Abstract
Hepatitis C virus (HCV) belongs to the Hepacivirus genus in the Flaviviridae family. Among the least known viral proteins in this family is the nonstructural protein NS4B, which has been suggested to be a part of the replication complex. Hydrophobicity plots indicate a common profile among the NS4B proteins from different members of the Flaviviridae family, suggesting a common function. In order to gain a deeper understanding of the nature of HCV NS4B, we have determined localization and topology of this protein by using recombinant HCV NS4B constructs. The protein localized to the endoplasmic reticulum (ER), but also induced a pattern of cytoplasmic foci positive for markers of the ER. Computer predictions of the membrane topology of NS4B suggested that it has four transmembrane segments. The N and C termini were anticipated to be localized in the cytoplasm, because they are processed by the cytoplasmic NS3 protein. By introducing glycosylation sites at various positions in HCV NS4B, we show that the C terminus is cytoplasmic and the loop around residue 161 is lumenal as predicted. Surprisingly, the N-terminal tail was translocated into the lumen in a considerable fraction of the NS4B molecules, most likely by a posttranslational process. Interestingly, NS4B proteins of the yellow fever and dengue viruses also have their N termini located in the ER lumen due to an N-terminal signal peptide not found in NS4B of HCV. A shared topology achieved in two different ways supports the notion of a common function for NS4B in Flaviviridae.
Hepatitis C virus (HCV) is a positive-stranded RNA virus and has been classified as a separate genus, Hepacivirus, in the Flaviviridae family, which also includes the genera Pestivirus (e.g., bovine viral diarrhea virus [BVDV]) and Flavivirus (e.g., Kunjin, dengue, and yellow fever viruses). The three genera in the Flaviviridae family all have a similar genetic organization, and the general natures of their polyproteins are similar, as indicated by the similarities of their hydrophobicity profiles (7, 8).
The genome of HCV encodes a long polyprotein of approximately 3,000 amino acids, which is subsequently processed co- and posttranslationally into at least 10 different proteins (C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The structural proteins Core, E1, and E2 are located in the N-terminal part of the polyprotein and are cut off by cellular proteases, as is p7, which has an unknown function. The NS2-NS3 junction is digested by the autoprotease consisting of NS2 and the N-terminal region of NS3. The other nonstructural proteins are cleaved by the NS3 serine protease together with its cofactor, NS4A. In addition to the two different protease activities, NS3 acts as a helicase and an NTPase. NS5B is the RNA-dependent RNA polymerase necessary for replication. No function has yet been assigned to either NS5A or NS4B. However, NS5A has been associated with differential sensitivity to interferon and is known to have different states of phosphorylation. (15).
Surprisingly little is known about HCV NS4B, least of all its function. The protein is hydrophobic and approximately 27 kDa in size. These features and its location in the genome are similar to those of the NS4Bs in other members in the Flaviviridae family (7, 8). However, the functions of NS4B in genera other than HCV are also unknown. NS4B has long been assumed to be a part of the replication complex of the Flaviviridae family, although this has never been clearly shown. Nonetheless, a single amino acid substitution in the HCV NS4B is incompatible with a substitution in NS5B that normally results in a more efficient replication. This indicates that NS4B at some point in the replication cycle has a function that is tightly coupled to that of the viral RNA polymerase (30).
NS4B is required for replication, both in HCV as well as in other members of the Flaviviridae family (Ralf Bartenschlager, personal communication). When using the BVDV replicon as a model system, both deletion of the entire NS4B and insertions in the sequence inhibited replication (13, 26). In neither case could addition of BVDV NS4B in trans restore replication. The same was evident when investigating in-frame deletions in the Kunjin virus (21).
There have been indications that NS4B may be part of a multiprotein complex. Lin et al., using coimmunoprecipitation, reported that HCV NS4A, NS3, and NS4B are associated when expressed in eucaryotic cells, with NS4A being necessary for the interaction between NS3 and NS4B (29). Additionally, NS4B has also been demonstrated to be chemically cross-linked to both NS3 and NS5A in BVDV, suggesting that they are associated (42).
The hyperphosphorylation of the HCV protein NS5A is dependent on NS4B as well as on NS3 and NS4A (23, 34). In these studies, neither of the proteins could be complemented in trans. Furthermore, Koch et al. showed that two different deletions in NS4B inhibited the hyperphosphorylation of NS5A, even though both deletions were designed not to interfere with either their predicted structure of NS4B or its processing (23).
HCV NS4B has also been shown to exhibit different kinds of activities in cell culture systems: transforming NIH 3T3 cells, activating NF-κB-associated signals, and suppressing translation (19, 20, 38). Moreover, a single mutation in NS4B of BVDV changed the phenotype of a cytopathic BVDV virus to a noncytopathic variant (42).
Current knowledge of NS4B is thus scarce and scattered. In order to improve our understanding of the HCV protein NS4B, in the present report, we have attempted to characterize its cellular location and molecular topology in detail. We find that when expressed in recombinant form, NS4B is mostly localized to endoplasmic reticulum (ER) membranes and even induces the formation of new ER-derived membrane structures, corroborating a recent report from Egger et al. (10). Furthermore, we demonstrate that it is an integral membrane protein in the ER. The C-terminal part of NS4B (residues 135 to 261) is composed of two transmembrane domains with the C-terminal tail localized in the cytoplasm. The topology of the N-terminal half of the protein cannot be unambiguously assigned from our data, but the N-terminal tail unexpectedly appears to be translocated to the lumenal side of the ER membrane.
MATERIALS AND METHODS
Plasmid constructs.
The infectious clone pCV/H77C, genotype 1a (a generous gift from Jens Bukh) (55), was used as a template for NS4B in all constructs described below.
The pNS4B-EGFP plasmid was made by amplifying NS4B cDNA by PCR with the primers NS4B-5 and NS4B-3B (Table 1), contributing the restriction sites HindIII and BamHI. After restriction endonuclease treatment and gel purification (Concert rapid gel extraction system; Gibco BRL) of the PCR product and the vector, pEGFP-N3 (Clontech), they were ligated and electrotransformed into Escherichia coli XL1-Blue. The resulting vector, pNS4B-EGFP, expresses the gene NS4B-EGFP under a cytomegalovirus promoter.
TABLE 1.
Sequences of the oligonucleotides used for mutagenesis and construction of different vectors
| Primer | Sequence (5′ → 3′)a |
|---|---|
| NS4B-5 | GAGTTCGATGAGATGAAGCTTATGTCTCAGCACTTACCGTACATC |
| NS4B-3B | GATGTCCCTTAGCCAGGATCCGGAGCATGGMGTGG |
| NS4B-5C | GAGTTCGATGAGATGGTACCTTCTCAGCACTTACCGTACATC |
| NS4B-3C | GTCCCAGATGTCCCTTAGCGCGGCCGCTTAGCATGGMGTGG |
| NS4B1a-5Nhe | CCAGGAGTTCGAGCTAGCCACCATGGGCTCTCAGCACTTACCGTACATCG |
| 3′-93-B-stop-Eco/AS | CCCACCCCCCCAAGAATTCTCCCTAGGATCCGCCGCCAGTGGTTAGTGGGCTGG |
| EC4 (sense) | AATTCTACACCCAGAAGCTGTCCGTGCCCGACGGCTTCAAGGTGTCCAACTCCT CCGCCCGGGGCTGGGTGATCCACCCCCTGGGCCTGCGGTCCG |
| EC4 (antisense) | AATTCGGACCGCAGGCCCAGGGGGTGGATCACCCAGCCCCGGGCGGAGGAGT TGGACACCTTGAAGCCGTCGGGCACGGACAGCTTCTGGGTGTAG |
| E33-Sb | CCGCGTCCCGCCATGCAGAATTCATCACCCCTGCTGTCCAGACC |
| E72-Sb | GCCTGTCAACGCTGCCTGGTGAATTCAACCCCGCCATTGC |
| E92-Sb | CCAGCCCACTAACCGAATTCCAAACCCTCCTCTTCAACATATTGG |
| E112-Sb | CCCAGCTCGCCGCCCCCGAATTCGGTGCCGCTACTGCC |
| E135-Sb | GCGTTGGACTGGGGGAATTCCTCGTGGACATTCTTGC |
| E161-Sb | GCATTCAAGATCATGAGCGGTGAATTCCCCTCCACGGAGGACCTGG |
| E210-Sb | GGATGAACCGGCTAATAGAATTCGCCTCCCGGGGGAACC |
| T7-NS4B 1a-f | TTATACGACTCACTATAGGGAGCCACCATGTCTCAGCACTTACCGTACATC |
| NS4B 1a-r | CTAGACTCGAGCGCTAGCTTAGCATGGCGTGG |
| T7-NSS-NS4B | TTATACGACTCACTATAGGGAGCCACCATGTCTCAGAACTCATCGTACATCGAG CCAGGGATG |
| pcDNA3-seqAS | GGCTGGCAACTAGAAGGCAC |
| EGFP-AS | CCCTCTAGACTCGAGCGGCCGCTTTACTTG |
Endonuclease restriction sites are underlined.
The mutation primers used to make the glycosylation mutants are only presented in the sense direction.
To make pHM-NS4B, the primers NS4B-5C and NS4B-3C (Table 1) were used for amplification; they included the restriction sites KpnI and NotI. After restriction endonuclease treatment and purification of the PCR product and the vector pcDNA4/HisMax A (Invitrogen), they were ligated and transformed into E. coli XL1-Blue.
To make p93-EGFP, the N-terminal part of NS4B was amplified with the primers NS4B1a-5Nhe and 3′-93-B-stop-Eco/AS (Table 1), contributing the restriction sites NheI to the 5′ end and BamHI and EcoRI to the 3′ end. After restriction endonuclease treatment with NheI and EcoRI and purification of the PCR product and vector, pcDNA3.1/zeo(+) (Invitrogen), they were ligated and amplified as described above. After sequencing the construct, this plasmid and pNS4B-EGFP were digested with BamHI and NotI. This linearized the plasmid and released the fragment of enhanced green fluorescent protein (EGFP). Both were purified in agarose gels, ligated, and amplified in E. coli XL1-Blue.
The seven constructs used for the glycosylation studies were all made in the same way, with the pHM-NS4B construct as a template. An EcoRI site was introduced by mutation at the site of interest by using the Stratagene QuickChange site-directed mutagenesis kit (for primers, see Table 1). The mutated plasmids were cut with EcoRI, purified in agarose gels, and allowed to ligate to a double-stranded oligonucleotide, EC4. The EC4 oligonucleotide contained the sequence of the EC4 loop (40), as well as the sequence of a cut EcoRI site at each end (Table 1). After ligation and subsequent amplification, the new plasmid preparations were sequenced to find clones in which EC4 had been inserted in the right direction.
All plasmid constructs have been sequenced at modified regions and have been determined to be as designed.
Cells, transfection, cell lysis, and Western blotting.
Cells were grown in Dulbecco's modified Eagle medium (Hep3B, HeLa, and Huh7 cells) or RPMI (CHO cells) supplemented with 10% fetal calf serum and 1% PEST (100 U of penicillin per ml, 100 μg of streptomycin per ml) (all reagents from Invitrogen).
Cells were transfected with Fugene 6 (Roche) and the appropriate amount of plasmid according to the manufacturer's instructions.
Cell lysis and phase separation with Triton X-114 were done as described by Bordier (4). Briefly, cells were lysed with 1% Triton X-114, 150 mM NaCl, and 10 mM Tris-HCl (pH 8.0) at 0°C. Nuclei were removed from the lysates by spinning at 2,500 × g for 5 min. Separation of the proteins was made on a cushion of 6% sucrose, 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.06% Triton X-114 at 30°C, after which the lysates were centrifuged at 300 × g. The aqueous phase was removed from the detergent phase and “washed” two times by addition of undiluted Triton X-114 and separation as described above. The aqueous and detergent phases, as well an input sample taken before separation, were analyzed by Western blotting.
Cell lysates were run in 4 to 12% NuPAGE gels with MES [2-(N-morpholino) ethane sulfonic acid] buffer (both from Invitrogen). Proteins were electroblotted onto a nitrocellulose membrane, which was blocked in 5% milk at 4°C overnight. The membrane was incubated with primary antibody for 2 h at room temperature, washed in phosphate-buffered saline with 0.05% Tween 20 (PBS-T) three times for 5 min, and incubated with alkaline phosphatase (AP)-conjugated secondary antibody for 1 h at room temperature. After the membrane had been washed as described above, enzyme activity was detected with BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium (NBT) (both from Sigma). The antibodies used were Living Colors peptide antibody (Clontech), anti-GFP (Abcam), anti-Xpress (Invitrogen), and AP-conjugated goat anti-rabbit and goat anti-mouse (both Pierce). All antibodies were diluted in 5% milk in PBS-T.
Indirect immunofluorescence.
Cells were grown on coverslips, transfected as described above, and fixed at the times indicated in the text. If immunostaining was performed, they were also permeabilized. When cells were treated with cycloheximide (Sigma), it was added to cells to a final concentration of 1.6 mM for 4 h before fixation.
For detection of PDI, EEA1, ERGIC, and the Xpress epitope, cells were fixed with 4% formaldehyde in PBS for 10 min and permeabilized with 1% Triton X-100 in PBS for 5 min. For detection of calnexin, cells were fixed and permeabilized with methanol-acetone (1:1) for 2 min. Immunostaining with the primary antibody against these epitopes was made at room temperature for 1 h, after which the cells were washed three times for 5 min with PBS. Secondary antibody incubation was performed at room temperature for 30 to 45 min, after which the cells were washed as described above. All antibodies were diluted in 3% bovine serum albumin (BSA), and cells were subsequently mounted in Mowiol (Calbiochem).
To detect p58 (anti-Golgi), cells were fixed in 4% formaldehyde in PBS for 20 min at 37°C and washed twice in PBS. After this, they were permeabilized, reduced, and denatured for 30 min at 37°C in PBS buffer containing 0.5% sodium dodecyl sulfate (SDS), 5% β-mercaptoethanol, and 10% fetal calf serum (FCS), and then cells were washed five times with PBS containing 4% FCS and 0.1% Triton X-100 (PFT buffer). Immunostaining and mounting were conducted as described above, with the exception that all dilutions and washes were made with PFT buffer. The method was adapted from that of Blum et al. (3).
The following antibodies were used: anti-PDI (Affinity Bioreagents), anti-EEA1 (BD biosciences), anti-Golgi 58 K (Sigma), and anti-Xpress (Invitrogen). Rabbit anti-ERGIC and rabbit anticalnexin antibodies were generous gifts from Ralf Pettersson (Ludwig Institute for Cancer Research, Stockholm Branch). The secondary antibodies anti-mouse immunoglobulin G (IgG) and anti-rabbit IgG were both raised in goats and conjugated with tetramethyl rhodamine isothiocyanate (TRITC) (Sigma).
Immunostained samples were analyzed by fluorescence microscopy (Nikon Eclipse E800). Photographs were taken using an Optronics Engineering DEI-750 video camera and Easy Image software (Bergström Instrument AB, Solna, Sweden). The images were merged with Adobe Photoshop 5.0 (Adobe).
In vitro transcription and translation.
Transcription was made with PCR products as templates, except for Lep, which was transcribed directly from the plasmid. The PCR amplification was done with either pHM-NS4B, one of the seven glycosylation constructs described above, pNS4B-EGFP, or p93-EGFP as a template. The appropriate primer combinations were used, with T7-NS4B1a-f or T7-NSS-NS4B as the forward primer and NS4B1a-r, pcDNA3-SeqAS, or EGFP-AS as the reverse primer (Table 1). PCR products were purified in agarose gels before transcription. The transcription mixtures contained the following: 0.5 to 1.0 μg of PCR product, 1 × T3/T7 buffer (Invitrogen), consisting of 0.04 mM Tris-HCl (pH 8.0), 8 mM MgCl2, 2 mM spermidine-(HCl)3, and 25 mM NaCl; 5 mM dithiothreitol (DTT); 1 mM (each) ATP, CTP, UTP, and GTP (Invitrogen); 50 U of RNasin (Promega); and 100 U of T7 polymerase (Invitrogen). Reaction mixtures were incubated at 37°C for 1 h. When Lep was transcribed, 1.8 μg of plasmid was used as a template, and 30 U of SP6 polymerase (Invitrogen) was used with the buffer provided by the manufacturer. Translation was performed in 13- or 14-μl reaction mixtures for 1 h at 30°C. The translation mixture contained the following: 1 μl of amino acid mix (without methionine), 1 μl of RNasin, 1 μl of [35S]methionine (Perkin-Elmer), 1 μl of mRNA, 0 or 1 μl of canine pancreas microsomes, and 9 μl of rabbit reticulocyte lysate (all from Promega except where indicated). The acceptor peptide, benzoyl-Asn-Leu-Thr-methylamide, was obtained from Quality Controlled Biochemicals (Hopkinton, Mass.) and was added to reaction mixtures to a final concentration of 0.4 mM. Since the peptide was dissolved in dimethyl sulfoxide (DMSO), the same volume of DMSO was added to the reaction mixtures containing only microsomal membranes and no acceptor peptide. This was done in order to ensure that the acceptor peptide, and not DMSO, inhibited the glycosylation of the protein.
Sodium carbonate washing of microsomes was done by adding 90 μl of 0.1 M Na2CO3 to a 10-μl translation mixture. This mixture was layered on top of a 50-μl sucrose cushion (0.2 M sucrose, 0.1 M Na2CO3) and centrifuged at 4°C for 15 min at 70,000 rpm in a TLA100 rotor (Beckman Instruments) to spin down the membranes. For the controls containing Triton X-100, this detergent was added to both solutions indicated above to a final concentration of 0.5% before centrifugation. The supernatant fraction was mixed with 15 μl of 100% trichloroacetic acid (TCA), incubated for 30 min on ice to precipitate proteins, and subsequently centrifuged at 20,800 × g. The pellet was washed with 100% acetone. Membrane pellet and pellet from precipitated supernatant were both solubilized with loading buffer, and equal volumes were loaded for SDS-polyacrylamide gel electrophoresis (PAGE) as described below.
Kinetics assay.
In vitro translation reactions were made in 4× the volume described above. After 1.5 min at 30°C, 1.2 μl of 3.5 mM aurintricarboxylic acid (ATA) (Sigma) was added to the reaction to inhibit translation initiation. Samples of 6 μl were taken from the translation mix at the time points indicated and moved to tubes containing 0.3 μl of 20% Triton X-100 to disrupt microsomes. The aliquots were reincubated at 30°C for a total of 60 min.
Analysis of in vitro translations.
Translation products were analyzed by SDS-PAGE with 12% NuPAGE gels with MOPS buffer [3-(N-morpholino)propane sulfonic acid] (both from Invitrogen), and gels were quantified with a Fuji BAS 1000 phosphoimager by using Image Gauge V3.45 software (Fuji Film Science Lab 99/2001). The glycosylation efficiency of a given mutant was calculated as the quotient between the intensity of the glycosylated band divided by the summed intensities of the glycosylated and nonglycosylated bands.
RESULTS
Localization in transfected cells.
As a first step in the characterization of NS4B, its cellular localization was determined with a recombinant NS4B with EGFP fused to the C terminal. Both the original EGFP plasmid and the NS4B-EGFP construct were transfected into different mammalian cell lines.
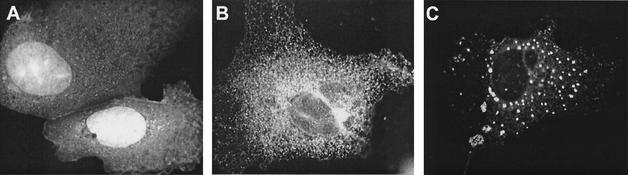
When EGFP was expressed alone, it was distributed throughout the entire cell in both the cytoplasm and the nucleus. However, when fused to NS4B, the pattern of EGFP localization changed dramatically. After 17 h of expression, the NS4B-EGFP fusion protein exhibited a distinct reticular pattern throughout the cytoplasm, which was somewhat more concentrated in the perinuclear region. Some cells also showed a pattern of small, but intense cytoplasmic foci as well as the reticular pattern (Fig. 1).
FIG. 1.
NS4B-EGFP exhibits different kinds of patterns when expressed in cells. Hep3B cells were transfected with pEGFP (A) or pNS4B-EGFP (B and C). The different patterns of NS4B can be seen in panel B, showing the typical reticular ER pattern, and panel C, exhibiting the cytoplasmic foci on top of the ER pattern.
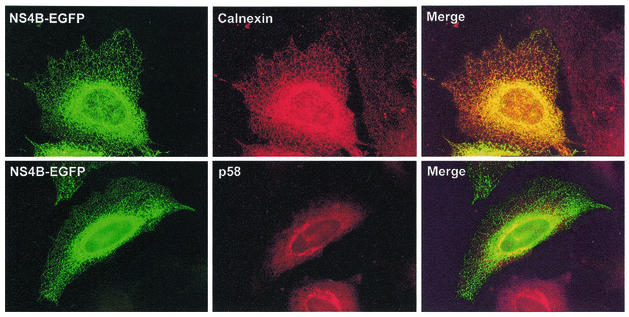
By immunostaining the transfected cells for specific cellular proteins, the precise localization of NS4B could be determined. Antibodies against both protein disulfide-isomerase (PDI; a soluble protein resident in the ER) (data not included) and calnexin (a membrane-bound protein resident in the ER), exhibited perfect colocalization with the reticular pattern described above (Fig. 2). The perinuclear staining of NS4B overlapped with both the ER markers (calnexin and PDI) and the Golgi marker (p58). The overlapping of NS4B and p58 could be either because of a true colocalization or the result of the ER containing NS4B being located under or above the Golgi apparatus, giving the impression of colocalization.
FIG. 2.
NS4B-EGFP colocalizes with the ER. Cells were transfected with pNS4B-EGFP and immunostained against ER (calnexin) or the Golgi apparatus (p58) 17 h posttransfection. Panels to the left depict NS4B-EGFP, and panels in the middle depict the immunostaining of the different proteins indicated in the separate pictures. Merged pictures are presented to the right. The cells stained for calnexin are Hep3B cells, and those stained for p58 are HeLa cells.
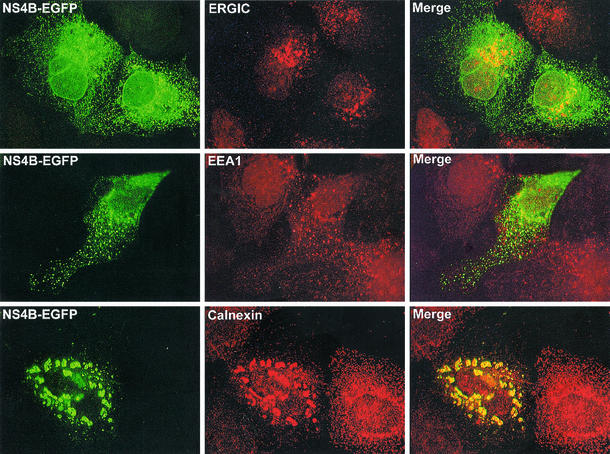
The cytoplasmic foci visible in some cells were not part of an expected ER pattern. To characterize the cellular origins of these structures, they were compared to the immunostaining patterns of different cell markers. The foci apparent with NS4B-EGFP were increased in cells that had expressed the protein for a longer time. Accordingly, cells were transfected and allowed to express the protein for 48 instead of 17 h. At this point, not only were many cells with intense foci in the cytoplasm evident, but there were also many cells with larger foci that were mainly assembled around the nucleus. The larger foci gave the impression of being aggregates of the smaller ones. Cells were immunostained for the intermediate compartment (ERGIC), early endosomes (EEA1), and ER (calnexin) and compared for colocalization. Only calnexin showed full colocalization with NS4B-EGFP in the focal structures—both with the smaller foci in the cytoplasm and the larger aggregates around the nucleus (Fig. 3, bottom panels). The pattern of ERGIC looked different from that of NS4B-EGFP, but there might have been a partial colocalization as well, especially close to the nucleus (Fig. 3, top panels). No colocalization was evident with the marker against early endosomes (Fig. 3, middle panels).
FIG. 3.
The cytoplasmic foci induced by NS4B-EGFP colocalize with the ER marker. Cells were transfected with pNS4B-EGFP and immunostained for ERGIC, EEA1, or calnexin 48 h posttransfection. Panels to the left represent NS4B-EGFP, and panels in the middle represent the immunostaining of the different proteins indicated in the separate pictures. Merged pictures are shown to the right. All cells are Hep3B cells. The cells stained for calnexin clearly show the larger aggregates visible in some cells.
To make sure that the fusion of EGFP to the NS4B C-terminal end did not alter its true localization, another construct was also made. This construct, pHM-NS4B, contained the same NS4B sequence, but instead of having EGFP in the C-terminal end, it carried an Xpress epitope (as well as a His tag and a linker: total length, 33 amino acids) in its N-terminal end. Cells transfected with this construct and immunostained against the Xpress epitope gave the same pattern as that described above, having both the network and the cytoplasmic foci (data not included). The same pattern of NS4B was recorded in all cell lines tested (Hep 3B, HeLa, CHO, and Huh-7), as well as after treating the cells with cycloheximide before fixation. Hence NS4B was retained in the ER, and this distribution was not cell type specific.
The foci and aggregates described above were only found in cells expressing NS4B, both when in fusion with EGFP and when having the N-terminal Xpress tag. To make certain that these patterns were not artifacts of overexpression of proteins in the ER membrane, cells were also made to express another ER-localized, membrane-bound protein under the same conditions. This construct contained a signal sequence (from antibody light chain) to locate it to the ER, a heterologous lumenal part (Tim protein), a transmembrane domain (from B71 protein), and EGFP expressed in the cytoplasm (D. Johansson and M. A. A Persson, unpublished observations). This control construct was expressed under the same promoter as the NS4B constructs and had an ER distribution, but without any of the foci apparent with NS4B (data not included).
Integration into the ER membrane.
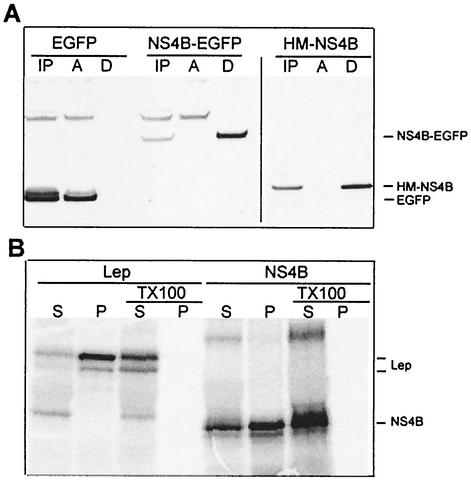
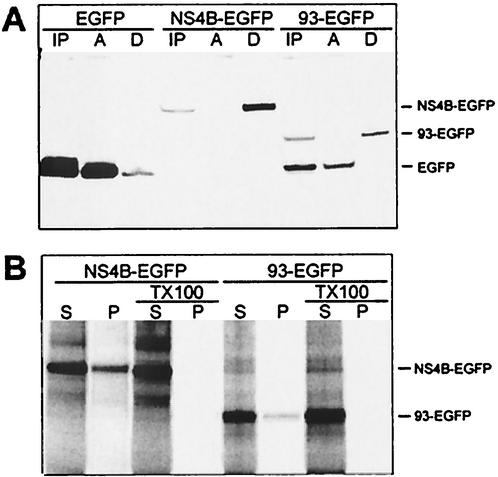
To study how NS4B is associated with the ER, a method based on phase separation with Triton X-114 was used. With this method, only membrane proteins will be present in the detergent phase after separation (4). HeLa cells were transfected with either pEGFP, pNS4B-EGFP, or pHM-NS4B and lysed after 40 h of expression, and the cell lysates were separated. When the samples were analyzed by Western blotting, EGFP was detected in the aqueous phase as expected. When fused to NS4B, however, it was only detected in the detergent phase, as was HM-NS4B (Fig. 4A). This indicated that NS4B is closely associated with the ER membrane. The result was also confirmed with sodium carbonate wash of in vitro-translated NS4B in the presence of microsomal membranes, indicating that it is an integral membrane protein (Fig. 4B). These results show concordance with those of previous studies (14, 16, 50). The sodium carbonate wash data indicated that NS4B integrated less efficiently in the microsomal membranes than our control protein, Lep (a known integral protein) (17), despite the clear ER localization of NS4B shown above by microscopy. Whether the slightly different results for NS4B and Lep from the sodium carbonate wash are a methodological artifact due to differences in behavior from in vivo and in vitro expression or have some other basis remains to be studied. When the microsomes were disrupted with Triton X-100, however, all proteins were present in the supernatant.
FIG. 4.
Separation of membrane-bound proteins. (A) Phase separation of cell lysates. HeLa cells were transfected with pEGFP, pNS4B-EGFP, or pHM-NS4B and lysed with Triton X-114. Phases were separated and analyzed by Western blotting with anti-EGFP or anti-Xpress as the detection antibody. Samples from before separation (IP), from the aqueous phase (A), and from the detergent phase (D) were loaded into the gel (approximately 1/10 of each preparation was loaded). (B) Sodium carbonate wash of the in vitro-translated proteins Lep and NS4B. Each protein was translated in two different reactions. Both were subjected to sodium carbonate wash, in which the wash solutions for one of also contained 0.5% Triton X-100 to disrupt the microsomal membranes. Equal volumes of supernatant (S) and membrane pellet (P) were loaded on the gel. Lep is apparent as two bands; the upper band is the glycosylated version of the lower band. NS4B has a strong main band and a weak lower band. This is due to an alternative start site in the protein sequence. Residues 11 and 12 are both methionines, giving this effect. In constructs lacking a methionine prior to the NS4B sequence, this lower band is the only one expressed (data not included).
Prediction of the topology of NS4B in the membrane.
To investigate the topology of NS4B in the ER membrane, a theoretical model of the protein was first made. It was based on the sequence of NS4B genotype 1a (pCV/H77C), which was analyzed with the programs TMHMM (24), HMMTOP (51), and MEMSAT (18). Since the NS3 protease, which is responsible for processing the polyprotein in the nonstructural region, is located on the cytoplasmic side of the ER membrane (1, 12, 14, 53, 54), we further assumed that both the N terminal and the C terminal of the protein were cytoplasmic.
All three programs predicted that NS4B has four transmembrane regions and that the N and C termini are located in the cytosol. TMHMM and HMMTOP also agreed in the placement of all four transmembrane domains, whereas MEMSAT indicated another location of the first transmembrane domain. Only the third and the fourth of the predicted transmembrane regions were clearly separated from each other by a loop (Fig. 5A). According to the assumptions described above, this loop should be located in the lumen of the ER. The N-terminal half of the protein was more difficult to interpret, however, as was indicated by the different results from the three prediction programs. Furthermore, recent studies have indicated that the reliability of a TMHMM prediction in a given region of a protein can be gauged by the value of the probability scores in the TMHMM analysis (Melén and von Heijne, unpublished data). The topology predicted for NS4B is thus more uncertain for the N-terminal half than for the C-terminal half of the protein.
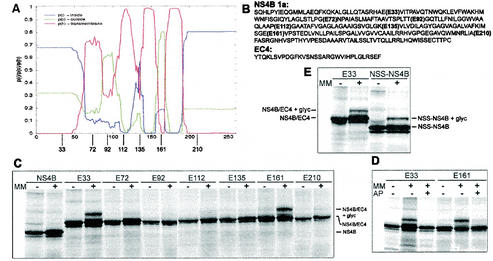
FIG. 5.
TMHMM analysis of NS4B genotype 1a and glycosylation patterns of mutants derived from the same sequence (A) Graphical output of the TMHMM analysis, with the 261 amino acid residues of NS4B indicated on the x axis. The red line describes the probability of that part of the protein to be a transmembrane domain, the blue line describes the probability of it being inside the membrane (cytoplasmic), and the green line describes the probability of it being outside the membrane (lumenal). The approximate sites of the EC4 insertions are depicted below the graph. (B) Amino acid sequences of NS4B genotype 1a and the EC4 loop. The different insertion sites of the EC4 loops in the NS4B sequence are indicated. (C to E) Glycosylation patterns of different constructs that had been in vitro transcribed and translated in the absence or presence of microsomal membranes (MM). The identities of the bands are indicated. An explanation for the weak bands below the main bands is given in the legend to Fig. 4. (C) NS4B and the seven glycosylation mutants containing the EC4 loop. (D) E33 and E161 translated in the presence of an acceptor peptide (AP) that competes for the glycosylation. (E) Control of the N-tail glycosylation. An NSS sequence was put directly in the N-terminal tail of the original NS4B sequence, changing the amino acids HLP to NSS.
Topology inferred from the pattern of glycosylation in vitro.
To experimentally evaluate the predicted model, we chose to investigate the glycosylation pattern of mutants of NS4B, into which glycosylation sites were introduced at different sites. Glycosylation of such sites would indicate their lumenal localization, and thus the location of that particular region of NS4B.
Seven different mutants were made, each with a 30-residue-long insert containing a glycosylation motif in the middle (Fig. 5B). The long insert was necessary, since the putative loops in NS4B may otherwise be too short to be glycosylated; a minimum distance of 12 to 14 amino acids from the membrane to the acceptor Asn is necessary for glycosylation to occur (35). The amino acid sequence of the insert, including the NSS glycosylation site, was taken from the lumenal EC4 loop in Band 3, the anion exchanger of human erythrocytes (40).
The sites for insertion of the EC4 loop were chosen based on the predicted topology. The mutants E92, E135, and E161 had their sites chosen based on the hydrophobicity profile, with an expected lumenal position of E161 and possibly also E92 (Fig. 5A). (The “E” indicates the EcoRI site that was made in the sequence and used for inserting the EC4 loop, and the number indicates the number of amino acids preceding this site in the NS4B sequence.) E72 and E112 were chosen to test whether the corresponding hydrophobic regions might form “helical hairpins” with tight lumenal turns defined by the helix-breaking motifs PGNP and PG, respectively (32). Finally, in mutants E33 and E210, the EC4 loop was placed in the N- and C-terminal tails, respectively.
The mutants were subjected to in vitro transcription and in vitro translation in the presence or absence of microsomal membranes. Only mutants with their glycosylation site in the lumen of the ER will be glycosylated when translated in the presence of microsomes, and glycosylation can be detected as an ∼2-kDa shift in SDS-PAGE (Fig. 5C).
The C-terminal mutant E210 was not glycosylated and therefore was in the cytosol, as expected. E161 was glycosylated and therefore was in the ER lumen, confirming the presence of the last transmembrane segment. The only other mutant that was glycosylated was, to our surprise, the N-terminal mutant E33, although the level of glycosylation was somewhat lower than that for E161 (Fig. 5C). Glycosylation of the EC4 loop in E33 and E161 was completely abolished in the presence of a competitive acceptor peptide, confirming that glycosylation was indeed the cause of the noted increase in molecular weight (Fig. 5D). Because the glycosylation of E33 was unexpected, we further demonstrated the lumenal localization of the N-terminal tail by testing a construct that had only an NSS glycosylation acceptor sequence in the N-terminal tail instead of the long EC4 insert (Fig. 5E). This construct (NSS-NS4B) was glycosylated to approximately the same extent as the E33 construct. All seven mutants with the EC4 loop inserted in the sequence had been successfully integrated in the microsomal membranes, as was apparent by sodium carbonate wash (data not included).
Kinetics of glycosylation.
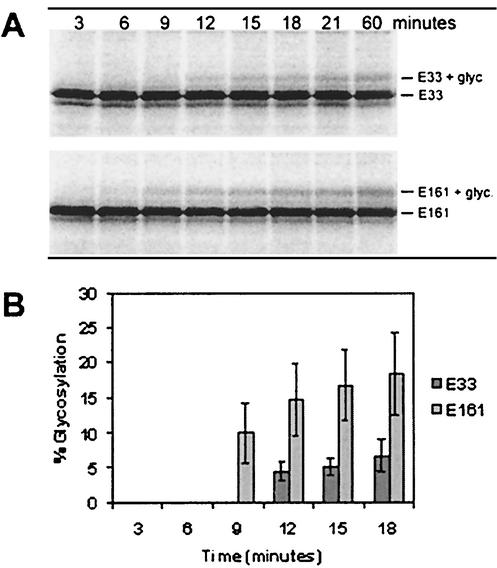
The time of glycosylation of the different sites of the protein was then investigated by testing the two glycosylated mutants, E33 and E161, in a kinetic assay (45). In this assay, the translation is synchronized by the addition of ATA, which at low concentrations inhibits new initiation of translation (by inhibiting the interaction between the mRNA and the ribosome). By adding ATA after the first 1.5 min of translation, it was possible to track the glycosylation of the peptide chains, the synthesis of which has been initiated during this time span. Detergent was added to aliquots taken at different time points from the translation mix. The addition of detergent disrupts the microsomal membranes and thereby inhibits any further glycosylation of proteins in the aliquot. E161 was clearly glycosylated after 9 min, as expected from combining the average in vitro translation rate (0.4 residues per s) with the anticipated time for the glycosylation motif to reach proper localization. Remarkably, E33 was only glycosylated after 12 min, even though the glycosylation site of E33 is located almost 130 residues upstream of the E161 site (Fig. 6).
FIG. 6.
Time of glycosylation of E33 and E161. (A) SDS-PAGE of aliquots taken during translation at the time points indicated. (B) Percentage of glycosylation of the aliquots, given as an average of three different experiments. Standard deviation is indicated by bars.
The N-terminal 93 amino acids are not sufficient for membrane integration.
During this study, it became evident that the prediction for the N-terminal half of NS4B was less reliable than that for the other half. Together with the unexpected finding of glycosylation of the E33 position, these results prompted us to try to determine where the first transmembrane domain of NS4B is located. Since both TMHMM and HMMTOP predicted that the first transmembrane domain is located upstream of the 93rd amino acid, we decided to test whether this part of the protein could be integrated into the ER membrane even without the downstream portions of NS4B. The first 93 amino acids of NS4B turned out to be very difficult to express, both in cells and in vitro. However, when its C-terminal end was fused to EGFP, it could readily be expressed (resulting in the construct 93-EGFP). HeLa cells were transfected with 93-EGFP, allowed to express for 24 h, and lysed with Triton X-114. The lysates were separated into aqueous and detergent phases as described above and subsequently were analyzed by Western blotting. 93-EGFP partitioned into the detergent phase, as did the full-length NS4B-EGFP (Fig. 7A). In contrast to the full-length protein, almost all of the in vitro-translated 93-EGFP was detected in the supernatant after sodium carbonate wash of the membranes (i.e., it was not integrated in the membrane) (Fig. 7B). Sodium carbonate wash of microsomes turns the microsomes to membrane sheets, leaving only proteins that were successfully integrated in the membrane in the pellet fraction. In contrast, the method based on lysis with Triton X-114 separates proteins based on their hydrophobicity and is not able to distinguish between membrane proteins that are correctly inserted in the membrane and those that are not. Our interpretation of these results is that the first 93 amino acids of NS4B may well associate with membranes, but that the protein is not properly integrated during translation; the implications of this are further discussed below. The sodium carbonate wash experiments indicated a varied degree of membrane insertion, even for the proteins evident by microscopy to clearly reside in the ER (i.e., NS4B and NS4B-EGFP) (Fig. 4B and 7B). This may indicate that the EGFP portion, for unknown reasons, influences the proportions of the molecule that will partition in the pellet and in the supernatant, respectively. There is still, however, a major difference between the results of NS4B-EGFP and 93-EGFP, reflecting their distinct localization apparent by microscopy.
FIG. 7.
Membrane association of 93-EGFP. (A) Phase separation of cell lysates in Triton X-114. HeLa cells were transfected with pEGFP, pNS4B-EGFP, or p93-EGFP and lysed with the detergent Triton X-114. Phases were separated and analyzed by Western blotting with anti-EGFP as detection antibody. Samples from before separation (IP), from the aqueous phase (A), and from the detergent phase (D) were loaded into the gel (approximately 1/10 of each preparation was loaded). When translating 93-EGFP, the cells use two different start sites: at the beginning of the 93-EGFP gene as well as the original start site at the beginning of EGFP. This effect has been seen in some other fusion proteins with EGFP used in our laboratory and can be abolished by destroying the second start site (D. Johansson, personal communication). This explains the presence of soluble EGFP in the samples from cells transfected with p93-EGFP. (B) Sodium carbonate wash of in vitro-translated NS4B-EGFP and 93-EGFP. Each protein was translated in two reactions. Both were subjected to sodium carbonate wash, in which the wash solutions for one of them also contained 0.5% Triton X-100 to disrupt the microsomal membranes. Equal volumes of supernatant (S) and membrane pellet (P) were loaded in the gel. The identity of the bands is indicated.
DISCUSSION
Localization and membrane association of NS4B in eucaryotic cells.
It was reported in earlier studies that the HCV protein NS4B tends to localize to the ER, with a small fraction possibly localizing to the Golgi when expressed in eucaryotic cells. This is the case when the protein is expressed both in the context of the other nonstructural proteins and when expressed alone (16, 22, 47). Our results confirm these findings, but we also observe new membrane structures visible by immunofluorescence in cells expressing NS4B. Besides the typical ER pattern evident in most cells, we find very intense foci that tend to be both larger and more common the longer the cells are allowed to express the recombinant protein. By immunostaining cells expressing these structures, we determined that they are positive for ER markers. Cytoplasmic foci induced specifically by HCV NS4B and seen with immunofluorescence have only been reported once before. As in our study, Kim et al. used recombinant NS4B fused to EGFP (22). The intense foci are not likely to be a result of the EGFP fusion, however, since we observed the same phenomena by using another construct having a much smaller epitope fused to the N terminus of NS4B. Thus, rearrangement of intracellular membranes seems to be an intrinsic property of NS4B, although we cannot rule out the possibility that the phenomenon is restricted to cells overexpressing the protein or that expression of the full-length HCV polyprotein would have given different results. We note that in many cases, positive-stranded RNA viruses induce new membrane structures, and these are often used as replication centers or places of viral assembly (2, 31, 39, 46, 52). Furthermore, there have been several reports concerning membrane rearrangements in cells expressing HCV proteins, but not until recently did Egger et al. pinpoint the protein(s) responsible for this effect (10, 33, 48, 49). By expressing the different HCV proteins in eucaryotic cells, they demonstrated that NS4B induced vesicles in a membranous matrix. This structure was called a “membranous web,” and all the other viral proteins investigated localized to these sites. A similar web-like structure was observed in the liver of an HCV-infected chimpanzee. Egger et al. used electron microscopy to detect the structural changes in cells expressing HCV NS4B, and our results indicate that membrane changes induced by this protein can also be detected by light microscopy.
Neither in HCV (a hepacivirus), Kunjin virus (a flavivirus), nor BVDV (a pestivirus) can NS4B be complemented in trans to rescue replication (13, 21, 26; R. Bartenschlager, personal communication), and it is likely that HCV, like other members of the Flaviviridae family, uses the self-induced membrane structures for replication, even though this has not been confirmed yet. The observation that NS4B seems to induce membrane alterations in cells may be the event tying these two facts together. In poliovirus, for example, the virus needs a coupled process involving viral translation, membrane modification, and viral RNA synthesis in order to assemble a functional replication complex (9). This may partly explain the need for NS4B to be translated in cis in order to allow replication.
Topology of NS4B in the ER membrane.
Computer analyses predicted four transmembrane domains in the NS4B protein from genotype 1a, with both the C- and the N-terminal tails located in the cytoplasm (Fig. 5A). Our experimental data are only partially consistent with this predicted topology. Only two out of seven engineered glycosylation sites were glycosylated, E33 and E161, indicating a lumenal orientation. The glycosylation of E161 and the lack of glycosylation of E135 and E210 agreed with the predicted topology. The glycosylation of the E33 construct (and the related NSS-NS4B construct) was more surprising, since both the predicted topology and the knowledge that the N terminus of NS4B is cleaved by the cytoplasmic NS3 protease indicate that the N-terminal tail should be located in the cytoplasm.
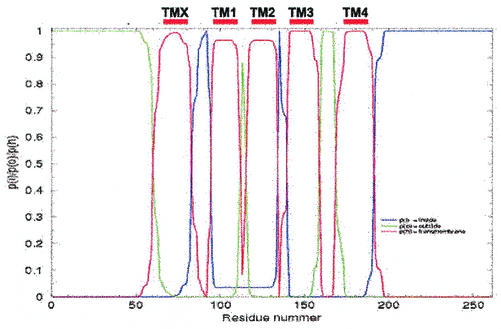
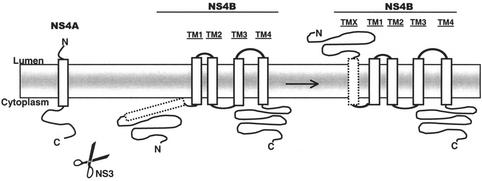
The results from the E33, E92, E135, E161, and E210 constructs were used as constraints in a new version of the TMHMM predictor that allows experimental information to be used as input to the program (K. Melén, A. Krogh, and G. von Heijne, unpublished data). With these constraints, NS4B is predicted to have five transmembrane segments with the N terminus in the lumen and the C terminus in the cytoplasm (Fig. 8 [please note our designations TMX and TM1 to -4]). As is apparent in Fig. 8, there is also a weak prediction for the part encompassing TM1 and TM2 to be in the cytoplasm (the calculated probability for cytoplasmic location between residues 95 to 135 is ∼0.05), which would result in an alternative topology with only three transmembrane segments. The latter possibility is consistent with the lack of glycosylation of the E72 and E112 constructs; however, E72 is placed in the middle of predicted transmembrane segment TMX and may thus yield artifactual results. E112 is placed in a very tight loop between predicted transmembrane segments TM1 and TM2. The insertion of the 30-residue-long EC4 segment at this position could easily disrupt critical interactions between the two helices TM1 and TM2 that may be required for proper membrane insertion (37). We thus favor the topology with five transmembrane segments (Fig. 9).
FIG. 8.
TMHMM analysis of NS4B with the prediction constrained by the glycosylation results for constructs E33 (lumenal), E92 (cytoplasmic), E135 (cytoplasmic), E161 (lumenal), and E210 (cytoplasmic). The 261 amino acid residues of NS4B are indicated on the x axis. The red line describes the probability of that part of the protein to be a transmembrane domain, the blue line describes the probability of it being inside the membrane (cytoplasmic), and the green line describes the probability of its being outside the membrane (lumenal). Our designations of the different transmembrane domains are presented above the figure (TMX and TM1 to -4).
FIG. 9.
Our current working model of the topology of HCV NS4B. The protease (NS3 with its cofactor NS4A) residing in the cytoplasm cuts the preprotein between NS4A and NS4B. After processing, a rearrangement of the NS4B occurs, giving it a fifth transmembrane region, resulting in a lumenal orientation of the N-terminal tail of the protein.
The glycosylation of the N-terminal tail of NS4B was an unexpected finding, since both the N terminus and the C terminus of NS4B are cleaved by the cytoplasmic NS3 protease during processing of the HCV polyprotein (14, 53, 54). We do not currently know if the topology of NS4B is the same when it is expressed alone and when expressed as part of the polyprotein. (Studies addressing this question are in progress.) If the topologies are not the same, this would indicate that the intrinsic preferred topology of NS4B is different from the one it assumes when expressed as part of the polyprotein. It has been shown that the NS3-catalyzed cleavage between NS4A and NS4B is a slow process compared with the other cleavages of the polyprotein (43). Furthermore, this cleavage requires the presence of microsomal membranes in vitro, even though the NS3 protease-processing site is cytoplasmic (14, 28). This indicates that NS4A-4B must be inserted in the membrane to have the correct folding, allowing cleavage. During viral assembly, it is thus possible that the slow processing between NS4A and NS4B is actually a device designed to prevent the NS4B N-terminal tail from translocating to the lumenal side.
If conversely the topology is the same, both when NS4B is expressed alone and in the polyprotein context, the slow cleavage between NS4A and NS4B suggests that the N-terminal tail of NS4B is translocated across the ER membrane by a posttranslational mechanism. Our observation that E161 is glycosylated before E33 strongly supports this model. The finding that the 93-EGFP segment does not integrate properly but shows some degree of membrane association may also be a sign that this portion of NS4B will only integrate after the neighboring transmembrane segments have been inserted in the membrane, as for the full-length NS4B.
Posttranslational reorientation of viral membrane proteins is not unprecedented. The hepatitis B virus L envelope protein has two different topologies in the ER membrane, with the N terminal (pre-S domain) on either the cytoplasmic or the lumenal side of the membrane. This is supposed to occur by a posttranslational translocation of the N-terminal tail from the cytoplasm to the lumen through an as yet unknown mechanism (5, 25, 36, 41). Another example of dual topology is the M protein from transmissible gastroenteritis corona virus. Two-thirds of the total number of M protein molecules in the virion keep their C-terminal tails inside the virion, while one-third expose it on the surface. Whether the mechanism behind this is due to a maturation process of the virion or a cotranslational diversity is not yet known (11, 44).
This is the first study that directly assesses the topology of HCV NS4B. Other studies have approached this question indirectly. Hügle et al. performed proteinase protection experiments with in vitro-translated NS4B. (As in our study, NS4B was expressed alone and not as part of a polyprotein.) They could not detect any protected fragments, as would be expected if any large part of the protein were on the lumenal side of the membranes. However, their level of expression of NS4B before the proteinase K digestion was low, and our results indicate that the possible N-terminal translocation event is rather inefficient (Fig. 6). These facts may explain our different conclusions as to which side of the membrane the N-terminal tail of NS4B resides.
The role of NS4B in the replication and life cycle of HCV is still not understood. It is clear that the protein is needed in the replication process, in which it may function, for example, as an anchor to secure part of the HCV replication apparatus to the ER membrane. Additionally, as our results indicate, it may rearrange intracellular membranes in order to optimize them for the replication or assembly process. The function of the N-terminal tail cannot yet be guessed, but similar to the hepatitis B virus L protein, it may have a dual role, one on each side of the membrane. Interestingly, the NS4B proteins of yellow fever virus and dengue virus, both flaviviruses, have been reported to have their N-terminal tails in the lumen of the ER. In these cases, however, translocation of the N-terminal tail is initiated by an N-terminal, cleavable signal peptide not found in HCV NS4B (6, 27). A shared topology achieved in two different ways supports the notion of a common function for NS4B in the Flaviviridae family.
Acknowledgments
We thank Daniel Johansson for providing the control construct in the immunofluorescence experiments, Hannah Lindström for primers used for the PCR amplification in the in vitro experiments, Karin Melén for TMHMM analysis, Robert A. Harris for linguistic advice, and Jens Bukh for the pCV/H77C plasmid. We are also grateful to Ralf Pettersson for sharing reagents and for helpful discussions during the progress of this work.
This work was supported by The Foundation for Strategic Research (I&V), The Torsten and Ragnar Söderberg Foundations, the Swedish Research Council, the Swedish Cancer Society, the Medical Faculty of Lund University, and the Cancer Foundation of Malmö University Hospital.
REFERENCES
- 1.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienz, K., D. Egger, and T. Pfister. 1994. Characteristics of the poliovirus replication complex. Arch. Virol. Suppl. 9:147-157. [DOI] [PubMed] [Google Scholar]
- 3.Blum, R., F. Pfeiffer, P. Feick, W. Nastainczyk, B. Kohler, K.-H. Schäfer, and I. Schulz. 1999. Intracellular localization and in vivo trafficking of p24A and p23. J. Cell Sci. 112:537-548. [DOI] [PubMed] [Google Scholar]
- 4.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 5.Bruss, V., X. Lu, R. Thomssen, and W. H. Gerlich. 1994. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein EMBO J. 13:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahour, A., B. Falgout, and C.-J. Lai. 1992. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J. Virol. 66:1535-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo, Q.-L., J. Han, A. J. Weiner, L. R. Overby, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Hepatitis C virus is a distant relative of the flaviviruses and pestiviruses, p. 47-52. In T. Shikata, R. H. Purcell, and T. Uchida (ed.), Viral hepatitis C, D and E. Elsevier Science Publishers B.V., Amsterdam, The Netherlands.
- 8.Choo, Q.-L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, A. Medina-Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egger, D., B. Wölk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escors, D., E. Camafeita, J. Ortego, H. Laude, and L. Enjuanes. 2001. Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core. J. Virol. 75:12228-12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grassman, C. W., O. Isken, N. Tautz, and S.-E. Behrens. 2001. Genetic analysis of the pestivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J. Virol. 75:7791-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kumura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. USA 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houghton, M. 1996. Hepatitis C viruses, p. 1035-1058. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott Raven Publishers, Philadelphia, Pa.
- 16.Hügle, T., F. Fehrmann, E. Bieck, M. Kohara, H.-G. Kräusslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology. 284:70-81. [DOI] [PubMed] [Google Scholar]
- 17.Johansson, M., I. Nilsson, and G. von Heijne. 1993. Positively charged amino acids placed next to a signal sequence block protein translocation more efficiently in Escherichia coli than in mammalian microsomes. Mol. Gen. Genet. 239:251-256. [DOI] [PubMed] [Google Scholar]
- 18.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 19.Kato, J., N. Kato, H. Yoshida, S. K. Ono-Nita, Y. Shiratori, and M. Omata. 2002. Hepatitis C virus NS4A and NS4B proteins suppress translation in vivo. J. Med. Virol. 66:187-199. [DOI] [PubMed] [Google Scholar]
- 20.Kato, N., H. Yoshida, S. K. Ono-Nita, J. Kato, T. Goto, M. Otsuka, K.-H. Lan, K. Matsushima, Y. Shiratori, and M. Omata. 2000. Activation of intracellular signaling by hepatitis B and C viruses: C-viral core is the most potent signal inducer. Hepatology 32:405-412. [DOI] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J.-E., W. K. Song, K. M. Chung, S. H. Back, and S. K. Jang. 1999. Subcellular localization of hepatitis C proteins in mammalian cells. Arch. Virol. 144:329-343. [DOI] [PubMed] [Google Scholar]
- 23.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2000. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 25.Lambert, C., and R. Prange. 2001. Dual topology of the hepatitis B large envelope protein. J. Biol. Chem. 276:22265-22272. [DOI] [PubMed] [Google Scholar]
- 26.Li, Y., and J. McNally. 2001. Characterization of RNA synthesis and translation of bovine viral diarrhea virus (BVDV). Virus Genes 23:149-155. [DOI] [PubMed] [Google Scholar]
- 27.Lin, C., S. M. Amberg, T. J. Chambers, and C. M. Rice. 1993. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 67:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, C., and C. M. Rice. 1995. The hepatitis C virus NS3 serine proteinase and NS4A cofactor: establishment of a cell-free trans-processing assay. Proc. Natl. Acad. Sci. USA 92:7622-7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, C., J.-W. Wu, K. Hsiao, and M. S.-S. Su. 1997. The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J. Virol. 71:6465-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacKenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monné, M., M. Hermansson, and G. von Heijne. 1999. A turn propensity scale for transmembrane helices. J. Mol. Biol. 288:141-145. [DOI] [PubMed] [Google Scholar]
- 33.Mottola, G., G. Cardinal, A. Cessacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 34.Nedderman, P., A. Clementi, and R. De Francesco. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J. Virol. 73:9984-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson, I., and G. von Heijne. 1993. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmatic reticulum membrane. J. Biol. Chem. 268:5798-5801. [PubMed] [Google Scholar]
- 36.Ostapchuk, P., P. Hearing, and D. Ganem. 1994. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 13:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ota, K., M. Sakaguchi, N. Hamasaki, and K. Mihara. 2000. Membrane integration of the second transmembrane segment of Band 3 requires a closely apposed preceding signal anchor sequence. J. Biol. Chem. 275:29743-29748. [DOI] [PubMed] [Google Scholar]
- 38.Park, J.-S., J. M. Yang, and M.-K. Min. 2000. Hepatitis C virus nonstructural protein NS4B transforms NIH3T3 cells in cooperation with the Ha-ras oncogene. Biochem. Biophys. Res. Commun. 267:581-587. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popov, M., L. Y. Tam, J. Li, and R. A. F. Reithmeier. 1997. Mapping of the ends of transmembrane segments in a polytopic membrane protein. J. Biol. Chem. 272:18325-18332. [DOI] [PubMed] [Google Scholar]
- 41.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu, L., L. K. McMullan, and C. M. Rice. 2001. Isolation and characterization of noncytopathic pestivirus mutants reveals a role for nonstructural protein NS4B in viral cytopathogenicity. J. Virol. 75:10651-10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed, K. E., and C. M. Rice. 1998. Molecular characterization of hepatitis C virus. Curr. Stud. Hematol. Blood Transfus. 62:1-37. [DOI] [PubMed] [Google Scholar]
- 44.Risco, C., I. M. Antón, C. Suné, A. M. Pedregosa, J. M. Martín-Alonso, F. Parra, J. L. Carrascosa, and L. Enjuanes. 1995. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J. Virol. 69:5269-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothman, J., and H. Lodish. 1977. Synchronised transmembrane insertion and glycosylation of a nascent membrane protein. Nature 269:775-780. [DOI] [PubMed] [Google Scholar]
- 46.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selby, M. J., Q.-L. Choo, K. Berger, G. Kuo, E. Glazier, M. Eckart, C. Lee, D. Chien, C. Kuo, and M. Houghton. 1993. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J. Gen. Virol. 74:1103-1113. [DOI] [PubMed] [Google Scholar]
- 48.Serafino, A., M. B. Valli, F. Andreola, G. Carloni, and L. Bertolini. 1998. Morphological modifications induced by HCV infection in the TOFE human lymphoblastoid cell line. Res. Virol. 149:299-305. [DOI] [PubMed] [Google Scholar]
- 49.Steffen, A.-M., P. Marianneau, C. Caussin-Schwemling, C. Royer, C. Schmitt, D. Jaeck, P. Wolf, J.-L. Gendrault, and F. Stoll-Keller. 2001. Ultrastructural observations in hepatitis C virus-infected lymphoid cells. Microbes Infect. 3:193-202. [DOI] [PubMed] [Google Scholar]
- 50.Tanji, Y., M. Hijikata, S. Satoh, T. Kaneko, and K. Shimotohno. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tusnády, G., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 52.Westaway, E. G., J. M. MacKenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wölk, B., D. Sansonno, H.-G. Kräusslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 1999. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan, Y., Y. Li, S. Munshi, V. Sardana, J. L. Cole, M. Sardana, C. Steinkuehler, L. Tomei, R. De Francesco, L. C. Kuo, and Z. Chen. 1998. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 Å resolution structure in a hexagonal crystal form. Protein Sci. 7:837-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]