 0.23 | 0.28 ± 0.05* | 0.68 ± 0.15 | 0.17 ± 0.04* |
| sctN | 1.28 ± 0.26 | 0.99 ± 0.31 | 3.78 ± 0.49b | 0.40 ± 0.12 | 0.51 ± 0.08 | 0.36 ± 0.07* | 0.27 ± 0.12* | 1.57 ± 0.12 | 0.79 ± 0.13 | 0.38 ± 0.23* | 0.68 ± 0.14 | 0.12 ± 0.06* |
| incA | 3.59 ± 1.08 | 1.11 ± 0.30 | 3.08 ± 0.50 | 3.96 ± 0.99 | 1.16 ± 0.33 | 0.99 ± 0.32 | 1.09 ± 0.21 | 0.32 ± 0.16* | 1.33 ± 0.38 | 0.42 ± 0.13 | 0.23 ± 0.14*b | 0.36 ± 0.06*b |
| PP2C (CT259) | 1.42 ± 0.84 | 1.92 ± 0.76 | 0.90 ± 0.26 | 0.15 ± 0.06* | 0.73 ± 0.14 | 0.74 ± 0.20 | 0.37 ± 0.07* | 3.77 ± 1.21 | 1.25 ± 0.34 | 0.28 ± 0.13* | 0.88 ± 0.07 | 0.15 ± 0.11* |
| pknD | 1.28 ± 0.24 | 1.27 ± 0.56 | 1.18 ± 0.45 | 0.36 ± 0.09*b | 0.73 ± 0.08 | 1.39 ± 0.21 | 0.81 ± 0.23 | 2.81 ± 0.77b | 0.88 ± 0.69 | 0.43 ± 0.16 | 0.97 ± 0.25 | 0.15 ± 0.07* |
| pkn1 | 1.61 ± 0.24 | 1.01 ± 0.32 | 6.97 ± 0.72 | 0.20 ± 0.05* | 0.45 ± 0.05* | 0.35 ± 0.16* | 0.88 ± 0.10 | 3.15 ± 0.89 | 1.52 ± 0.27 | 0.97 ± 0.10 | 1.20 ± 0.29 | 0.80 ± 0.26 |
| pkn5 | 1.26 ± 0.21 | 1.31 ± 0.20 | 1.20 ± 0.49 | 0.33 ± 0.24 | 0.53 ± 0.19 | 0.28 ± 0.11*b | 0.28 ± 0.11*b | 1.46 ± 0.25 | 0.56 ± 0.19 | 0.27 ± 0.12* | 0.41 ± 0.03*b | 0.38 ± 0.07* |
Expression ratios are for DAM, penicillin G, or IFN-γ versus acute infection ± SD and are normalized for the 16S rRNA gene. PenG, penicillin G; boldface type, significant upregulation (by a factor of >2.0); *, significant downregulation (by a factor of >2.0).
Differential regulation was significant with 16S rRNA normalization but not significant with gyrA normalization.
At 48 h p.i., transcription of virtually all genes, including cell division factor ftsW, the apoptosis-associated chlamydia protein associating with death domains (CADD) gene, and sctN and incA of the type III secretion system, was reduced or shut down in DAM and IFN-γ experiments. In contrast, the penicillin G model showed steady upregulation of stress response genes at early and late stages; this was particularly the case for grpE and groES, whose expression was not shut down even at 48 h p.i. Also, ompA remained twofold upregulated at 48 h p.i. Another distinctive feature of penicillin G-induced persistence was a secondary peak in mRNA expression of euo, pkn1, and PP2C (significant upregulation) as well as groES, efp2 and pknD (measurable, though not significant, upregulation) at 36 h p.i.
Statistical analysis confirmed the absence of significant differences between mRNA expression ratios from two independent infection experiments (Student's t test). The analysis of variance test revealed significant variations in mRNA expression along the time axis in those cases highlighted in Table 1.
DISCUSSION
This study was undertaken to elucidate three different in vitro models for changes in mRNA expression during the transition of C. psittaci to the persistent state and to work out common and distinctive features of chlamydial persistence by comparisons with published data on Chlamydia trachomatis and C. pneumoniae. While the phenotypical criteria were in line with observations from the literature, i.e., aberrant morphology of reticulate bodies, loss of cultivability, and rescue of infectivity upon removal of inducers, the transcriptional response of C. psittaci to persistence-inducing factors was found to display several new and distinctive features.
The first persistence model, DAM-induced iron depletion, resembles the local microenvironment of inflamed infected tissues (1). Under our experimental conditions, it appeared to be the most resilient model, having the highest recovery rate from persistence at 24 h p.i. At the same time, we observed the lowest total number of upregulated genes among the models we tested, which indicates a relatively straight transition to the persistent state.
Exposure to 240 U/ml IFN-γ caused the most intensive transcriptional response of chlamydial cells at the early stage, where 8 of the 27 genes were significantly upregulated. At the late stage, the most complete shutdown of mRNA expression among the three models occurred, which is in line with the observation that the highest loss of infectivity occurred in the IFN-γ trials (Fig. 1). Many studies in the literature were based on the IFN-γ model (3, 6, 27), which is thought to reflect locally elevated levels of this cytokine during chlamydial infection in vivo. In this context, persistence can be regarded as an alternative branch of the chlamydial developmental cycle designed to evade the host's immune response (5). Susceptibility of C. psittaci to IFN-γ may be expected, as this species has no functional tryptophan synthesis operon (G. S. A. Myers, personal communication). Early studies of avian Chlamydia psittaci had already shown inhibition of intracellular replication and inclusion development as a result of IFN-γ treatment (7, 33). Current data on stress response factors and surface proteins are showing good agreement with recent findings on C. pneumoniae (which also lacks the trp operon) in IFN-γ and iron depletion models (39). In contrast, Chlamydophila caviae (formerly Chlamydia psittaci GPIC strain) harbors a complete tryptophan biosynthesis operon consisting of six genes, trpRDCFBA, whose expression is regulated by the amino acid. This set of genes enables C. caviae to recycle tryptophan and thus accounts for the IFN-γ-resistant phenotype displayed in indolamine dioxygenase-expressing host cells (41). Similarly, genital strains of Chlamydia trachomatis were found to possess functional trpBA genes, which enable them to utilize exogenous indole for tryptophan biosynthesis (12). This characteristic difference provides an explanation for our observation that the late shutdown of essential genes in C. psittaci with IFN-γ-induced persistence was more comprehensive than that in Chlamydia trachomatis D strains (5).
Penicillin-induced persistence imitates in vivo conditions of inadequate antibiotic treatment. The fact that the response pattern of this model was markedly different from the other two should be due to the distinct mechanism of action, i.e., targeting cell wall synthesis rather than chlamydial metabolic pathways. Furthermore, Timms (39) pointed to an overlap between IFN-γ and iron depletion models, as IFN-γ also downregulates transferrin receptors, which results in lower host cell iron levels.
One of the most interesting findings of the present study is the conspicuous upregulation at 12 h p.i. of late genes coding for membrane proteins (omcA, pomp91B, omcB) and for proteins involved in DNA condensation (hctA), RB-EB differentiation (ctcB), type III secretion system (incA), signal transduction (pkn1), and transcriptional regulation (rpoN), particularly in the IFN-γ model. This seems to illustrate that, in response to challenge by the host immune system, the chlamydial cell tries to accelerate its developmental cycle by prematurely initiating processes that would normally occur at a later stage and/or enhance already ongoing expression of genes that are essential for long-term survival. Nonetheless, this transient increase in mRNA abundance does not result in measurable EB formation, as one would expect from the massive upregulation of hctA and some membrane proteins.
Another characteristic feature of the present persistence models is downregulation of the vast majority of genes at 48 h p.i. Notably, the group of the most intensely downregulated genes includes omcA, omcB, pomp91B, hctA, and ctcC, all of which were transiently upregulated at 12 h p.i. This mRNA expression pattern underlines the hypothesis that the processes prematurely initiated and probably not completed can no longer be maintained by the chlamydial cell at the late stage, and it further illustrates the idea that mRNA expression rates at 48 h p.i. do not merely reflect a general shutdown but show differential regulation.
It is also important that the persistent state of C. psittaci can be extended considerably. Our observation that a sizeable proportion of chlamydial cells remained visibly persistent and viable at least until 96 h p.i. with all three models (data not shown) additionally illustrates the chronic aspect of this phenomenon.
Summarizing the qrt-PCR data, we postulate that downregulation of genes coding for membrane proteins, transcription regulators, cell division factors, and EB-to-RB differentiation factors is associated with establishment and maintenance of the persistent state in vitro, independent of the inducing reagent. The downregulation of genes and proteins that are specifically expressed late in the productive developmental cycle most likely reflects the inhibited RB-to-EB differentiation. This is a common observation with persistence models, and it led to the late gene shut-down hypothesis (16).
Regarding regulation of individual genes during transition to persistence, the present data do not support the suggestion that a decrease in the ompA-to-groEL ratio is a universal persistence marker. No steady decline of the ratio was observed for any of the C. psittaci models as mRNA expression rates of both genes were oscillating along the time axis, and clear downregulation at the late stage was observed only with IFN-γ-induced persistence. Recent studies on C. pneumoniae also reported data showing that this ratio was not suitable as a marker for all chlamydial persistence systems (16, 22, 24).
Cell division protein FtsW is assumed to be involved in cell wall formation through septum peptidoglycan synthesis (37). The present study demonstrates the association of low mRNA expression levels of ftsW with persistence of C. psittaci, which is in agreement with reports of downregulation in several in vitro persistence models of Chlamydia trachomatis (5, 13) and C. pneumoniae (8). Consistent downregulation of this gene from 24 h p.i. onward is in accordance with the observation that, in persistent infection, chromosome partitioning still takes place, whereas cell division is inhibited (8).
Interestingly, the CADD gene was found to be downregulated at the last stage of the present IFN-γ experiments after having been transiently upregulated at 24 h p.i. This is in contrast to data from IFN-γ-induced persistence of Chlamydia trachomatis, where it was downregulated at 24 h p.i. and upregulated at 48 h (5). CADD shares homology with the death domains of tumor necrosis factor family receptors and induces apoptosis when transiently transfected to noninfected cells (36). Although it is not known at present whether CADD is the dominant protein governing apoptosis, a recent study reporting the crystal structure of this protein (35) demonstrated its capability to induce in vivo cell death of HeLa cells. Both catalytic activity and death domain binding were found to be required for its complete biological function. As this redox protein toxin was shown to necessitate two Fe2+ ions per molecule, an iron-depleted environment during persistence may contribute to the inhibition of apoptosis, thus further supporting the hypothesis that apoptosis inhibition is an integral part of persistent infection or at least concurrent with transition to that state (11, 28).
We believe that consistent and severe downregulation of the ctcB-ctcC gene pair also represents an interesting observation. It was predicted to encode highly conserved proteins with homology to bacterial two-component regulatory systems, and a possible role in late gene regulation during RB-to-EB transition was suggested (21). As both ctcB and ctcC were downregulated in our persistence models from 24 h p.i. onward, and RB-to-EB differentiation was obviously not taking place, it is conceivable that the genes and/or proteins are involved in this late differentiation process during acute infection.
The considerable divergence among data from the different model systems suggests that, at least in vitro, there is no persistence model per se, but rather a number of different ways for chlamydiae to reach this state. Similarly, cytokine-mediated inhibition of intracellular chlamydial growth was found to involve multiple mechanisms, such as nitric oxide induction, tryptophan catabolism, and iron deprivation (19). On the other hand, the effect of persistent chlamydial infection on the host cell response was also found to be dependent on the inducing reagent. While IFN-γ and penicillin treatment led to inhibition of C. pneumoniae-induced gene expression in HeLa cells, continuous and even increasing responses were observed with the iron depletion model (29).
Being the first molecular study on C. psittaci persistence, the present work demonstrates the capability of this pathogen to respond to simulated host challenges by altering mRNA expression of specific genes, which led mostly to strong inhibition or complete shutdown of mRNA expression in the persistent state. The reversibility of this phenomenon illustrates the efficiency of the chlamydial response and provides a principal clue for the explanation of a number of peculiarities, including chronicity of chlamydioses, failure of antibiotic therapy and immunoprophylaxis, and practical difficulties in isolating and culturing viable strains from infected tissue.
Supplementary Material
Acknowledgments
We thank Matthias Horn, Vienna, Austria, for providing TTSS operon sequences of environmental chlamydiae and David Longbottom, Edinburgh, United Kingdom, for providing primer sequences for pmp genes. We are grateful to Petra Reinhold, Jena, Germany, for advice concerning statistical data analysis. The technical assistance of Sabine Scharf and Sabine Lied is gratefully acknowledged.
S.G. received a monthly grant from H. Wilhelm Schaumann Stiftung, Hamburg, Germany. The study was financially supported by Deutsche Forschungsgemeinschaft grant Sa 597/4-1 and is an integral part of the European COST Action 855, “Animal chlamydioses and the zoonotic implications.”
Editor: J. B. Bliska
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Al-Younes, H. M., T. Rudel, V. Brinkmann, A. J. Szczepek, and T. F. Meyer. 2001. Low iron availability modulates the course of Chlamydia pneumoniae infection. Cell Microbiol. 3:427-437. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, A. A., and D. Vanrompay. 2000. Avian chlamydiosis. Rev. Sci. Tech. 19:396-404. [PubMed] [Google Scholar]
- 3.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. USA 90:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58:686-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belland, R. J., D. E. Nelson, D. Virok, D. D. Crane, D. Hogan, D. Sturdevant, W. L. Beatty, and H. D. Caldwell. 2003. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. USA 100:15971-15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, J., and G. Entrican. 1996. Interferon-gamma mediates long-term persistent Chlamydia psittaci infection in vitro. J. Comp. Pathol. 115:373-383. [DOI] [PubMed] [Google Scholar]
- 7.Byrne, G. I., L. K. Lehmann, and G. J. Landry. 1986. Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect. Immun. 53:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrne, G. I., S. P. Ouellette, Z. Wang, J. P. Rao, L. Lu, W. L. Beatty, and A. P. Hudson. 2001. Chlamydia pneumoniae expresses genes required for DNA replication but not cytokinesis during persistent infection of HEp-2 cells. Infect. Immun. 69:5423-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadevall, A., and L. A. Pirofski. 2000. Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect. Immun. 68:6511-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coles, A. M., D. J. Reynolds, A. Harper, A. Devitt, and J. H. Pearce. 1993. Low-nutrient induction of abnormal chlamydial development: a novel component of chlamydial pathogenesis? FEMS Microbiol. Lett. 106:193-200. [DOI] [PubMed] [Google Scholar]
- 11.Dean, D., and V. C. Powers. 2001. Persistent Chlamydia trachomatis infections resist apoptotic stimuli. Infect. Immun. 69:2442-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehlner-Gardiner, C., C. Roshick, J. H. Carlson, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-26903. [DOI] [PubMed] [Google Scholar]
- 13.Gerard, H. C., J. Freise, Z. Wang, G. Roberts, D. Rudy, B. Krauss-Opatz, L. Kohler, H. Zeidler, H. R. Schumacher, J. A. Whittum-Hudson, and A. P. Hudson. 2002. Chlamydia trachomatis genes whose products are related to energy metabolism are expressed differentially in active vs. persistent infection. Microbes Infect. 4:13-22. [DOI] [PubMed] [Google Scholar]
- 14.Haranaga, S., H. Ikejima, I. Yamaguchi, H. Friedman, and Y. Yamamoto. 2002. Analysis of Chlamydia pneumoniae growth in cells by reverse transcription-PCR targeted to bacterial gene transcripts. Clin. Diagn. Lab. Immunol. 9:313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan, R. J., S. A. Mathews, A. Kutlin, M. R. Hammerschlag, and P. Timms. 2003. Differential expression of genes encoding membrane proteins between acute and continuous Chlamydia pneumoniae infections. Microb. Pathog. 34:11-16. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, R. J., S. A. Mathews, S. Mukhopadhyay, J. T. Summersgill, and P. Timms. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 72:1843-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hotzel, H., A. Berndt, F. Melzer, and K. Sachse. 2004. Occurrence of Chlamydiaceae spp. in a wild boar (Sus scrofa L.) population in Thuringia (Germany). Vet. Microbiol. 103:121-126. [DOI] [PubMed] [Google Scholar]
- 18.Hotzel, H., and K. Sachse. 1998. Improvement and acceleration of the diagnosis of contagious bovine pleuropneumonia by direct detection of the microbe using polymerase chain reaction (PCR). Berl. Munch. Tierarztl. Wochenschr. 111:268-272. (In German.) [PubMed] [Google Scholar]
- 19.Igietseme, J. U., G. A. Ananaba, D. H. Candal, D. Lyn, and C. M. Black. 1998. Immune control of chlamydial growth in the human epithelial cell line RT4 involves multiple mechanisms that include nitric oxide induction, tryptophan catabolism and iron deprivation. Microbiol. Immunol. 42:617-625. [DOI] [PubMed] [Google Scholar]
- 20.Kaleta, E. F., and E. M. Taday. 2003. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 32:435-462. [DOI] [PubMed] [Google Scholar]
- 21.Koo, I. C., and R. S. Stephens. 2003. A developmentally regulated two-component signal transduction system in Chlamydia. J. Biol. Chem. 278:17314-17319. [DOI] [PubMed] [Google Scholar]
- 22.Mathews, S., C. George, C. Flegg, D. Stenzel, and P. Timms. 2001. Differential expression of ompA, ompB, pyk, nlpD and Cpn0585 genes between normal and interferon-gamma treated cultures of Chlamydia pneumoniae. Microb. Pathog. 30:337-345. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto, A., and G. P. Manire. 1970. Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J. Bacteriol. 101:278-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molestina, R. E., J. B. Klein, R. D. Miller, W. H. Pierce, J. A. Ramirez, and J. T. Summersgill. 2002. Proteomic analysis of differentially expressed Chlamydia pneumoniae genes during persistent infection of HEp-2 cells. Infect. Immun. 70:2976-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moulder, J. W., N. J. Levy, and L. P. Schulman. 1980. Persistent infection of mouse fibroblasts (L cells) with Chlamydia psittaci: evidence for a cryptic chlamydial form. Infect. Immun. 30:874-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholson, T. L., L. Olinger, K. Chong, G. Schoolnik, and R. S. Stephens. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 185:3179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pantoja, L. G., R. D. Miller, J. A. Ramirez, R. E. Molestina, and J. T. Summersgill. 2001. Characterization of Chlamydia pneumoniae persistence in HEp-2 cells treated with gamma interferon. Infect. Immun. 69:7927-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perfettini, J. L., T. Darville, A. Dautry-Varsat, R. G. Rank, and D. M. Ojcius. 2002. Inhibition of apoptosis by gamma interferon in cells and mice infected with Chlamydia muridarum (the mouse pneumonitis strain of Chlamydia trachomatis). Infect. Immun. 70:2559-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters, J., S. Hess, K. Endlich, J. Thalmann, D. Holzberg, M. Kracht, M. Schaefer, G. Bartling, and A. Klos. 2005. Silencing or permanent activation: host-cell responses in models of persistent Chlamydia pneumoniae infection. Cell. Microbiol. 7:1099-1108. [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raulston, J. E. 1997. Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect. Immun. 65:4539-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothermel, C. D., B. Y. Rubin, and H. W. Murray. 1983. Gamma-interferon is the factor in lymphokine that activates human macrophages to inhibit intracellular Chlamydia psittaci replication. J. Immunol. 131:2542-2544. [PubMed] [Google Scholar]
- 34.Salisch, H., K. V. Malottki, M. Ryll, and K. H. Hinz. 1996. Chlamydial infections of poultry and human health. World's Poultry Sci. J. 52:279-308. [Google Scholar]
- 35.Schwarzenbacher, R., F. Stenner-Liewen, H. Liewen, H. Robinson, H. Yuan, E. Bossy-Wetzel, J. C. Reed, and R. C. Liddington. 2004. Structure of the Chlamydia protein CADD reveals a redox enzyme that modulates host cell apoptosis. J. Biol. Chem. 279:29320-29324. [DOI] [PubMed] [Google Scholar]
- 36.Stenner-Liewen, F., H. Liewen, J. M. Zapata, K. Pawlowski, A. Godzik, and J. C. Reed. 2002. CADD, a Chlamydia protein that interacts with death receptors. J. Biol. Chem. 277:9633-9636. [DOI] [PubMed] [Google Scholar]
- 37.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 38.Szeredi, L., H. Hotzel, and K. Sachse. 2005. High prevalence of chlamydial (Chlamydophila psittaci) infection in fetal membranes of aborted equine fetuses. Vet. Res. Commun. 29:37-49. [DOI] [PubMed] [Google Scholar]
- 39.Timms, P. 2005. The zoonotic potential of chlamydial infections—a molecular analysis, p. 75-86. Proceedings of the 3rd Workshop of COST 855 on Diagnosis and Pathogenesis of Animal Chlamydioses, 22 to 23 September 2005, Siena, Italy. Bononia University Press, Bologna, Italy.
- 40.Vanrompay, D., T. Geens, A. Desplanques, T. Q. Hoang, L. De Vos, M. Van Loock, E. Huyck, C. Mirry, and E. Cox. 2004. Immunoblotting, ELISA and culture evidence for Chlamydiaceae in sows on 258 Belgian farms. Vet. Microbiol. 99:59-66. [DOI] [PubMed] [Google Scholar]
- 41.Wood, H., C. Roshick, and G. McClarty. 2004. Tryptophan recycling is responsible for the interferon-gamma resistance of Chlamydia psittaci GPIC in indoleamine dioxygenase-expressing host cells. Mol. Microbiol. 52:903-916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.