Abstract
A spectrum of in vivo-expressed Staphylococcus epidermidis antigens was identified by probing a bacteriophage lambda library of S. epidermidis genomic DNA with human serum from infected and uninfected individuals. This analysis resulted in identification of 53 antigen-encoding loci. Six antigenic polypeptides were expressed from these loci and purified. These polypeptides were the propeptide, mature amidase, and repeat sequence domains of the major autolysin AtlE, GehD (lipase), and two members of a conserved family of surface proteins (ScaA [AaE] and ScaB). AtlE, ScaA, and ScaB all exhibit human ligand binding capacity. Screening a bank of human serum samples revealed that there were significant increases in the amounts of reactive immunoglobulin G in infected individuals compared to the amounts in healthy individuals for the repeat sequence and mature amidase domains of AtlE, ScaB, and GehD. Vaccination of mice with recombinant antigens stimulated an immune response which in vitro opsonized S. epidermidis. In this study we identified prospective candidate antigens for prophylaxis or immunotherapy to control disease.
Staphylococcus epidermidis is a common cause of nosocomial infections, which have significant rates of morbidity and mortality (26) and can be difficult to treat with conventional antibiotics (39). An inhabitant of human skin, this bacterium is the most common organism causing infection of cerebrospinal fluid shunts and endocarditis of prosthetic valves and catheters (20). Resistance to a wide range of antibiotics is now common in S. epidermidis, and many isolates are resistant to erythromycin, clindomycin, chloramphenicol, tetracycline, and methicillin (2, 3, 18, 34). Recently, isolates with reduced susceptibility to vancomycin have been observed (19). Thus, there have been renewed efforts to identify suitable immunotherapeutic or prophylactic targets. Production of an effective subunit vaccine or monoclonal antibody requires identification of surface antigens that are recognized by the immune system and, critically, are expressed during infection.
Identification of suitable targets for opsonizing anti-S. epidermidis antibodies provides a potential alternative to antimicrobial chemotherapy. Different techniques have been used to identify candidate antigens in a range of pathogenic bacteria, including the closely related organism Staphylococcus aureus.
Bioinformatics have been used to identify putative surface and secreted proteins in Streptococcus pneumoniae and Neisseria meningitidis (37, 44). However, this method does not eliminate the need for expression and purification of suitable amounts of protein in a heterologous system. In a study of S. pneumoniae carriage using an experimental human colonization model McCool et al. identified a serum immunoglobulin G (IgG) and secretory IgA response to pneumococcal surface protein A as a result of colonization; individuals who did not become colonized after inoculation had preexisting antibodies to this protein (31).
Using randomly fragmented DNA of S. aureus in a bacterial expression library, which was subsequently screened with serum from infected patients, resulted in identification of a range of antigenic proteins. However, this required appropriate cloning of all expressed genes in frame with the fusion protein, and the target proteins had to be amenable to expression in Escherichia coli (11). More recently, antigenic components of S. epidermidis have been identified by Western blotting of cellular extracts with serum from experimentally infected rabbits (40).
In a recent study Clarke et al. (9) used a bacteriophage lambda expression library of S. aureus DNA, which did not require in-frame cloning into vector DNA, that was screened with convalescent patient serum to identify candidate antigens for prophylaxis. Recombinant polypeptides were then used to determine specific antibody titers in a large collection of human sera by an enzyme-linked immunosorbent assay (ELISA). Significantly higher titers of antibodies to several proteins were observed in individuals who were not nasal carriers of S. aureus at the time of serum collection than in individuals who were carriers. This led to the hypothesis that an increased antibody titer protected against nasal carriage, which was confirmed by protection studies performed with cotton rats.
In recent years, studies of surface proteins of S. aureus have led to development of several potentially efficacious immunological therapeutic and prophylactic strategies for control of this organism (15). A donor serum with a high anti-ClfA IgG titer has been shown to have the potential to reduce sepsis caused by S. aureus and mortality in infants with very low birth weights (6, 42). Currently, there is much research in which workers are trying to develop humanized anti-ClfA monoclonal antibodies for treatment of S. aureus infections (10, 21, 36), which could be used for passive immunotherapy. The collagen adhesin, Cna, has been proposed as a target for anti-adhesive antibody therapy (43) and as a vaccine component (33). It has also been suggested that FnBPA and FnBPB could be targets for passive immunotherapy (16, 38).
In this study we identified antigenic components of S. epidermidis using a bacteriophage lambda expression library screened with sera from patients with confirmed S. epidermidis and S. aureus infections. The relative titers of serum IgG reactive to the antigens were determined by ELISA. Polyclonal antibodies generated from these recombinant polypeptides were shown to be opsonic in vitro, demonstrating their potential therapeutic efficacy.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in Luria-Bertani medium, using selection with ampicillin (100 μg/ml) or kanamycin (50 μg/ml) when appropriate. S. epidermidis was grown in brain heart infusion medium (Oxoid).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| S. epidermidis strains | ||
| 138 | Clinical isolate | 32 |
| Hay | ATCC 55133 | ATCC |
| E. coli strains | ||
| XL-1 Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac (F′ proAB lacIqZΔM15 Tn5 [Tcr]) | Stratagene |
| XLOLR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA thi-1 recA1 gyrA96 relA1 lac (F′ proAB lacIqZΔM15 Tn5 [Tcr]) Su− λr | Stratagene |
| BL21(DE3) | F−ompT gal (dcm) (lon) hsdSB (rB− mB−) | Novagen |
| Plasmids | ||
| pET21-a | His6-tagged overexpression vector; Apr | Novagen |
| pET24-d | His6-tagged overexpression vector; Kmr | Novagen |
| pMA | pET24-d containing internal fragment encoding AtlE amidase domain; Kmr | This study |
| pML | pET24-d containing internal fragment encoding GehD; Kmr | This study |
| pRSN | pET21-a containing internal fragment encoding AtlE repeat sequence; Apr | This study |
| pPP | pET24-d containing internal fragment encoding AtlE propeptide; Kmr | This study |
| pScaA | pET24-d containing internal fragment encoding ScaA; Kmr | This study |
| pScaB | pET24-d containing internal fragment encoding ScaB; Kmr | This study |
Construction of S. epidermidis expression libraries.
Genomic DNA of S. epidermidis 138 was partially digested with Sau3A to generate 2- to 10-kb fragments which were ligated into bacteriophage lambda DNA and packaged into phage particles by using a Lambda ZAP Express kit (Stratagene) according to the manufacturer's instructions.
Human sera.
Eighteen sera were used to screen the genomic expression library. Since one element of this study was to identify antibodies which cross-react with proteins from different species of staphylococci, a variety of sources of sera were used. Of the 18 samples used in the screening process, 4 were obtained from hospital workers, 4 were obtained from community volunteers, 4 were obtained from patients with S. epidermidis infections, and 6 were obtained from patients with S. aureus infections. All samples were collected at the Royal Hallamshire Hospital, Sheffield, United Kingdom; the individuals gave informed consent, and the study was approved by the local IRB (SSREEC/02/299). For ELISA experiments, serum samples from 15 individuals with confirmed S. epidermidis infections, collected at Aston University, Birmingham, United Kingdom, were used along with 10 samples from community volunteers collected at the Royal Hallamshire Hospital, Sheffield, United Kingdom.
Screening of expression libraries with human sera.
Bacteriophage were propagated, and plaque lifts were made on Immobilon-NC membranes (Millipore) and probed according to the manufacturer's instructions with human sera diluted 1:1,000 and then with alkaline phosphatase-conjugated anti-human IgG gamma chain-specific monoclonal antibodies (Sigma) diluted 1:50,000. Additional rounds of screening resulted in pure phages from which phagemids were excised (according to the manufacturer's instructions).
Phagemid DNA was purified using QIAprep spin miniprep columns, and DNA was sequenced using primers T3 and T7. BLAST-N searches using sequences from either end of the insert were carried out against the S. epidermidis ATCC 12228 genome (http://www.tigr.org).
E. coli XLOLR containing excised phagemids was grown overnight in the presence of 10 μM isopropyl-β-d-thiogalactopyranoside (IPTG), cells were boiled in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) buffer, and the proteins were separated by 12% (wt/vol) SDS-PAGE prior to Western blotting and probing with human serum, which were carried out by using standard laboratory methods, as described previously (8).
Cloning and overexpression of recombinant polypeptides.
Oligonucleotides were used to PCR amplify antigen-encoding genes (Table 2). DNA fragments were cloned into pET24-d or pET21-a. Clones were verified by DNA sequencing. His6-tagged recombinant polypeptides were expressed, purified, and verified by N-terminal sequencing.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| T3 | AATTAACCCTCACTAAAGGG |
| T7 | GTAATACGACTCACTATAGGG |
| Amidase F | GCGCGCCCATGGTATCTAGTCAAAAAACATCATC |
| Amidase R | GCGCGCCTCGAGTGATGGTGCAGTTAAATAGTA |
| RSN F | GCGCGCCATATGACAACATCTACAAAACCGTC |
| RSN R | GCGCGCCTCGAGTGATGGTGCAGTTAAATAGTA |
| ML F | GCGCGCCCATGGCGCAAGCTCAATATAA |
| ML R | GCGCGCCTCGAGCTTACGTGTAATACCATCTAACTC |
| ScaA F | GCGCGCCCATGGCAACAACGCATACAGT |
| ScaA R | GCGCGCCTCGAGATGAATAAATTTGTAATTTCTCACT |
| ScaB F | GCGCGCCCATGGCTTCTACACAACATAAGGTTC |
| ScaB R | GCGCGCCTCGAGGTGGATAAATGCATATGAAGA |
Restriction endonuclease sites are underlined.
ELISA.
Total IgG contents in serum were determined by serial dilution (1:100 to 1:100,000) in phosphate-buffered saline (PBS). Purified human IgG (1 mg/ml to 10 ng/ml; Calbiochem) was used as the standard. One hundred microliters of a diluted sample was coated onto wells of a 96-well microtiter plate at 4°C overnight. The plates were washed three times with PBST (PBS containing 0.05% [vol/vol] Tween 20) and blocked with 5% (wt/vol) bovine serum albumin in PBST for 2 h at room temperature). After three washes with PBST, alkaline phosphatase-conjugated anti-human IgG gamma chain-specific monoclonal antibodies (Sigma) diluted 1:50,000 in PBST-bovine serum albumin were added, and the preparations were incubated for 1 h at room temperature. After three washes with PBST, antibodies were detected using the Sigma Fast p-nitrophenyl phosphate system (Sigma) at A405 (Victor plate reader [Wallac]).
To quantify specific antibodies to each antigen in serum samples, 100-μl portions (10 μg/ml in PBS) of recombinant protein were added to 96-well microtiter plates (4°C, overnight). The plates were washed and blocked as described above. One hundred microliters of a randomized 1:100 dilution serum sample (consistently within a linear range) was added to each well (1 h) at room temperature. The plates were developed as described above. The results were standardized for total IgG concentration in each serum sample.
Opsonization of S. epidermidis.
Antibodies were raised by inoculating 8-week-old female BALB/c mice with appropriate recombinant antigens. The opsonophagocytic (bacterial killing) activities of specific antisera were determined with an assay by using S. epidermidis Hay in the presence of polymorphonuclear leukocytes (PMNs) and complement, as described previously (14). Briefly, PMNs were isolated from freshly obtained human blood using PMN separation medium (Robbins Scientific) according to manufacturer's instructions. Reference antibody was mixed with ca. 2 × 106 PMNs, human complement, and ca. 3 × 104 bacteria in a sterile 96-well plate in a final volume of 100 μl. Immediately after mixing a sample of bacteria was removed from each well and plated onto tryptic agar plates with 5% (vol/vol) sheep blood (T0 sample). The assay plate was incubated for 2 h at 37°C on an orbital shaker. A second sample was removed from each well and plated as described above (T2 sample). The plates were incubated at 37°C for 20 to 24 h, and the percentage of bacterial killing was calculated using the following formula: (T0 − T2/T0) × 100.
Ligand binding assays.
Ligand binding was assayed by Western blotting, using biotinylated human serum proteins as described previously (8). Human fibrinogen, human lactoferrin, and bovine submaxillary mucin were purchased from Sigma. Human fibronectin was obtained from ICN.
RESULTS AND DISCUSSION
Identification of S. epidermidis antigens.
In order to identify S. epidermidis antigens expressed during human infection, a bacteriophage lambda expression library of S. epidermidis 138 was probed with sera collected at the Royal Hallamshire Hospital (Sheffield, United Kingdom). Expression does not necessarily require correct in-frame cloning of insert DNA and allows production of insoluble proteins or proteins toxic to E. coli. Eighty-seven clones were isolated, and they corresponded to 53 different loci (comprising a contiguous DNA region from single or overlapping clones) containing 121 known or putative open reading frames of S. epidermidis (Table 3).
TABLE 3.
Identification of S. epidermidis loci encoding antigens recognized by human seraa
| Locus | Locus coordinates | Gene in locus (cloned region amino acids) | Designation, putative function, and/or homology |
|---|---|---|---|
| 1 | 735135-739142 | SE0750 (45-1053) | AtlE, major autolysin |
| 2 | 1281499-1283139 | SE1250 (1-137) | Conserved hypothetical protein |
| SE1251 (1-127) | Conserved hypothetical protein | ||
| SE1252 (1-85) | Glycyl-tRNA synthetase | ||
| 3 | 2430659-2432743 | SE2349 (1-422) | Xanthine permease |
| SE2350 (1-192) | Xanthine phosphoribosyltransferase | ||
| 4 | 2142357-2144445 | SE2098 (40-350) | Alcohol dehydrogenase |
| SE2099 (1-255) | Conserved hypothetical protein | ||
| 5 | 469921-471793 | SE0477 (1-368) | CsbB stress response protein |
| SE0478 (190-322) | Histidine kinase | ||
| 6 | 428348-430455 | SE0432 (113-336) | Low-affinity inorganic phosphate transporter |
| SE0433 (1-266) | Secretory antigen SsaA-like protein | ||
| 7 | 2401382-2403540 | SE2318 (343-501) | Poly(glycerol-phosphate) alpha-glucosyltransferase |
| SE2319 (1-324) | Aae, autolysin | ||
| 8 | 1431038-1433713 | SE1394 (1-156) | Conserved hypothetical protein |
| SE1396 (1-200) | 30S ribosomal protein S4 | ||
| 9 | 2421512-2423462 | SE2337 (1-342) | PfoR, regulatory protein |
| SE2339 (1-103) | Conserved hypothetical protein | ||
| 10 | 2192554-2194685 | SE2146 (1-151) | Transcription regulator |
| SE2147 (1-378) | Amino acid transporter | ||
| 11 | 1640739-1643296 | SE1587 (315-511) | High-affinity proline permease |
| SE1588 (1-402) | Conserved hypothetical protein | ||
| 12 | 465589-468072 | SE0473 (1-390) | N-Acetylglucosamine-6-phosphate deacetylase |
| SE0474 (1-139) | Conserved hypothetical protein | ||
| 13 | 1979057-1981257 | SE1945 (1-439) | l-Lactate permease LctP-like protein |
| SE1946 (1-178) | Conserved hypothetical protein | ||
| 14 | 1925763-1928363 | SE1889 (37-295) | Conserved hypothetical protein |
| SE1890 (1-474) | Phosphotransferase system sucrose-specific IIBC component | ||
| SE1891 (1-124) | RpiR, transcription regulator | ||
| 15 | 980757-978852 | SE0971 (309-514) | Conserved hypothetical protein |
| SE0972 (1-121) | Conserved hypothetical protein | ||
| SE0973 (1-164) | Conserved hypothetical protein | ||
| 16 | 1941916-1943617 | SE1905 (1-293) | Dehydrogenase |
| SE1906 (198-372) | Amino acid amidohydrolase | ||
| 17 | 129349-135120 | SE0137 (1-581) | Cap5B, capsular polysaccharide synthesis enzyme |
| SE0138 (1-115) | ArsD, arsenic resistance operon | ||
| SE0139 (1-106) | Arsenic resistance operon repressor-like protein | ||
| SE0140 (1-294) | Putative membrane protein | ||
| 18 | 1513472-1516321 | SE1464 (1-365) | O-Succinylbenzoic acid-coenzyme A ligase |
| SE1466 (1-167) | Conserved hypothetical protein | ||
| 19 | 175644-179837 | SE0185 (155-643) | GehD |
| SE0186 (1-132) | Conserved hypothetical protein | ||
| SE0187 (1-86) | Conserved hypothetical protein | ||
| SE0188 (1-293) | Cobalamin synthesis-related protein | ||
| 20 | 756962-759182 | SE0768 (1-494) | PurF |
| SE0769 (1-207) | PurM | ||
| 21 | 804022-806137 | SE0812 (153-372) | Conserved hypothetical protein |
| SE0813 (1-355) | Pyruvate carboxylase | ||
| 22 | 1658497-1660726 | SE1602 (132-309) | Manganese-dependent inorganic pyrophosphatase |
| SE1603 (1-459) | Aldehyde dehydrogenase | ||
| 23 | 2248831-2250776 | SE2197 (374-489) | Alkaline phosphatase III precursor |
| SE2198 (23-514) | Conserved hypothetical protein | ||
| 24 | 334901-337600 | SE0331 (933-1065) | Fbe |
| SE0332 (1-500) | Poly(glycerol-phosphate) alpha-glucosyltransferase | ||
| 25 | 1837430-1839195 | SE1779 (1-348) | Alkanal monooxygenase alpha chain |
| SE1780 (157-289) | Conserved hypothetical protein | ||
| 26 | 2283238-2285302 | SE2228 (105-646) | Phosphotransferase system fructose-specific II component |
| SE2229 (1-141) | Mannose-6-phosphate isomerase | ||
| 27 | 1602487-1603234 | SE1548 (79-328) | Glutamate-1-semialdehyde aminotransferase |
| 28 | 2086731-2089154 | SE2044 (1-452) | Gluconate permease |
| SE2045 (326-513) | Gluconokinase | ||
| 29 | 2345106-2346500 | SE2272 (230-695) | Cell division protein |
| 30 | 2376544-2377407 | SE2303 (1-61) | Spermidine acetyltransferase |
| 31 | 263229-263653 | SE0263 (165-306) | Drp35 protein |
| 32 | 2492025-2494409 | SE2411 (1-127) | Conserved hypothetical protein |
| SE2412 (1-119) | Conserved hypothetical protein | ||
| SE2413 (1-53) | Hypothetical protein | ||
| SE2414 (1-279) | DNA-binding Spo0J-like protein | ||
| SE2415 (125-239) | Glucose-inhibited division protein B | ||
| 33 | 2454565-2456586 | SE2380 (143-391) | Putative cystathionine beta-lyase |
| SE2381 (1-439) | Conserved hypothetical protein | ||
| 34 | 2481293-2483748 | SE2400 (201-252) | VraD |
| SE2401 (1-626) | VraE | ||
| SE2402 (1-63) | Conserved hypothetical protein | ||
| 35 | 1369989-1372476 | SE1340 (138-186) | DNA-3-methyladenine glycosidase |
| SE1341 (1-355) | Conserved hypothetical protein | ||
| SE1342 (156-427) | Glutamate-1-semialdehyde 2,1-aminomutase | ||
| 36 | 585999-587077 | SE0594 (1-18) | Thioredoxin |
| SE0595 (1-117) | Arsenate reductase | ||
| SE0596 (1-118) | Glycine cleavage system protein H | ||
| 37 | 1247787-1250204 | SE1211 (1-310) | Glutamate N-acetyltransferase |
| SE1212 (1-341) | N-Acetylglutamate gamma-semialdehyde dehydrogenase | ||
| SE1213 (88-185) | Elongation factor EF-P | ||
| 38 | 2387434-2389445 | SE2309 (1-311) | Conserved hypothetical protein |
| SE2310 (1-273) | LysR-type transcriptional regulator | ||
| 39 | 1940963-1942972 | SE1903 (1-66) | Conserved hypothetical protein |
| SE1904 (1-203) | Conserved hypothetical protein | ||
| SE1905 (4-293) | Dehydrogenase | ||
| 40 | 465589-467653 | SE0472 (591-650) | Phosphotransferase system fructose-specific IIABC component |
| SE0473 (1-390) | N-Acetylglucosamine-6-phosphate deacetylase | ||
| 41 | 813245-815819 | SE0819 (1-308) | Glycerophosphoryl diester phosphodiesterase |
| SE0820 (1-39) | Hypothetical protein | ||
| SE0821 (1-83) | Conserved hypothetical protein | ||
| SE0822 (1-129) | Conserved hypothetical protein | ||
| 42 | 177914-179837 | SE0187 (46-86) | Conserved hypothetical protein |
| SE0188 (1-293) | CobW | ||
| SE0189 (193-460) | Ferrous iron transporter protein B | ||
| SE0190 (1-61) | Hypothetical protein | ||
| 43 | 469456-469924 | SE0475 (113-269) | Plant metabolite dehydrogenase |
| 44 | 152788-156004 | SE0165 (1-252) | Two-component response regulator |
| SE0166 (1-512) | Two-component sensor histidine kinase | ||
| SE0167 (24-324) | BitC | ||
| 45 | 2044647-2046972 | SE2007 (282-505) | Beta-lactamase |
| SE2008 (1-278) | Lipoprotein precursor | ||
| SE2009 (133-310) | 2-Dehydropantoate 2-reductase | ||
| 46 | 972873-974051 | SE0966 (125-264) | Conserved hypothetical protein |
| SE0967 (1-215) | Putative 2-oxoacid ferredoxin oxidoreductase alpha subunit | ||
| 47 | 240928-242180 | SE0244 (38-207) | Conserved hypothetical protein |
| SE0245 (1-124) | Lipase precursor | ||
| 48 | 1845438-1846232 | SE1786 (1-153) | Galactose-6-phosphate isomerase LacB subunit |
| SE1787 (36-142) | Galactose-6-phosphate isomerase LacA subunit | ||
| 49 | 1016461-1018676 | SE1011 (1-306) | Homoserine kinase |
| SE1012 (1-268) | Conserved hypothetical protein | ||
| 50 | 169874-172149 | SE0180 (1-451) | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase |
| SE0181 (1-301) | 8-Amino-7-oxononanoate synthase | ||
| 51 | 890136-893481 | SE0894 (1-247) | Putative PP2C protein phosphatase |
| SE0895 (1-667) | Protein kinase | ||
| 52 | 2070338-2072937 | SE2032 (1-272) | Glucose 1-dehydrogenase |
| SE2033 (1-353) | Exopolyphosphatase | ||
| 53 | 319239-321290 | SE0318 (1-358) | Branched-chain amino acid aminotroansferase-like protein |
| SE0319 (1-85) | Putative 4-diphosphocytidyl-2C-methyl-d-erythritol synthase |
The library was screened with four sera obtained from patients with confirmed S. epidermidis bacteremia, six sera obtained from patients with confirmed S. aureus bacteremia, four sera obtained from hospital workers, and four sera obtained from community volunteers. The locus, clone, and gene coordinates correspond to the coordinates in the S. epidermidis ATCC 12228 genome (http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org_search=&org=ntse02). Functions were assigned according to the ATCC 12228 genome database.
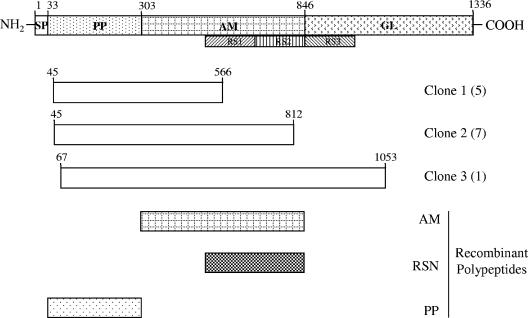
Antigenic regions of AtlE, the major autolysin of S. epidermidis, were identified by using overlapping clones due to the random nature of library construction (Fig. 1). Information concerning the genes in each clone is shown in Table 3. Corresponding DNA can be obtained by using clone coordinates from the S. epidermidis ATCC 12228 genome (http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org_search=&org=ntse02).
FIG. 1.
Organization of atlE clones isolated during the screening process. The full-length nascent protein with putative signal peptide (SP), propeptide (PP), amidase (AM), and glucosaminidase (GL) domains is shown, and repeat sequence (RS) areas are also indicated. The clones isolated (with numbers in parentheses) and recombinant peptides are also indicated. The numbers indicate the positions of the relevant amino acid residues.
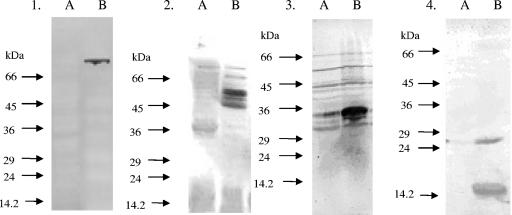
In order to select suitable antigens for further study, E. coli phagemid clones that were representative of each individual locus were screened by Western blotting using the serum that originally identified the clone. This revealed four representative clones that exhibited high levels of reactivity (Fig. 2), corresponding to loci 1, 6, 7, and 19. In these loci there were three likely candidate antigens due to the predicted surface or secreted location, and these antigens were used for further study (AtlE, GehD, ScaA [AaE], and ScaB). All atlE clones isolated from the screen contained at least some of the amidase domain, the repeat sequences, and the propeptide region (Fig. 1). For this reason, recombinant peptides corresponding to the amidase domain, repeat sequences, and propeptide were generated and used in Western blotting experiments with patient serum. The amidase domain and the repeat sequence, but not the propeptide, reacted with serum (results not shown). Thus, the amidase domain and repeat sequence were used in ELISA. Clones expressing GehD represented the entire mature lipase domain and part of the propeptide (Fig. 3); hence, the mature lipase was overexpressed and used in further experiments.
FIG. 2.
Western blot analysis of antigen-encoding clones. All membranes were blotted with human sera, which identified the original clone to confirm its suitability for overexpression. In all blots, lane A contained a negative control (E. coli XLOLR/pBK-CMV). The proteins encoded by the clones in the blots were Atl Clone3 (blot 1), GehD (blot 2), ScaA (blot 3), and ScaB (blot 4).
FIG. 3.
Structural organization of GehD. The signal peptide (SP), propeptide (PP), and mature lipase (ML) domains are indicated, along with their lengths in amino acid (aa) residues (adapted from reference 30). Region A is the region that is present in the three identical clones identified in the serum screen. Region B encompasses the mature lipase which was used for overexpression.
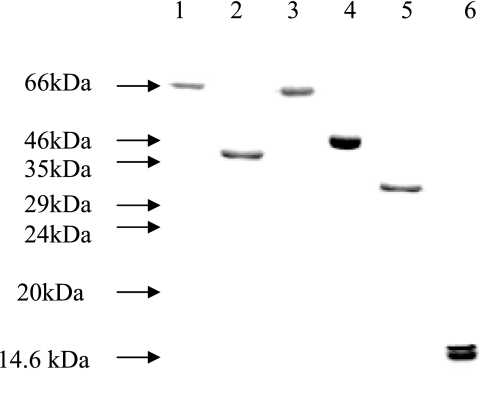
AtlE, the major autolysin of S. epidermidis, exhibits significant structural homology with Atl, Aas, and AtlC, the major autolysins of S. aureus, Staphylococcus saprophyticus, and Staphylococcus caprae, respectively, and consists of both amidase and glucosaminidase domains (1, 17, 22, 25, 35). Interestingly, a recent study showed that the glucosaminidase domain of Atl is associated with S. aureus infection (9). In this study, we did not identify the glucosaminidase domain of AtlE as an antigen, although this may have been due to the cloning strategy employed. Thus, we confined our study to the propeptide, amidase, and central repeat sequence domains (22). In order to determine the importance of the three domains, the propeptide, mature amidase, and repeat sequence domains were overexpressed and purified (Fig. 4).
FIG. 4.
Coomassie blue-stained 12% (wt/vol) SDS-PAGE gel containing the six purified polypeptides. Lane 1, mature amidase domain of AtlE; lane 2, repeat sequence domain of AtlE; lane 3, propeptide domain of AtlE; lane 4, GehD; lane 5, ScaA; lane 6, ScaB.
The N-terminal sequences for the recombinant proteins were checked and were found to be as follows: VSSQKT (AtlE amidase), TTSTKP (AtlE repeat sequence), AEQPQNQS (AtlE propeptide), ATTHTVK (ScaA), and AQAQYKN (GehD). All sequences were as predicted, confirming the fidelity of the cloning process. However, the N-terminal sequence of ScaB was SGGTATQ and was 127 amino acids downstream of the translation start of the recombinant protein. This corresponds to a truncated (ca. 16-kDa) form of the protein which is produced (Fig. 4) and is most likely the result of proteolytic processing in E. coli.
scaA and scaB are members of a multigene family.
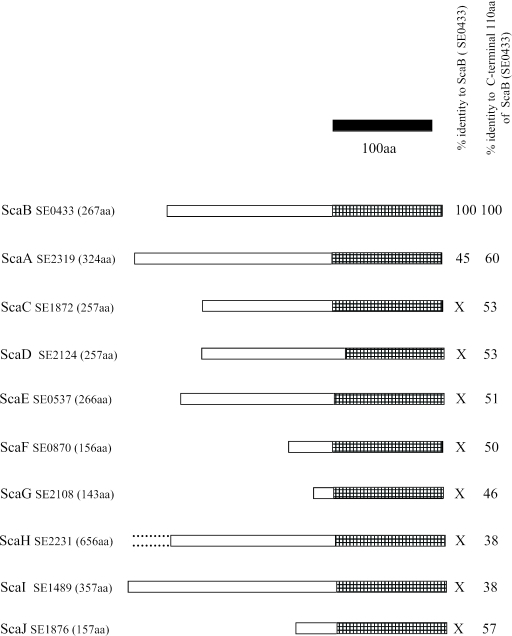
Screening of the library with human sera resulted in identification of two proteins, SE0433 (locus 6) and SE2319 (locus 7), which are closely related to each other. Sequence analysis using CLUSTALW (http://npsa-pbil.ibcp.fr) revealed 45% identity for the entire length of both proteins and more than 60% identity in the 110-amino-acid C-terminal domains. Both proteins have apparent signal peptides, which result in mature proteins whose molecular masses are 25 kDa (SE0433) and 32 kDa (SE2319). Interestingly, these two proteins are both members of a 10-protein family in S. epidermidis, based on homology of the ca. 110-amino-acid C-terminal domain. We designated these proteins staphylococcal conserved antigen A (ScaA), ScaB, ScaC, ScaD, ScaE, ScaF, ScaG, ScaH, ScaI, and ScaJ (Fig. 5). SE2319 and SE0433 are ScaA and ScaB, respectively. ScaA, ScaB, and ScaE all contain one or more LysM domains, which were originally identified in enzymes that degrade bacterial peptidoglycan but are also found in many other bacterial proteins that are associated with cell walls (5). The LysM domain has a peptidoglycan-binding fold (27), and although a potential binding site in a shallow groove on the surface of the protein has been identified, the precise component to which it binds remains unknown (4).
FIG. 5.
Comparison of the ScaA-J family of S. epidermidis. The hatched bars indicate the 110-amino-acid (aa) C-terminal domain. The solid bar indicates 100 amino acids. X, significant homology (≥20% identity) occurs only in the C-terminal domain.
ScaB exhibits 56% identity with SsaA of S. epidermidis. In S. aureus, SsaA has a virulence role specific to the pathogenesis of staphylococcal infections (29).
During this study it was shown that SE2319 (ScaA) is a novel autolysin, which has been designated Aae (23), and its paralogue in S. aureus has also been identified (24) and has been shown to be a virulence factor (28). While it is not essential that vaccine or antibody targets have such characteristics, these are attractive attributes as it is important that antigens are expressed on the bacterial cell surface during the infectious process so that the immune system of the host can mount a response. S. aureus possesses a 10-member Sca protein family with homology in the C-terminal domain. Open reading frames encoding homologous C-terminal domains are also to be found in the genomes of S. pneumoniae, Streptococcus pyogenes, Streptococcus agalactiae, Streptococcus mutans, Enterococcus faecalis, Lactococcus lactis, and Bifidobacterium longum (results not shown). Vaccination against, or therapeutic antibodies reactive to, such proteins may provide targets for use against a broad spectrum of gram-positive bacteria.
Human antibody response to S. epidermidis antigens.
An antibody response to a given antigen is indicative of its expression in vivo and of its potential for use in a subunit vaccine or as a target in passive immunotherapy or prophylaxis. The immune response against the selected antigens was quantified by ELISA using four recombinant proteins and 30 serum samples, including 20 samples of disease-associated sera (15 S. epidermidis infections and 5 S. aureus infections) and 10 serum samples from healthy blood donors (5 non-S. aureus nasal carriers and 5 S. aureus nasal carriers). Two of the original six polypeptides (Fig. 4) were dropped from the study; AtlE propeptide was not included as it did not react with serum in the Western blotting stage, and ScaA was not included because of its homology to ScaB.
The statistical significance of differences in serum antibody levels between the diseased and healthy patient groups was tested using the Kruskall-Wallis test. A comparison of the results for diseased and healthy individuals revealed that all four recombinant antigens had significantly higher (P ≤ 0.05) reactive IgG titers in serum samples obtained from diseased individuals than in serum samples obtained from healthy individuals (Table 4). This indicates that all four proteins are produced in vivo and are thus immunological markers of infection that have potential diagnostic utility.
TABLE 4.
Statistical analysis of specific antibody levels in S. epidermidis patient sera and community sera
| Protein | Median absorbance at 450 nm (interquartile range)a
|
P value | |
|---|---|---|---|
| S. epidermidis infection (n = 15) | Healthy (n = 10) | ||
| AtlE amidase | 0.650 (0.110-1.004) | 0.260 (0.002-0.342) | 0.004 |
| AtlE repeat sequence | 0.546 (0.152-0.999) | 0.165 (0-0.2329) | 0.002 |
| GehD | 0.071 (0.030-0.310) | 0.032 (0.014-0.048) | 0.0001 |
| ScaB | 0.137 (0.017-0.260) | 0.058 (0-0.130) | 0.027 |
The interquartile range indicates the minimum and maximum values for reactions of sera with each antigen.
Opsonization of S. epidermidis.
Different dilutions of serum from immunized mice were used to obtain measurable killing of S. epidermidis Hay (Table 5). Antibodies raised against AtlE or ScaB are opsonic in vitro. The antibodies raised against GehD, a lipase with collagen binding activity (7, 30), did not opsonize S. epidermidis. The mature processed form of GehD is found predominantly in the extracellular milieu, and it has been proposed that some of this form is present on cell walls (7). Thus, a relatively small amount of the protein on the cell wall may result in an insufficient amount of antigen binding to trigger opsonization. Therefore, it is tempting to speculate that either AtlE or ScaB or both could be efficacious vaccines or passive immunotherapy targets and thus be useful for prevention or treatment of infections caused by many staphylococcal species due to the presence of paralogues in these organisms. Historically, polyclonal antibodies developed for use in passive immunotherapy of S. aureus infections have been limited by serotype specificity (12, 13) and recognize only 75% to 80% of all S. aureus clinical isolates (41). It is unlikely that targets such as AtlE or ScaB would pose such a problem, since several staphylococcal species possess AtlE paralogues and since ScaB is conserved in many gram-positive species.
TABLE 5.
Opsonization using antibodies raised against recombinant antigensa
| Antigen | Dilution | Avg % killed | SD |
|---|---|---|---|
| AtlE amidase | 1/20 | 80 | 15 |
| AtlE repeat sequence | 1/20 | 55 | 41 |
| GehD | 1/10 | 0.4 | 1 |
| ScaB | 1/10 | 42 | 35 |
S. epidermidis Hay was mixed with PMNs isolated from freshly obtained human blood and complement. After incubation for 2 h at 37°C, the surviving bacteria were counted, and the percentage of killed bacteria was determined. The results indicate the average killing efficacy of serum obtained from five animals.
Ligand binding activity of antigenic surface proteins.
Bacterial colonization is a crucial step in pathogenicity, and its onset is determined by an interaction between the invading organism and its host. To study the possible involvement of the purified recombinant proteins in adhesion, these proteins were analyzed by performing binding assays with a selection of plasma proteins (fibrinogen, fibronectin, mucin, and lactoferrin), Western blotting, and probing with biotinylated ligands (Fig. 6 and Table 6).
FIG. 6.
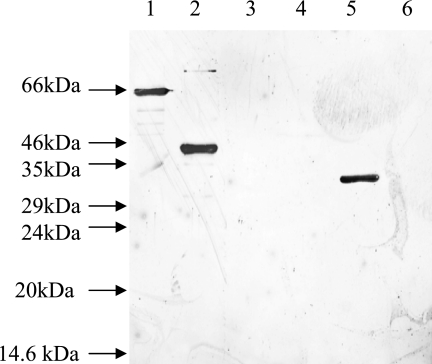
Screening for fibrinogen binding by purified recombinant polypeptides, using the Western affinity blotting technique. All lanes contained ca. 1 μg of protein. Lane 1, mature amidase domain of AtlE; lane 2, repeat sequence domain of AtlE; lane 3, propeptide domain of AtlE; lane 4, GehD; lane 5, ScaA; lane 6, ScaB.
TABLE 6.
Ligand binding capabilities of recombinant proteins (fibrinogen, fibronectin, lactoferrin, and mucin)
| Antigen | Ligand binding capability of:
|
|||
|---|---|---|---|---|
| Fibrinogen | Fibronectin | Mucin | Lactoferrin | |
| AtlE mature amidase | + | + | + | + |
| AtlE repeat sequence | + | + | + | + |
| AtlE propeptide | − | − | − | − |
| GehD | − | − | − | − |
| ScaA | + | + | + | + |
| ScaB | − | + | − | + |
The mature amidase fragment of AtlE bound all four plasma proteins. Interestingly, the repeat sequences of mature amidase exhibited such binding, which corresponds to findings for Aas and AtlC, whose central repeat sequences have been shown to mediate fibronectin binding (1, 25). Here we found that in AtlE, the repeat sequences also mediated binding to fibrinogen, mucin, and lactoferrin. The propeptide fragment of AtlE did not exhibit binding to any of the plasma proteins. ScaA bound all four of the plasma proteins, but the very highly homologous molecule ScaB bound to only fibronectin and lactoferrin, although a reduction in ligand binding capacity may have resulted from the proteolytic processing of recombinant ScaB. Alternatively, sequence differences between the two proteins may provide clues concerning the binding properties. Previous studies have shown that ScaA (AaE) binds fibrinogen and fibronectin (23) and that its S. aureus paralogue, AaA, binds fibrinogen, fibronectin, and vitronectin (24). Although GehD has been shown to bind to collagen (7), it did not bind any plasma proteins used in this study.
In this study we identified many antigenic S. epidermidis proteins. Some of these proteins are potential targets for immunotherapy, which provides a novel strategy for control of S. epidermidis infection, and could potentially reduce the toll of infection and have a significant impact on human health. Interestingly, a subset of S. epidermidis proteins are also conserved in S. aureus (9) and in other genera. These antigens may have important functions in disease.
Acknowledgments
This work was funded by the Iranian Government (M.R.P.) and Biosynexus (S.R.C.).
Oligonucleotide synthesis and N-terminal sequencing were carried out by Arthur Moir, University of Sheffield, Sheffield, United Kingdom. Human serum samples were kindly provided by Robert Read, Royal Hallamshire Hospital, Sheffield, United Kingdom, and Peter Lambert, Aston University, Birmingham, United Kingdom. S. epiderimidis 138 was kindly supplied by Paul Williams, University of Nottingham, United Kingdom.
Editor: D. L. Burns
REFERENCES
- 1.Allignet, J., S. Aubert, K. G. Dyke, and N. El Sohl. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, G. L., and E. Pennell. 1990. Detection of methicillin resistance in staphylococci by using a DNA probe. Antimicrob. Agents Chemother. 34:1720-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, G. L., and M. W. Climo. 1994. Antimicrobial susceptibility of coagulase-negative staphylococci. Antimicrob. Agents Chemother. 38:2231-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., and M. Bycroft. 2000. The structure of LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 5.Birkeland, N. K. 1994. Cloning, molecular characterization, and expression of the genes encoding lytic functions of lactococcal bacteriophage phi LC3: a dual lysis system of modular design. Can. J. Microbiol. 40:658-665. [DOI] [PubMed] [Google Scholar]
- 6.Bloom, B., R. Schelonka, T. Kueser, W. Walker, E. Jung, D. Kaufman, K. Kesler, D. Robertson, J. Patti, and S. Hetherington. 2005. Multicenter study to assess safety and efficacy of INH-A21, a donor selected human staphylococcal immunoglobulin, for prevention of nosocomial infections in very low birth weight infants. Pediatr. Infect. Dis. J. 24:858-866. [DOI] [PubMed] [Google Scholar]
- 7.Bowden, M. G., L. Visai, C. M. Longshaw, K. T. Holland, P. Speziale, and M. Höök. 2002. Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J. Biol. Chem. 277:43017-43023. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, S. R., L. G. Harris, R. G. Richards, and S. J. Foster. 2002. Analysis of Ebh, a 1.1.-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect. Immun. 70:6680-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke, S. R., K. J. Brummell, M. J. Horsburgh, P. W. McDowell, S. A. Syed Mohamad, M. R. Stapleton, J. Acevedo, R. C. Read, N. J. P. Day, S. J. Peacock, J. J. Mond, J. F. Kokai-Kun, and S. J. Foster. 2006. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J. Infect. Dis. 193:1098-1108. [DOI] [PubMed] [Google Scholar]
- 10.Domanski, P. J., P. R. Patel, A. S. Bayer, L. Zhang, A. E. Hall, P. J. Syribeys, E. L. Gorovits, D. Bryant, J. H. Vernachio, J. T. Hutchins, and J. M. Patti. 2005. Characterization of a humanized monoclonal antibody recognizing clumping factor A expressed by Staphylococcus aureus. Infect. Immun. 73:5229-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etz, H., D. B. Minh, T. Henics, A. Dryla, B. Winkler, C. Triska, A. P. Boyd, J. Sollner, W. Schmidt, U. von Ahsen, M. Buschle, S. R. Gill, J. Kolonay, H. Khalak, C. M. Fraser, A. von Gabain, E. Nagy, and A. Meinke. 2002. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 99:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fattom, A. I., and R. Naso. 1996. Staphylococcal vaccines: a realistic dream. Ann. Med. 28:43-46. [DOI] [PubMed] [Google Scholar]
- 13.Fattom, A. I., J. Sarwar, A. Ortiz, and R. Naso. 1996. A Staphylococcus aureus capsular polysaccharide (CP) vaccine and CP-specific antibodies protect mice against bacterial challenge. Infect. Immun. 64:1659-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, G. W., T. J. Cieslak, S. R. Wilson, L. E. Weisman, and V. G. Hemming. 1994. Opsonic antibodies to Staphylococcus epidermidis: in vitro and in vivo studies using human intravenous immune globulin. J. Infect. Dis. 169:324-329. [DOI] [PubMed] [Google Scholar]
- 15.Flock, J.-I. 1999. Extracellular-matrix-binding proteins as targets for the prevention of Staphylococcus aureus infections. Mol. Med. Today 5:532-537. [DOI] [PubMed] [Google Scholar]
- 16.Flock, J.-I., and F. Brennan. 1999. Antibodies that block adherence of Staphylococcus aureus to fibronectin. Trends Microbiol. 7:140-141. [DOI] [PubMed] [Google Scholar]
- 17.Foster, S. J. 1995. Molecular characterization and functional analysis of a major autolysin of Staphylococcus aureus 8325/4. J. Bacteriol. 177:5723-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froggatt, J. W., J. L. Johnston, D. W. Galetto, and G. L. Archer. 1989. Antimicrobial resistance in nosocomial isolates of Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 33:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrett, D. O., E. Jochimsen, K. Murfitt., B. Hill, S. McAllister, P. Nelson, R. V. Spera, R. K. Sall, F. C. Tenover, J. Johnston, B. Zimmer, and W. R. Jarvis. 1999. The emergence of decreased susceptibility to vancomycin in Staphylococcus epidermidis. Infect. Control Hosp. Epidemiol. 20:167-170. [DOI] [PubMed] [Google Scholar]
- 20.Giamarellou, H. 2002. Nosocomial cardiac infections. J. Hosp. Infect. 50:91-105. [DOI] [PubMed] [Google Scholar]
- 21.Hall, A. E., P. J. Domanski, P. R. Patel, J. H. Vernachio, P. J. Syribeys, E. L. Gorovits, M. A. Johnson, J. M. Ross, J. T. Hutchins, and J. Patti. 2003. Characterization of a prospective monoclonal antibody recognizing Staphylococcus aureus MSCRAMM protein clumping factor A. Infect. Immun. 71:6864-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heilmann, C., M. Hussain, G. Peters, and F. Götz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 23.Heilmann, C., G. Thumm, G. S. Chhatwal, J. Hartlieb, A Uekotter, and G. Peters. 2003. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 149:2769-2778. [DOI] [PubMed] [Google Scholar]
- 24.Heilmann, C., J. Hartlieb, M. Hussain, and G. Peters. 2005. The multifunctional Staphylococcus aureus autolysin Aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect. Immun. 73:4793-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hell, W., H. G. Meyer, and S. G. Gattermann. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871-881. [DOI] [PubMed] [Google Scholar]
- 26.Huebner, J., and D. A. Goldmann. 1999. Coagulase-negative staphylococci: role as pathogens. Annu. Rev. Med. 50:223-236. [DOI] [PubMed] [Google Scholar]
- 27.Joris, B., S. Englebert, C. P. Chu, R. Kariyama, L. Daneo-More, G. D. Shockman, and J. M. Ghuysen. 1992. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 70:257-264. [DOI] [PubMed] [Google Scholar]
- 28.Kajimura, J., T. Fujiwara, S. Yamada, Y. Suzawa, T. Nishida, Y. Oyamada, I. Hayashi, J. Yamagishi, H. Komatsuzawa, and M. Sugai. 2005. Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 58:1087-1101. [DOI] [PubMed] [Google Scholar]
- 29.Lang, S., M. A. Livesley, P. A. Lambert, W. A. Littler, and T. S. Elliot. 2000. Identification of a novel antigen from Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 29:213-220. [DOI] [PubMed] [Google Scholar]
- 30.Longshaw, C. M., A. M. Farrell, J. D. Wright, and K. T. Holland. 2000. Identification of a second lipase gene, gehD, in Staphylococcus epidermidis: comparison of sequence with those of other staphylococcal lipases. Microbiology 146:1419-1427. [DOI] [PubMed] [Google Scholar]
- 31.McCool, T. M., T. R. Cate, G. Moy, and J. N. Weiser. 2002. The immune response to pneumococcal proteins during experimental human carriage. J. Exp. Med. 195:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modun, B., D. Kendall, and P. Williams. 1994. Staphylococci express a receptor for human transferrin: identification of a 42-kilodalton cell wall transferrin-binding protein. Infect. Immun. 62:3850-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson, I.-M., J. Patti, T. Bremell, M. Höök, and A. Tarkowski. 1998. Vaccination with a recombinant fragment of collagen adhesin provides protection against Staphylococcus aureus-mediated septic death. J. Clin. Investig. 101:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Hara, D. M., C. R. Harrington, and P. E. Reynolds. 1989. Immunological detection of penicillin binding protein 2′ in clinical isolates of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. FEMS Microbiol. Lett. 57:97-104. [DOI] [PubMed] [Google Scholar]
- 35.Oshida, T., M. Sugai, H. Komatusuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetlymuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patti, J. M. 2004. A humanized monoclonal antibody targeting Staphylococcus aureus. Vaccine 22S:S39-S43. [DOI] [PubMed] [Google Scholar]
- 37.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Stroni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandini, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-organism sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 38.Rozalska, B., and T. Wadstrom. 1993. Protective opsonic activity of antibodies against fibronectin-binding proteins (FnBPs) of Staphylococcus aureus. Scand. J. Immunol. 37:575-580. [DOI] [PubMed] [Google Scholar]
- 39.Rupp, M. E., and G. L. Archer. 1994. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19:231-243. [DOI] [PubMed] [Google Scholar]
- 40.Sellman, B. R., A. P. Howell, C. Kelly-Boyd, and S. M. Baker. 2005. Identification of immunogenic and serum binding proteins of Staphylococcus epidermidis. Infect. Immun. 73:6591-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinefield, H., S. Black, A. Fattom, G. Horwith, S. Rasgon, J. Ordonez, H. Yeoh, D. Law, J. B. Robbins, R. Schneerson, L. Muenz, S. Fuller, J. Johnson, B. Fireman, H. Alcorn, and R. Naso. 2002. Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N. Engl. J. Med. 346:491-496. [DOI] [PubMed] [Google Scholar]
- 42.Vernachio, J., A. S. Bayer, T. Le, Y.-L. Chai, B. Prater, A. Schneider, B. Ames, P. Syribeys, J. Robbins, and J. Patti. 2003. Anti-clumping factor A immunoglobulin reduces the duration or methicillin-resistant Staphylococcus aureus bacteremia in an experimental model of infective endocarditis. Antimicrob. Agents Chemother. 47:3400-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visai, L., Y. Xu, F. Casolini, S. Rindi, M. Höök, and P. Speziale. 2000. Monoclonal antibodies to CNA, a collagen-binding microbial surface component recognizing adhesive matrix molecules, detach Staphylcoccus aureus from a collagen substrate. J. Biol. Chem. 275:39837-39845. [DOI] [PubMed] [Google Scholar]
- 44.Wizemann, T. M., J. H. Heinrichs, J. E. Adamou, A. L. Erwin, C. Kunsch, G. H. Choi, S. C. Barasch, C. A. Rosen, H. R. Masure, E. Tuomanen, A. Gayle, Y. A. Brewah, W. Walsh, P. Barren, R. Lathigra, M. Hanson, S. Langermann, S. Johnson, and S. Koenig. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]