Abstract
Centromeres of most organisms are embedded within constitutive heterochromatin, the condensed regions of chromosomes that account for a large fraction of complex genomes. The functional significance of this centromere–heterochromatin relationship, if any, is unknown. One possibility is that heterochromatin provides a suitable environment for assembly of centromere components, such as special centromeric nucleosomes that contain distinctive histone H3-like proteins. We describe a Drosophila H3-like protein, Cid (for centromere identifier) that localizes exclusively to fly centromeres. When the cid upstream region drives expression of H3 and H2B histone–green fluorescent protein fusion genes in Drosophila cells, euchromatin-specific deposition results. Remarkably, when the cid upstream region drives expression of yeast, worm, and human centromeric histone–green fluorescent protein fusion proteins, localization is preferentially within Drosophila pericentric heterochromatin. Heterochromatin-specific localization also was seen for yeast and worm centromeric proteins constitutively expressed in human cells. Preferential localization to heterochromatin in heterologous systems is unexpected if centromere-specific or site-specific factors determine H3-like protein localization to centromeres. Rather, the heterochromatic state itself may help localize centromeric components.
Keywords: centromeres, heterochromatin, Drosophila
For well over a century, the centromere has been recognized as the unique position on a chromosome that mediates its movement to the poles at anaphase (1). What determines this position? In Saccharomyces cerevisiae, a distinctive, ≈125-bp sequence uniquely specifies a centromere (2). However, in multicellular eukaryotes, there is no candidate unique sequence found at centromeres. Rather, centromeres consist of highly repetitive satellite sequences not obviously different from noncentromeric sequences (3–5). It is unlikely that satellite repeats obscure a unique centromere specification sequence, because mutagenesis studies define the minimum size of centromeres as hundreds of kilobases (6–8). Furthermore, the lack of evolutionary conservation of satellite repeats seems inconsistent with the absolute requirement for one and only one centromere on every chromosome.
Compounding the enigma of centromere identity is the discovery of human neocentromeres that lack satellite sequences (9–11). Even complete sequencing of one such centromere failed to detect satellite sequences and did not reveal any differences between the functional centromere and the same chromosomal region on the parental chromosome that could plausibly account for centromere behavior (12). Therefore, satellites must not be necessary for organization of a functional kinetochore. To explain how such neocentromeres can arise, it has been proposed that an epigenetic mark can be acquired that converts a region that was not a centromere into a stable centromere (13–16) or that replication timing distinguishes centromeres from noncentromeres (3). However, none of these hypotheses are accompanied by molecular mechanisms that can explain how a mark or a replication timing difference can lead to precise localization of a centromere.
A possible resolution of the centromere identity problem comes from the discovery that centromeres contain special nucleosomes with distinctive H3-like molecules. In contrast to the lack of obvious common attributes when centromeric DNA sequences are compared, such special nucleosomes are found both at the complex centromeres of mammals (17, 18) and at the simple centromeres of baker's yeast (19–21). Thus, it is possible that localization of special nucleosomes identifies the centromere. Support for this possibility comes from studies of the behavior of CENPA-H3 chimeras in human cells, which demonstrate that specific regions of CENP-A are responsible for localization (18, 22). However, the process whereby CENP-A-containing nucleosomes localize to centromeres remains unknown, and models ranging from recognition of arrays of nucleosomal contact sites (22) to recognition of an epigenetic mark (15) have been advanced.
If nucleosomes containing centromere-specific, H3-like proteins recognize the centromere, then it would be illuminating to ask about the behavior of centromere-specific H3-like proteins introduced into heterologous organisms, because these proteins have evolved to interact with centromeres in their native genomes. For instance, sequence differences between mammalian and baker's yeast centromeres are so profound that if sequence-based recognition is important, then yeast Cse4p will not localize to mammalian chromosomes. Alternatively, if centromeres from such different organisms share a common feature, then Cse4p indeed might localize to animal centromeres. To distinguish these possibilities, we have introduced H3-like proteins from vastly different organisms into Drosophila and human cells. Surprisingly, neither expectation is fulfilled. Rather, H3-like heterologs from yeast and worms localize to pericentric heterochromatic regions surrounding fly and human centromeres.
Materials and Methods
Computational Sequence Analysis.
Predicted amino acid sequences were detected with blastp and tblastn (23) in searches of GenBank and Genpept by using as query the histone H3 COBBLER sequence from the Blocks Database, version 11.0 (24). A clustalw (25) alignment was converted to a tree by using the multiple alignment processor at the Blocks web site (http://www.blocks.fhcrc.org). Multiple alignment display was performed by using Boxshade with default parameters (http://www.ch.embnet.org/software/BOX_form.html).
Plasmid Construction.
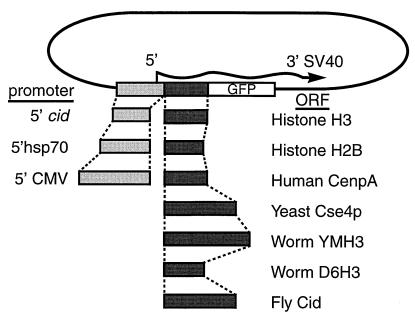
A bright green fluorescent protein (GFP) mutant variant (26) with EcoRI and SpeI ends was directionally cloned into the Drosophila transformation vector pPUAST (FlyBase) after digestion with EcoRI and XbaI to yield pPUgB. Digestion of pPUgB with Bsu36I and EcoRI allowed insertion of three tandem fragments produced by PCR: a 276-bp fragment with Bsu36I and EagI ends encoding the C terminus of GFP, a 388-bp fragment with EagI and XbaI ends encoding Drosophila histone H2B, and a 491-bp fragment with XbaI and EcoRI ends with the Drosophila HSP70 heat shock promoter, yielding pPgH2Bhs (Fig. 3). In transient and germ-line transformations using pPgH2Bhs, the H2B-GFP fusion protein produced under heat shock control was found to uniformly decorate Drosophila chromosomes (unpublished results) as had been shown for HeLa cell transfections (27). Digestion of pPgH2Bhs with EagI and XbaI allowed insertion of cid, upstream cid, and the cid coding region under hsp70 control, each produced by PCR and cleaved with NotI and XbaI. cid was amplified from Drosophila melanogaster (Amherst) genomic DNA by using primers 5′-GCTCTAGACGACATGGCTGTATCTTCAGTGCTCTGC- 3′ and 5′-ATAAGAATGCGGCCGCTGAAATTGCCGACC- CCGGTCGC-3′ for cid or 5′-AATGCGGCCGCGCACTA-GTAAATTTTCGGTATTTGCTTAAAATATTTTTAAAG- 3′ for upstream cid, where the latter provides a unique SpeI site for subsequent cloning of coding regions flanked by 5′ XbaI or SpeI and 3′ EagI or NotI ends. Each coding region was amplified either from genomic (H3, Cse4p, Cid, D6H3) DNA or from cDNA pools (CENP-A or HCP-3) as described previously (28). In each case, primers were designed to amplify the full-length coding region as was done for H2B in pPgH2Bhs. For HeLa cell transfection experiments, selected plasmid derivatives of pPgH2Bhs were digested with EcoRI and either XbaI or SpeI to accept cytomegalovirus promoter fragments amplified from pCDNA3 (Invitrogen) by using primers 5′-GGATTCTCGAGTGTACGGGCCAGATATACGCGTTG-3′ and 5′-GCTCTAGAAATTTCGATAAGCCAGTAAGCAGTGGG-3′. Amplifications were performed by using either Ampli-Taq Gold (Perkin–Elmer) or KlenTaq (CLONTECH) protocols, and cloned PCR-generated insertions were sequenced to check for misincorporations.
Figure 3.
Plasmid constructs used for transient transfection. In each case, the full protein-coding sequence is fused to a 6-aa spacer followed by the N terminus of GFP (27).
Cell-Line Transfections and Cytological Preparations.
Transfection of Kc167 cells was performed as described (29). Cells were grown on coverslips in Insect-XPRESS medium (BioWhittaker) at 25°C for 48 hr. For induction of the Hsp70 promoter, the dish was placed in an air incubator at 37°C for 1 hr and returned to 25°C for 4 hr before further processing. Colcemid (0.1 μg/ml) was added to the medium for 2 hr, after which cells were swelled in 0.5% sodium citrate for 20 min. Coverslips then were spun in a swinging-bucket tabletop centrifuge for 1 min at 4,000 rpm, and the cells were fixed in 4% paraformaldehyde in PBS supplemented with 0.3% Triton X-100 for 20 min. Transfection, fixation, and staining of HeLa cells, as well as the preparation and staining of mitotic spreads, was performed as described (30). DNA was stained with 4′,6-diamidino-2-phenylindole. Images were collected by using a DeltaVision deconvolution microscope (Applied Precision, Issaquah WA). Projections of deconvolved three-dimensional images and initial adjustments were performed by using deltavision software, and subsequent adjustments for printing were made using Adobe photoshop.
Immunolocalizations.
Rabbit antibody was raised against the peptide acetyl-CAKRAPRPSANNSKSPNDD-amide (Quality Controlled Biochemicals, Hopkinton, MA), which includes amino acids 7–24 of Cid. Cytological detection utilized a Texas Red- or FITC-conjugated goat anti-rabbit antibody. Drosophila kinetochores were detected with anti-POLO mouse antibody mAb 81 (31) followed by a Texas Red-conjugated rabbit anti-mouse antibody. Antibody staining was performed as described previously (32) by using the anti-POLO antibody at 1:10 dilution, but with the addition of a 2-min fixation with 2% formaldehyde in buffer after washing out the secondary antibody and omission of the alcohol dehydration. Human centromeres were detected with anticentromere antibody (ACA) serum (33) diluted 1:20,000 followed by a Texas Red-conjugated donkey anti-human antibody. All secondary antibodies were obtained from Jackson ImmunoResearch.
Results
A Drosophila H3-Like Protein Localizes Precisely to Centromeres.
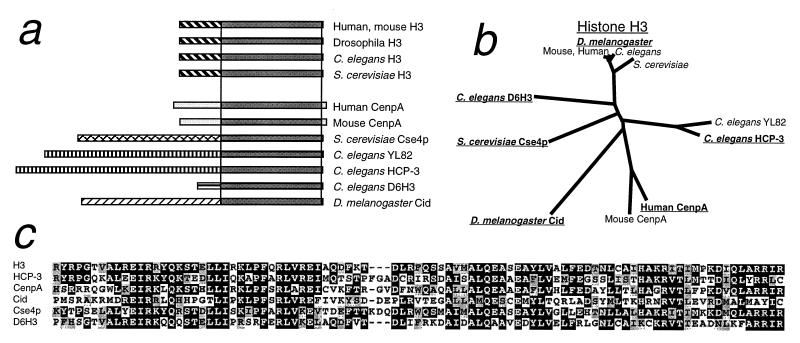
CENP-A and Cse4p share with histone H3 the ≈100-aa nucleosomal core, but have N-terminal tails that are completely dissimilar from one another and from H3 (Fig. 1a). In CENP-A, centromeric localization maps to the core (22), which is about equally divergent from both Cse4p and H3 (Fig. 1 b and c). Except for CENP-A and Cse4p, only three GenBank entries were found to have comparably diverged H3-like cores and dissimilar N-terminal tails (Fig. 1a). In addition, we found a 225-aa ORF within a “sequencing in progress” database entry (AC005652) deposited by the Berkeley Drosophila Genome Project that also fits this profile. We considered the possibility that this ORF encodes a centromeric protein. We raised an antiserum against a peptide predicted from the ORF and used it for immunocytochemistry to D. melanogaster Kc cells. Intense, point-like signals were observed over interphase nuclei and exclusively over centromeric constrictions of all Drosophila mitotic chromosomes in both Kc tissue culture cells (Fig. 2a) and larval neuroblasts (Fig. 2b). Similar results were obtained with antibody raised against a peptide predicted from another N-terminal tail region of the ORF (data not shown). Therefore, these epitopes are present on a normal centromere component that is present throughout the cell cycle. Because the 225-aa protein behaves as a Drosophila homolog of CENP-A and Cse4p, we have named the gene cid (for centromere identifier).
Figure 1.
Comparative sequence analysis of histone H3-like proteins used in this study. (a) Cartoon depicting nucleosomal core regions and unaligned N-terminal tails. Boxes representing the N-terminal tails are shaded differentially to depict the absence of sequence similarity between H3-like proteins. (b) Phylogenetic tree for the core region. Proteins used in this study are underlined. (c) Boxshade multiple alignment for the core region of the proteins used in this study.
Figure 2.
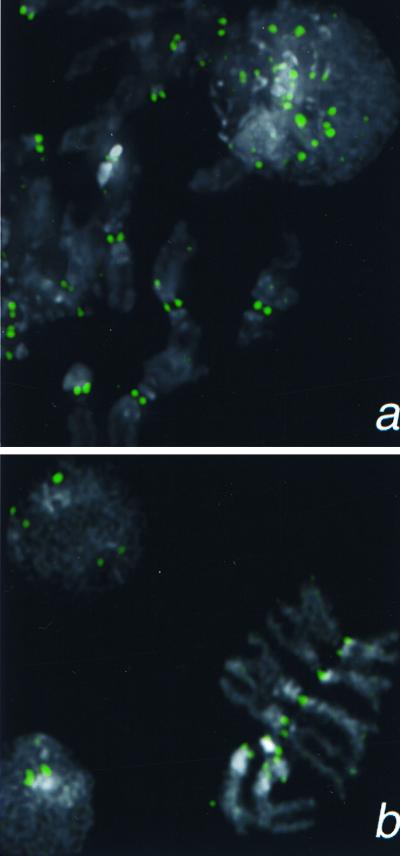
Detection of a Cid epitope at centromeric constrictions and in an interphase nucleus by using anti-Cid antiserum (1:5,000). (a) Kc167 cells. (b) Larval neuroblasts. Cid signal is shown in green 4′,6-diamidino-2-phenylindole staining in gray.
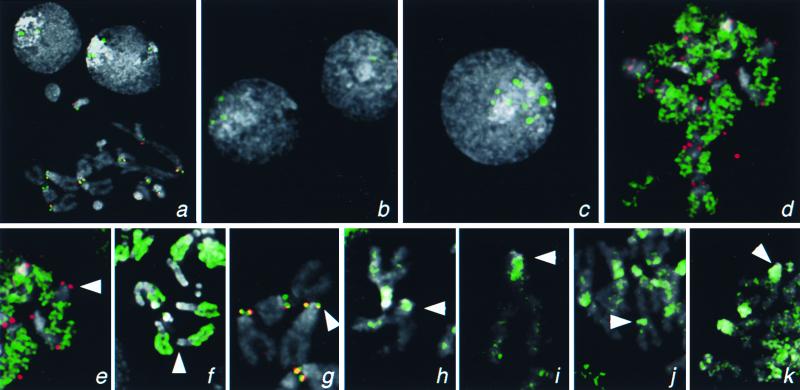
The cid gene appears to have a very short transcriptional regulatory region, because only 406 bp upstream separate the ORF from an oppositely oriented ORF that is represented in the expressed sequence tag database. We cloned this ORF and the upstream region into a GFP fusion construct (Fig. 3) and introduced plasmid into D. melanogaster Kc cells by transient transfection. Cytological spread preparations were examined by fluorescence microscopy. As with anti-Cid antibody, Cid-GFP localized to intense, point-like signals in interphase nuclei (Fig. 4a). In metaphase chromosomes, centromeres were found to be specifically labeled, based on precise colocalization with primary constrictions and colocalization with anti-POLO kinase antibody (Fig. 4 a and g), which decorates Drosophila kinetochores of metaphase chromosomes (31). The slight offset of Cid-GFP and POLO kinase may reflect the offset of the centromere from the kinetochore.
Figure 4.
Localization of H3-like protein-GFP fusions to centromeres of Drosophila. Kc167 cells were transfected with GFP fusion constructs driven by the cid promoter (a and d–k) or by basal (b) or induced (c) expression from the Hsp70 promoter. (a) Localization of Cid-GFP to the centromeres in a metaphase spread and to intense foci in interphase nuclei. Similar foci are seen in many interphase nuclei when Cid-GFP is expressed from the Hsp70 promoter (b and c) and are 10-fold more intense (185 ± 61 vs. 2,085 ± 979 pixel intensity) when this promoter has been induced (c). (d) Euchromatic labeling by cid-driven H3-GFP. Localization of GFP fusions on representative X chromosomes from mitotic figures with Drosophila H3 (e), Drosophila H2B (f), Drosophila Cid (g), human CENP-A (h), S. cerevisiae Cse4p (i), C. elegans HCP-3 (SWISS-PROT YMH3_CAEEL) (j), and C. elegans D6H3 (k) (GenBank accession no. AAB37052). GFP fluorescence was consistently lower with the H3-like heterolog-GFP fusion proteins than with the Cid- or the histone-GFP fusions. The X centromere is indicated with an arrowhead. The X chromosome is acrocentric, and the heterochromatin coheres well in metaphase spreads; thus, the position of the centromere can be identified by morphology alone. Identification of centromeres in metaphase spreads was confirmed by detection of the POLO kinetochore epitope. GFP signal is shown in green, the POLO epitope is shown in red (a, d, e, and g), and 4′,6-diamidino-2-phenylindole staining is shown in gray. Coincidence of GFP and POLO is yellow.
Cid-GFP localizes to centromeres even when it is driven by a heterologous promoter. When we substituted the Drosophila hsp70 promoter for the cid promoter, we found sharp interphase spots indicative of centromeric localization in uninduced cells (Fig. 4b). Expression is probably due to low-level constitutive activity of the hsp70 promoter (34). Upon heat shock induction, spots increased 10-fold in intensity (Fig. 4c).
Euchromatin-Specific Deposition of Core Histones Produced from the cid Promoter.
A report of cell cycle-limited expression of mRNA encoding CenpA (35) led us to test for comparable behavior of cid mRNA. However, cid mRNA appears to be rare, as its cDNA is not represented among the 80,000 D. melanogaster expressed sequence tags found in GenBank, and we were unable to detect cid mRNA in situ (unpublished results). To characterize the cid promoter, we used it to drive expression of Drosophila histones fused to GFP. Because histones are deposited at newly replicated DNA, cell cycle-limited expression might be revealed by restricted deposition of histone-GFP fusion protein observed at metaphase. When H2B-GFP was synthesized constitutively under control of the heat shock promoter, uniform GFP localization was seen consistently in mitotic figures of transiently transfected cells (data not shown). This confirms that histone-GFP fusions can be deposited throughout chromatin, as observed previously (27). However, H3- and H2B-GFP constructs driven by the cid promoter showed more limited localization: the euchromatic arms were labeled, but pericentric heterochromatin was not (Fig. 4 d–f). Because constitutive expression gives uniform deposition, cid-driven expression must produce H2B-GFP and H3-GFP early in the cell cycle, when euchromatin is replicating, but these histones must have been used up by the time that pericentric heterochromatin replicated. Thus, the cid promoter drives early S phase-limited expression. This conclusion differs from the report that cell cycle-limited expression of CenpA mRNA occurs much later in synchronized HeLa cells (35) and may reflect differences between Kc and HeLa cells or differences in procedures used to assess cell cycle-dependent expression.
H3-Like Heterologs Localize to Drosophila Pericentric Heterochromatin.
A serendipitous observation, that the Caenorhabditis elegans HCP-3 histone H3-like protein expressed from an uncharacterized genomic fragment displayed subnuclear localization in Kc cells (data not shown), led us to consider that any H3-like heterolog might localize in an interesting manner. As was done for Cid, H3 and H2B, a full-length ORF from each of the other branches of the H3 phylogenetic tree (Fig. 1b), was fused to GFP and driven by the cid promoter (Fig. 3). These ORFs are Cse4p, human CENP-A, C. elegans HCP-3, and C. elegans D6H3. In striking contrast to the euchromatic pattern seen for histone-GFP constructs driven by the cid promoter, all four H3-like heterologs expressed from the same promoter localized preferentially to pericentric heterochromatin. To identify heterochromatin in metaphase chromosomes, we utilized its appearance in standard cytological preparations: heterochromatin remains attached between sister chromatids and so can be readily distinguished from euchromatic arms, which display no sister chromatid cohesion. On the acrocentric X chromosome, which is heterochromatic for the entire proximal one-half, the peak of localization was observed consistently over a subset of heterochromatin near the centromere (Fig. 4 h–k). Localization of heterologs was seen to extend into pericentric regions of all chromosomes, sometimes with ubiquitous low-intensity labeling throughout the chromosomes. Preferential pericentric localization was seen over an ≈10× range of intensities (data not shown); this range is presumably due to cell-to-cell differences in plasmid copy number.
Pericentric localization of H3-like proteins from yeast, worms, and humans is unlikely to result from sequence recognition, because any presumptive sequence target is not thought to be in common between the centromeres of these organisms and the pericentric regions of flies. Preferential localization is also unlikely to be due to recognition of a preexisting centromeric determinant, because the labeling encompasses a region that is much broader than the centromere itself.
H3-Like Heterologs Localize to Human Centromeric Regions.
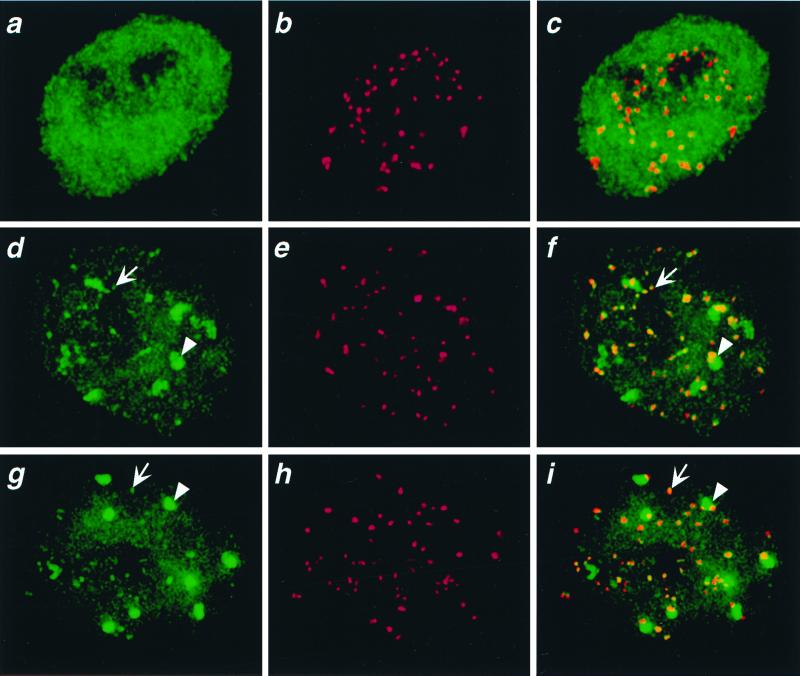
To test the generality of preferential pericentric localization in Drosophila, we introduced H3-like heterologs into human cells. GFP fusion constructs of Cse4p and one of the worm H3-like proteins, HCP-3, were driven by the constitutive cytomegalovirus promoter after transient transfection (Fig. 3). As a control, we transfected an H3-GFP construct, which gave a uniform nuclear pattern of fluorescence as expected for general chromatin localization (Fig. 5a). However, both yeast Cse4p-GFP and worm HCP-3-GFP displayed many small and six to nine large spots of localization over a weaker chromatin background (Fig. 5 d and g). These spots were observed consistently over an ≈10× range of intensities (data not shown) corresponding to a range of plasmid copy numbers that typically are obtained in transient transfection experiments (35). Using an ACA, we confirmed that most centromeres were associated with GFP spots. Many of the small GFP spots were coincident with the ACA spots, and each large GFP spot typically encompassed at least one ACA spot (Fig. 5 d–i). Therefore, heterologous H3-like proteins from yeast and worms localize on human chromatin to regions that include centromeres.
Figure 5.
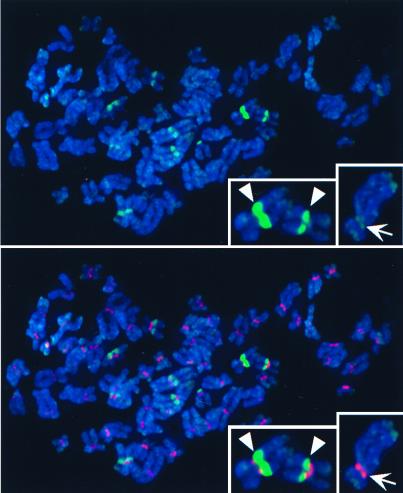
Colocalization of H3-like proteins with centromeres in human interphase nuclei. HeLa cells were transfected with GFP fusions of D. melanogaster H3 (a–c), C. elegans HCP-3 (d–f), or S. cerevisiae Cse4p (g–i) driven by the constitutive cytomegalovirus promoter. Cells were fixed and stained with ACA serum against centromeres (red); the GFP signal is shown in green. Representative interphase nuclei are shown. We chose the 60 brightest GFP spots in d and g; 70% of these spots colocalized with the ≈60 ACA spots (f and i). Arrows point to representative small spots and arrowheads point to large spots.
Localization of heterologous H3-like proteins to human centromeres was confirmed by examination of mitotic figures (Fig. 6). Overall, GFP signals were less intense at metaphase than at interphase, perhaps because of the more condensed state of mitotic chromosomes. Mitotic spreads revealed that the large, intense spots correspond to pericentric regions of a few specific chromosomes. ACA labeling is confined to the primary constriction, whereas large-spot GFP fluorescence is seen to extend into adjacent regions. This consistent pattern of pericentric localization suggests that the large spots are sites of human classical satellites, which show similar pericentric localization (36). Certain additional regions occasionally were seen to be labeled, including telomeres. We conclude that, in contrast to H3 itself, yeast and worm H3-like heterologs show preferential pericentric localization in both human and Drosophila cells.
Figure 6.
Colocalization of HCP-3-GFP with heterochromatin in human metaphase chromosomes. See legend to Fig. 5. 4′,6-Diamidino-2-phenylindole staining of DNA is shown in blue. Insets show two chromosomes with strong pericentric HCP-3-GFP signals (Left) and one chromosome with centromeric as well as telomeric HCP-3-GFP localization (Right).
Discussion
We have found that sequence features can predict whether a histone H3-like protein will be a centromere protein. A fly protein, Cid, that is distantly related to histone H3 but has a dissimilar N-terminal tail localized precisely to fly centromeres. In related work, C. elegans HCP-3, which also fits this profile, was found to be an essential component of holocentric centromeres (37). Identical pericentric localization in Drosophila cells was found for yeast, worm, and human H3-like proteins. Their localization behavior contrasts with that of histones H3 and H2B, which were deposited preferentially at euchromatic regions when driven by the cid promoter. Thus, when using the cid promoter, our cytological assay revealed three distinct patterns of localization: histones are deposited in euchromatin, native H3-like protein in centromeres and heterologous H3-like proteins from widely different organisms in heterochromatin. Deposition in heterochromatin appears to be a general feature of centromeric H3-like proteins in heterologous systems, because H3-like centromeric proteins from yeast and worms also localize to heterochromatic regions when they are constitutively expressed in human cells. This unexpected behavior of heterologs contradicts expectations based on specific recognition of centromere-specific or sequence-specific determinants. We conclude that localization to heterochromatin must be a general property of centromeric H3-like proteins.
Pericentric localization behavior is especially surprising given the divergence of these proteins from one another in the core region. Yeast and worm H3-like proteins are no more similar (in fact, marginally less similar) to native fly and mammalian centromeric H3-like proteins than they are to H3 itself (Fig. 1b). Yet, H3 displayed contrasting localization behavior when expressed identically to H3-like proteins in both fly and human cells. Therefore, preferential heterochromatic localization behavior in heterologous systems is not attributable to similarities that are shared with native centromere proteins and distinguish them from H3 itself.
Heterochromatin comprises ≈10–50% of the genomes of complex eukaryotes, and, yet, its function remains enigmatic, although the consistent presence of centromeres in heterochromatin suggests a mitotic role. However, deletion studies (7, 38) have not distinguished between a mitotic requirement for specific DNA or protein determinants vs. a requirement for heterochromatin per se. Our demonstration that in both Drosophila and human cells heterologous centromeric histones localize to heterochromatic regions suggests that the heterochromatic state directly facilitates the localization of centromere proteins.
In light of the identical heterochromatic localization of diverse heterologous H3-like proteins, the precise localization of the native proteins presents a paradox: what prevents Cid and CenpA from also localizing broadly to heterochromatin? Precise localization to centromeres occurs even when Cid-GFP is induced at high levels (Fig. 4c), so it seems unlikely that the failure to localize more broadly is due to limiting amounts of protein. It is conceivable that native H3-like proteins are actively prevented from depositing in heterochromatin; however, this hypothesis leaves the preferential deposition of heterologs unexplained.
This paradox is resolved if we suppose that both native and heterologous H3-like proteins deposit broadly to heterochromatin, but only native proteins come together to form a single, coherent structure that organizes a kinetochore. Such coming together of dispersed subunits was proposed previously for mammalian centromeres (39) and was thought to involve large-scale looping within pericentric heterochromatin not detectable in standard cytological preparations. Support for this model comes from the observation that in C. elegans, dispersed HCP-3 comes together at prophase to form a ribbon-like centromere (37). The subunit model is consistent with evidence that centromere competence may be a general feature of satellite sequences (40) and asserts that the mapping of centromeres to regions of a few hundred kilobases of satellite sequences within larger expanses of heterochromatin (6–8) simply maps the location of the highest concentration of centromere subunits. By this model, the machinery responsible for the coming together of subunits would discriminate between native and heterologous H3-like proteins, and the in situ assay described here may be used to probe the biochemical nature of this process.
Acknowledgments
We thank C. Sunkel, J. Rattner, and L. Moore for providing antibodies, Mark Roth for HeLa cells and C. elegans nucleic acids, Lenora Loo for Kc cells, and members of our lab for stimulating discussions and critical comments on the manuscript.
Abbreviations
- GFP
green fluorescent protein
- ACA
anticentromere antibody
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AAB37052).
References
- 1.Flemming W. Zellsubstanz, Kern und Zelltheilung. Leipzig, Germany: F. C. W. Vogel; 1882. [Google Scholar]
- 2.Wiens G R, Sorger P K. Cell. 1998;93:313–316. doi: 10.1016/s0092-8674(00)81157-5. [DOI] [PubMed] [Google Scholar]
- 3.Csink A K, Henikoff S. Trends Genet. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- 4.Willard H F. Curr Op Genet Dev. 1998;8:219–225. doi: 10.1016/s0959-437x(98)80144-5. [DOI] [PubMed] [Google Scholar]
- 5.Murphy T D, Karpen G H. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- 6.Brown K E, Barnett M A, Burgtorf C, Shaw P, Buckle V J, Brown W R. Hum Mol Genet. 1994;3:1227–1237. doi: 10.1093/hmg/3.8.1227. [DOI] [PubMed] [Google Scholar]
- 7.Murphy T D, Karpen G H. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaszas E, Birchler J A. Genetics. 1998;150:1683–1692. doi: 10.1093/genetics/150.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.du Sart D, Cancilla M R, Earle E, Mao J I, Saffery R, Tainton K M, Kalltsis P, Martyn J, Barry A E, Choo K H. Nat Gen. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 10.Depinet T W, Zackowski J L, Earnshaw W C, Kaffe S, Seknon G S, Stallard R, Sullivan B A, Vance G H, Van Dyke D L, Willard H F, et al. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- 11.Tyler-Smith C, Gimelli G, Giglio S, Floridia G, Pandya A, Terzoli G, Warburton P E, Earnshaw W C, Zuffardi O. Am J Hum Genet. 1999;64:1440–1444. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barry A E, Howman E V, Cancilla M R, Saffery R, Andy Choo K H. Hum Mol Genet. 1999;8:217–227. doi: 10.1093/hmg/8.2.217. [DOI] [PubMed] [Google Scholar]
- 13.Brown W, Tyler-Smith C. Trends Genet. 1995;11:337–339. doi: 10.1016/s0168-9525(00)89100-3. [DOI] [PubMed] [Google Scholar]
- 14.Dobie K W, Hari K L, Maggert K A, Karpen G H. Curr Op Genet Dev. 1999;9:206–217. doi: 10.1016/S0959-437X(99)80031-8. [DOI] [PubMed] [Google Scholar]
- 15.Karpen G H, Allshire R C. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 16.Choo K H. Nat Gen. 1998;18:3–4. doi: 10.1038/ng0198-3. [DOI] [PubMed] [Google Scholar]
- 17.Palmer D K, O'Day K, Wener M H, Andrews B S, Margolis R L. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer D K, O'Day K, Trong H L, Charbonneau H, Margolis R L. Proc Natl Acad Sci USA. 1991;88:3734–3748. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoler S, Keith K C, Curnick K E, Fitzgerald-Hayes M. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 20.Meluh P B, Yang P, Glowczewski L, Koshland D, Smith M M. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz J, Stemmann O, Rank S, Lechner J. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan K F, Hechenberger M, Masri K. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henikoff J G, Henikoff S, Pietrokovski S. Nucleic Acids Res. 1999;27:226–228. doi: 10.1093/nar/27.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siemering K R, Golbik R, Sever R, Haseloff J. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 27.Kanda T, Sullivan K F, Wahl G M. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 28.Henikoff S, Comai L. Genetics. 1998;149:307–318. doi: 10.1093/genetics/149.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherbas L, Moss R, Cherbas P. In: Methods in Cell Biology. Goldstein L S B, Fyrberg E A, editors. Vol. 44. San Diego: Academic; 1994. pp. 161–179. [DOI] [PubMed] [Google Scholar]
- 30.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 31.Logarinho E, Sunkel C E. J Cell Sci. 1998;111:2897–2909. doi: 10.1242/jcs.111.19.2897. [DOI] [PubMed] [Google Scholar]
- 32.Platero J S, Csink A K, Quintanilla A, Henikoff S. J Cell Biol. 1998;140:1297–1306. doi: 10.1083/jcb.140.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rattner J B, Mack G J, Fritzler M J. Mol Biol Rep. 1998;25:143–155. doi: 10.1023/a:1016523013819. [DOI] [PubMed] [Google Scholar]
- 34.Di Nocera P P, Dawid I B. Proc Natl Acad Sci USA. 1983;80:7095–7098. doi: 10.1073/pnas.80.23.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shelby R D, Vafa O, Sullivan K F. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almeida A, Kokalj-Vokac N, Lefrancois D, Viegas-Pequignot E, Jeanpeirre M, Dutrillaux B, Malfoy B. Hum Genet. 1993;91:538–546. doi: 10.1007/BF00205077. [DOI] [PubMed] [Google Scholar]
- 37.Buchwitz J G, Ahmad K, Moore L L, Roth M B, Henikoff S. Nature (London) 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 38.Wines D R, Henikoff S. Genetics. 1992;131:683–691. doi: 10.1093/genetics/131.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zinkowski R P, Meyne J, Brinkley B R. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platero J S, Ahmad K, Henikoff S. Mol Cell. 1999;4:995–1004. doi: 10.1016/s1097-2765(00)80228-2. [DOI] [PubMed] [Google Scholar]