Abstract
Gene products required for in vivo growth and survival of Staphylococcus aureus and other pathogens represent new targets for antimicrobial chemotherapy. In this study we created a Staphylococcus aureus yjeQ deletion strain and tested its virulence using a mouse kidney abscess infection model. The yjeQ deletion strain was compromised for growth in vitro and severely attenuated for virulence. We concluded that yjeQ is an attractive and novel new drug target.
The gram-positive bacterium Staphylococcus aureus is responsible for a wide variety of diseases in humans involving all organ systems and ranging from localized skin infections to life-threatening systemic infections (1). A large natural reservoir of S. aureus is the asymptomatic carriage by approximately 20% of the human population, and this provides continuous exposure to a number of antibiotics (20). Indeed, this is reflected by reports that up to 50% of isolates from intensive care units are resistant to methicillin and reports of a growing number of vancomycin-resistant isolates from around the world (19, 30). The continuing persistence of this organism as a dominant pathogen and its ability to cause such a wide range of infections are primarily due to its extensive number of virulence factors. These factors include capsular polysaccharides (26) and protein A (15), which assist in avoiding host defenses; surface protein adhesins (MSCRAMMs), which bind to fibronectin, fibrinogen, and collagen (13) and aid in bacterial colonization; lipases (16), nucleases, and proteases (27), which assist in tissue invasion; and toxins, such as exfoliative toxin (21), toxic shock syndrome toxin, and enterotoxin (1). Other well-characterized virulence determinants are biofilm formation (12) and iron acquisition (9, 29). In the majority of studies on the mechanisms of staphylococcal pathogenesis the workers have focused mainly on the virulence factors mentioned above or on regulatory mechanisms that control virulence factor gene expression. Key cellular processes, such as basic biosynthetic, metabolic, and transport pathways that contribute to the overall survival of S. aureus, have been less well studied and are prospective new drug targets with potential as virulence determinants during in vivo infection.
More than 50% of methicillin-resistant S. aureus strains are also resistant to macrolides, lincosamides, fluoroquinolones, and aminoglycosides, and another 30% are resistant to trimethoprim-sulfamethoxazole (1). These statistics indicate that S. aureus vaccines and new or improved antimicrobial agents should be developed. The widespread resistance to currently available drugs implies that new chemical classes of antimicrobial agents and new targets for the development of novel agents are needed. Attractive targets for the development of new antimicrobial agents are proteins having unknown functions that are essential or important for the survival of the bacterium. Genes encoding proteins having unknown functions account for one-third of most bacterial genomes, and many of these proteins are highly conserved and have critical roles (31). These proteins are an uncharted class of potential new drug targets; in particular, the P-loop GTPases represent a large group of uncharacterized proteins that are often indispensable and play central roles in bacterial physiology (4).
The YjeQ protein is a unique, circularly permuted GTPase that is broadly conserved in bacterial species but is not present in eukaryotes, which makes it an ideal candidate target. YjeQ contains an N-terminal oligonucleotide/oligosaccharide binding (OB-fold) domain, a central circularly permuted GTPase domain (G4-G1-G2-G3), and a C-terminal zinc finger (8, 23, 28). Escherichia coli YjeQ has been shown to possess slow GTPase activity that is stimulated by ribosomes (7, 18). Depletion of both YjeQ and its Bacillus subtilis orthologue, YloQ, results in accumulation of 30S and 50S ribosomal subunits (5, 18), and the B. subtilis mutant also exhibits filamentation (5). yjeQ and yloQ are dispensable in E. coli and B. subtilis, respectively (5, 14, 18); however, deletion strains have altered growth rates, implying that the proteins have a central role in fitness and the general ability of the bacteria to survive. In this study we examined the potential of the YjeQ protein as a virulence determinant in the pathogenic bacterium S. aureus.
Generation of S. aureus yjeQ deletions.
The orthologue of E. coli yjeQ and B. subtilis yloQ in the S. aureus COL genome was identified by sequence comparison as the SAcol1234 gene. The genetic context of yjeQ in all seven S. aureus genomes that have been sequenced is identical to that of B. subtilis yloQ; a gene encoding ribulose phosphate 3-epimerase is located downstream of yjeQ, and a gene encoding a phosphatase-kinase pair is located upstream of yjeQ. The two orthologues exhibit 45% identity with one another and 26 to 31% identity with the gene encoding the gram-negative E. coli protein. To create a precise deletion of S. aureus yjeQ, an allelic replacement strategy was employed. A 1,100-bp fragment upstream of yjeQ was amplified using primers SA1234-A and SA1234-B (Table 1). Similarly, a 1,300-bp downstream fragment was amplified with primers SA1234-C and SA1234-D. The downstream fragment included the last 160 bp of the yjeQ coding sequence in order to maintain any promoter or upstream regulatory elements for the ribulose phosphate 3-epimerase gene (SAcol1235). Primers SA1234-B and SA1234-C contained regions that were complementary to an erythromycin resistance cassette for use in a crossover PCR. The erythromycin resistance cassette was amplified from plasmid pDG1664 (17) using primers Erm-F and Erm-R. The three PCR products were purified and used in a final reaction with primers SA1234-A and SA1234-D. The resulting DNA fragment contained approximately 1.1 kb of sequences flanking the yjeQ gene with an erythromycin resistance cassette between them. This fragment was cloned into the EcoRV site of pSAKO and transformed into SA178RI (10). Integrants were selected on Mueller-Hinton agar supplemented with kanamycin and erythromycin. Single integrants were confirmed by PCR. To select for excision, which resulted in generation of either wild-type or deletion strains, the cells were grown on Mueller-Hinton agar supplemented with sucrose (5%, wt/vol). Strain EBII59 was confirmed by PCR to be a yjeQ deletion strain.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or oligonucleotide | Description | Source or reference |
|---|---|---|
| E. coli Novablue | General E. coli cloning strain | Novagen, Madison, Wis. |
| S. aureus strains | ||
| EBII42 | SA178RI, RN4220 derivative containing T7 RNA polymerase | 10 |
| EBII59 | SA178RI yjeQ::erm | This study |
| EBII61 | Wild-type Newman strain | 11 |
| EBII74 | Newman yjeQ::erm | This study |
| EBII83 | Newman yjeQ::erm/pLI50-yjeQ | This study |
| Plasmids | ||
| pDG1664 | Source for a erythromycin resistance cassette, used for gene replacement | 17 |
| pSAKO | E. coli replicating vector containing sacB[BamP]W29 and kanamycin resistance cassette | 10 |
| pLI50 | E. coli-S. aureus shuttle plasmid; Ap (E. coli), Cm (S. aureus) | 22 |
| Oligonucleotides | ||
| SA1234-A | 5′-GGGAAACTCGAGCAATGGCAATGTTTGGTAATAAATACG-3′ | |
| SA1234-B | 5′-GATACTGCACTATCAACACACTCTTAAGTTTGCTTCTAAGGGCACCTCTCGATTAATTTTACTGC-3′ | |
| SA1234-C | 5′-AGCTTCCAAGGAGCTAAAGAGGTCCCTAGACTCTAGACCCATCGATATGGTGAAACATGTAAGTTTAGGAATTGTAATC-3′ | |
| SA1234-D | 5′-GGGAAACTCGAGGGTATAAATGAAATGTACGGATAAC-3′ | |
| SA1234-F | 5′-GATAAAGATAATGACGGTTC-3′ | |
| SA1234-R | 5′-GGGAAACTCGAGGATAATAATGATGGATATAGTTTTG-3′ | |
| Erm-F | 5′-CTTAGAAGCAAACTTAAGAGTG-3′ | |
| Erm-R | 5′-GGGTCTAGAGTCTAGGGACC-3′ |
Newman yjeQ::erm mutants grow slowly in laboratory media.
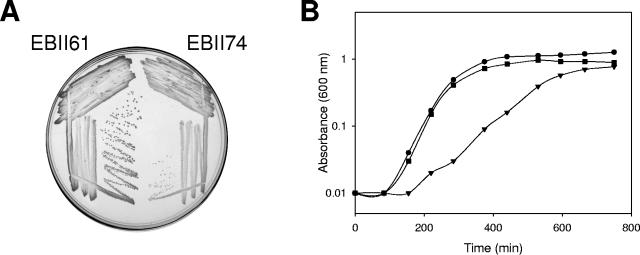
To assess the effect of deletion of yjeQ in a virulent S. aureus strain, the deletion in SA178RI was transduced into the Newman strain with phage 80α, using procedures described previously (25), to create strain EBII74. The growth phenotype of EBII74 was examined using both solid and liquid media. The Newman strain (EBII61) exhibited good growth after incubation for 24 h at 37°C and formed large single colonies on Trypticase soy agar (Fig. 1A), while the yjeQ deletion strain (EBII74) exhibited slow growth and produced single colonies that were smaller than the wild-type colonies. If a culture was incubated for an additional 24 h, the size of the colonies was similar to the size of wild-type colonies (data not shown). This result is analogous to previous results obtained for the yjeQ orthologue in B. subtilis (yloQ) (5).
FIG. 1.
Effect of yjeQ deletion on the growth of S. aureus. (A) Strains were grown on Trypticase soy agar for 24 h at 37°C. The left side of the plate contained EBII61 (strain Newman), and the right side of the plate contained EBII74 (yjeQ deletion strain). (B) Strains were grown in Trypticase soy broth at 37°C with shaking at 250 rpm. The growth curves of S. aureus Newman strain (EBII61) (•), EBII83 (Newman yjeQ deletion strain containing pLI50-yjeQ) (▪), and EBII74 (Newman yjeQ deletion strain) (▾) are shown.
To ensure that the slow-growth phenotype observed was a result of deletion of yjeQ and not due to a polar effect on a surrounding gene, yjeQ was PCR amplified using primers SA1234-F and SA1234-R, which included a native promoter, and cloned into plasmid pLI50 at the SmaI site. Complementation by this plasmid was assessed by analysis of the growth phenotype in liquid media.
Figure 1B shows growth curves in liquid media for wild-type Newman strain (EBII61), the yjeQ deletion strain (EBII74), and the strain having the yjeQ deletion complemented with pLI50-yjeQ (EBII83). The growth curve of the yjeQ deletion strain was distinctly different than that of the wild-type Newman strain. The lag time of the yjeQ deletion strain was increased by approximately 70 min, and the exponential growth rate was lower. The growth phenotype of the complemented deletion strain was similar to that of the wild-type strain, indicating that the growth phenotype observed was attributable to the deletion of yjeQ and was not the result of a polar effect.
Deletion of yjeQ from S. aureus resulted in a phenotype that mirrored the phenotypes observed with equivalent deletions in E. coli and B. subtilis, and the growth rate was markedly deceased. To ascertain if deletion of yjeQ compromised bacterial survival and fitness enough to alter the infectivity of S. aureus, we examined the virulence of this strain in mouse models.
S. aureus ΔyjeQ mutants are attenuated in a mouse kidney abscess model.
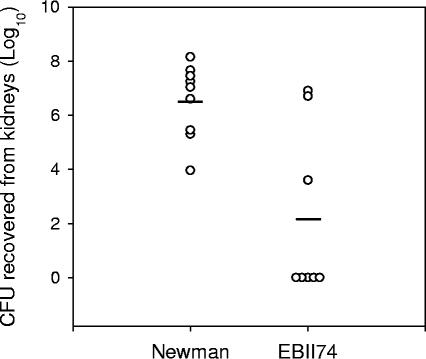
To determine whether the pathogenesis of S. aureus yjeQ deletion strains was altered, the ability of the yjeQ mutant to colonize mice was compared with the ability of its isogenic parent. Female Swiss-Webster mice (Charles River Laboratories Canada Inc., Montreal, Quebec) were inoculated intravenously in the tail vein with 1 × 107 CFU of S. aureus Newman or the yjeQ mutant, EBII74. In a blinded fashion, University of Western Ontario Animal Care and Veterinary Services personnel scored mice throughout the experiment for alertness, activity, and coat condition as a measure of morbidity. In each of the three categories, a score of 0 was normal, a score of 1 was slightly abnormal, and a score of 2 was very abnormal. At 5 days postinfection, the group of mice challenged with S. aureus Newman displayed overt signs of disease; the average clinical score was 3.1, and the average weight loss was 21.2% (Table 2). In contrast, for the group of mice challenged with S. aureus EBII74 the average clinical score was 0.4 and the average weight loss was 8.8%, indicating that these mice were significantly less moribund than the Newman-infected group. To examine the bacterial loads in the kidneys of infected animals, mice were euthanized on day 5, and the kidneys were aseptically removed. The kidneys were then homogenized in sterile phosphate-buffered saline containing 0.1% Triton X-100 using a PowerGen 700 homogenizer. The homogenates were serially diluted and plated on Trypticase soy agar to enumerate the bacteria recovered. Figure 2 shows that the number of CFU recovered from the kidneys of EBII74-challenged mice was significantly lower than the number of CFU obtained from Newman-challenged animals. Strikingly, in five of the eight mice infected with EBII74 there were no detectable CFU. For the three mice in which bacteria were present, disease symptoms were also evident. These mice exhibited the greatest disease progression (an average weight loss of 14.8% and a clinical score of 5.7, compared with 5.8% and 0, respectively, for the five healthy mice), implying that loss of yjeQ impairs the ability of S. aureus to colonize and that in the minority of cases in which colonization occurs, disease progression is possible. Colonization was likely prevented by the slow growth of the yjeQ deletion strain. Taken together, these results indicate that YjeQ contributes to the fitness of S. aureus and plays a vital role in the pathogenesis of this organism.
TABLE 2.
Clinical characteristics of mice infected with S. aureus Newman or Newman yjeQ (EBII74)
| Infecting strain | Clinical scorea | Weight loss (%) | Log10 CFUb |
|---|---|---|---|
| Newman (n = 9) | 3.1 ± 1.2 | 21.2 ± 6.4 | 6.5 ± 1.4 |
| EBII74 (n = 8) | 0.4 ± 0.9c | 8.8 ± 5.9c | 2.2 ± 3.1c |
Clinical score on day 5.
Total bacterial counts for two kidneys from each mouse.
P < 0.001 compared with the parental strain.
FIG. 2.
yjeQ deletion is compromised in a murine kidney abscess model. Two groups of mice were inoculated in the tail vein with 107 CFU. One group received S. aureus Newman (n = 9), while the other group was infected with the yjeQ deletion strain (n = 8). The data are the numbers of CFU recovered from the kidneys of mice at 5 days postinfection. Each circle represents the staphylococcal count for the kidneys of one animal, and the solid lines indicate the average numbers of CFU recovered. Statistical significance was determined using the Student unpaired t test, and differences were found to be highly significant (P < 0.001).
A number of genome-scale studies have been conducted with S. aureus to identify genes that are required for full virulence (2, 3, 6, 24). A survey of these studies indicated that in each study the workers identified a number of metabolic and biosynthetic pathway genes, such as the genes required for amino acid biosynthesis, response regulators, replication, cofactor biosynthesis, cell envelope biosynthesis, and purine and pyrimidine synthesis. Also identified in these studies were a number of proteins having unknown functions, which further highlights the utility of investigating this subset of proteins as virulence determinants. These new potential targets shift the focus from traditional virulence factors that directly contribute to pathogenesis toward identification of genes that encode proteins that affect in vivo growth and persistence, such as the dispensable protein YjeQ. This study is the first demonstration that a member of the GTPase superfamily is important for the in vivo survival of S. aureus, and the results indicate that YjeQ is a potential new target for antistaphylococcal therapies.
Editor: V. J. DiRita
REFERENCES
- 1.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 2.Bae, T., A. K. Banger, A. Wallace, E. M. Glass, F. Aslund, O. Schneewind, and D. M. Missiakas. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA 101:12312-12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benton, B. M., J. P. Zhang, S. Bond, C. Pope, T. Christian, L. Lee, K. M. Winterberg, M. B. Schmid, and J. M. Buysse. 2004. Large-scale identification of genes required for full virulence of Staphylococcus aureus. J. Bacteriol. 186:8478-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, E. D. 2005. Conserved P-loop GTPases of unknown function in bacteria: an emerging and vital ensemble in bacterial physiology. Biochem. Cell Biol. 83:738-746. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, T. L., D. M. Daigle, and E. D. Brown. 2005. Characterization of the Bacillus subtilis GTPase YloQ and its role in ribosome function. Biochem. J. 389:843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulter, S. N., W. R. Schwan, E. Y. Ng, M. H. Langhorne, H. D. Ritchie, S. Westbrock-Wadman, W. O. Hufnagle, K. R. Folger, A. S. Bayer, and C. K. Stover. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30:393-404. [DOI] [PubMed] [Google Scholar]
- 7.Daigle, D. M., and E. D. Brown. 2004. Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro. J. Bacteriol. 186:1381-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daigle, D. M., L. Rossi, A. M. Berghuis, L. Aravind, E. V. Koonin, and E. D. Brown. 2002. YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry 41:11109-11117. [DOI] [PubMed] [Google Scholar]
- 9.Dale, S. E., A. Doherty-Kirby, G. Lajoie, and D. E. Heinrichs. 2004. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 72:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Elia, M. A., M. P. Pereira, Y.-S. Chung, W. Zhao, A. Chau, T. J. Kenney, M. C. Sulavik, T. A. Black, and E. D. Brown. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 188:4183-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 12.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2005. The genetics of staphylococcal biofilm formation—will a greater understanding of pathogenesis lead to better management of device-related infection? Clin. Microbiol. Infect. 11:967-973. [DOI] [PubMed] [Google Scholar]
- 13.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 14.Freiberg, C., B. Wieland, F. Spaltmann, K. Ehlert, H. Brotz, and H. Labischinski. 2001. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J. Mol. Microbiol. Biotechnol. 3:483-489. [PubMed] [Google Scholar]
- 15.Gomez, M. I., A. Lee, B. Reddy, A. Muir, G. Soong, A. Pitt, A. Cheung, and A. Prince. 2004. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10:842-848. [DOI] [PubMed] [Google Scholar]
- 16.Gotz, F., H. M. Verheij, and R. Rosenstein. 1998. Staphylococcal lipases: molecular characterisation, secretion, and processing. Chem. Phys. Lipids 93:15-25. [DOI] [PubMed] [Google Scholar]
- 17.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 18.Himeno, H., K. Hanawa-Suetsugu, T. Kimura, K. Takagi, W. Sugiyama, S. Shirata, T. Mikami, F. Odagiri, Y. Osanai, D. Watanabe, S. Goto, L. Kalachnyuk, C. Ushida, and A. Muto. 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 32:5303-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 20.Kluytmans, J., A. van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladhani, S. 2003. Understanding the mechanism of action of the exfoliative toxins of Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 39:181-189. [DOI] [PubMed] [Google Scholar]
- 22.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 23.Levdikov, V. M., E. V. Blagova, J. A. Brannigan, L. Cladiere, A. A. Antson, M. N. Isupov, S. J. Seror, and A. J. Wilkinson. 2004. The crystal structure of YloQ, a circularly permuted GTPase essential for Bacillus subtilis. J. Mol. Biol. 340:767-782. [DOI] [PubMed] [Google Scholar]
- 24.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 25.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 26.O'Riordan, K., and J. C. Lee. 2004. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17:218-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw, L., E. Golonka, J. Potempa, and S. J. Foster. 2004. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150:217-228. [DOI] [PubMed] [Google Scholar]
- 28.Shin, D. H., Y. Lou, J. Jancarik, H. Yokota, R. Kim, and S.-H. Kim. 2004. Crystal structure of YjeQ from Thermotoga maritima contains a circularly permuted GTPase domain. Proc. Natl. Acad. Sci. USA 101:13198-13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skaar, E. P., M. Humayun, T. Bae, K. L. DeBord, and O. Schneewind. 2004. Iron-source preference of Staphylococcus aureus infections. Science 305:1626-1628. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan, A., J. D. Dick, and T. M. Perl. 2002. Vancomycin resistance in staphylococci. Clin. Microbiol. Rev. 15:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]