Abstract
Campylobacter jejuni, a major human enteric pathogen, exhibits significant strain-to-strain differences which result in differences in pathogenic potential. C. jejuni 81-176 is a highly virulent strain that exhibits unique pathogenic features and is used by many research laboratories. We have determined the nucleotide sequence of its genome and compared it to the genomes of other sequenced C. jejuni strains. We identified a number of unique genetic features which may confer specific metabolic and pathogenic properties on this strain. We have also identified regions of the C. jejuni genome that are hot spots for the integration of horizontally acquired genetic material. This information should help the understanding of the pathogenesis of C. jejuni and, in particular, the unique features of this highly pathogenic strain.
Campylobacter jejuni is one of the most common causes of food-borne illness in developed countries (2). Although it can colonize a variety of warm-blooded animals asymptomatically, this pathogen can also cause diseases in humans ranging from self-limiting gastroenteritis to more serious systemic infections (68). In addition, a small proportion of infected patients can develop a serious neurodegenerative complication known as Guillain-Barré syndrome (46).
Despite its importance, remarkably little is known about the mechanisms by which C. jejuni causes disease. A number of studies have demonstrated the importance of motility, as well as a number of surface structures and metabolic adaptations, in the pathogenesis of this bacterium (6, 17, 20, 22, 24, 29, 30, 44). The genome sequences of two C. jejuni isolates have provided important clues about its metabolism, as well as a catalogue of phase-variable surface proteins and determinants involved in the assembly or modification of highly variable surface structures such as, lipooligosaccharide (LOS) and capsule (16, 50). This genomic information, coupled with a number of comparative studies (13, 32, 35, 36, 51, 55), has revealed a remarkable degree of diversity among different C. jejuni isolates. This diversity is thought to be largely responsible for the different pathogenic properties of different isolates of this enteropathogen.
Many research laboratories use in their studies a strain of C. jejuni, 81-176, which exhibits unique pathogenic features. Originally isolated from a patient during an outbreak of C. jejuni campylobacteriosis (33), this strain has been shown to be highly pathogenic in monkeys (58), as well as in two human trials (9, 56). Furthermore, this strain exhibits high levels of invasion of tissue culture cells (25, 48), a property that has been correlated with virulence (4, 14, 70). Although a number of studies have described some of the unique genetic features of C. jejuni 81-176 (19, 31, 37, 63), the complete nucleotide sequence of its genome is not available. With a recently developed nucleotide sequencing technology based on the microfabricated high-density PicoTiterPlate (41), we have determined the entire nucleotide sequence of the genome of C. jejuni 81-176. The genome sequence revealed several new genetic loci, as well as a number of deletions, which may explain the unique pathogenic features of this strain.
MATERIALS AND METHODS
Bacterial strains, cell lines, and culture conditions.
C. jejuni 81-176 was obtained from Patricia Guerry, Naval Medical Research Center, Silver Spring, MD. Bacterial strains were grown routinely on brucella broth agar plates or on tryptic soy agar plates supplemented with 5% defibrinated sheep blood in a 10% CO2 atmosphere at 37°C. Brain heart infusion (BHI) medium was used to grow C. jejuni in liquid culture. For growth under anaerobic condition, C. jejuni was cultivated in BBL anaerobic jars (BD) with BBL GasPak Plus envelopes (BD). Escherichia coli strains were grown on Luria-Bertani agar plates or in Luria-Bertani liquid medium supplemented with ampicillin (100 mg/liter), chloramphenicol (30 mg/liter), or kanamycin (50 mg/liter), as appropriate. To determine growth curves, C. jejuni cultures were adjusted to an optical density at 600 nm of 0.1 in 3 ml of BHI medium and grown under an atmosphere of 10% CO2 in a rotating wheel (50 rpm). Alternatively, cultures (optical density at 600 nm of 0.1 in 25 ml of BHI medium in 50-ml Erlenmeyer flasks) were incubated under anaerobic conditions with orbital shaking (200 rpm). T84 cells, a human intestinal epithelial cell line, were obtained from the American Type Culture Collection (Manassas, Va.). Cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum containing penicillin (100 U ml−1) and streptomycin (50 μg ml−1).
Nucleotide sequencing and genome sequence analysis.
Total DNA of C. jejuni 81-176 was isolated with the DNeasy tissue kit (QIAGEN) according to the manufacturer's protocol. Sample preparation and genome sequencing in microfabricated high-density picoliter reactors were performed as described before (41), at the 454 Life Sciences Measurement Service Center (Branford, CT). Predictions of open reading frames (ORFs) in regions specific for C. jejuni 81-176 were performed with ARTEMIS (http://www.sanger.ac.uk/Software/Artemis) with a cutoff of 100 bp and BLASTX (http://www.ncbi.nlm.nih.gov/BLAST/). Homology searches were performed with BLASTP/N and PSI- and PHI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST). CampyDB (http://campy.bham.ac.uk/) was used for specific comparison of the C. jejuni 81-176 contigs with the published genome sequences of C. jejuni and H. pylori. Analysis of secondary structural properties and detection of specific domains or motifs in the predicted proteins were carried out with SMART (http://smart.embl-heidelberg.de), SCANPROSITE (http://www.expasy.org/tools/scanprosite/), and MOTIF (http://motif.genome.ad.jp).
Construction of recombinant plasmids and generation of C. jejuni mutants.
DNA manipulations were performed according to standard protocols (40). Plasmid isolation was carried out with the QIAGEN QIAprep Spin Miniprep kit according to the manufacturer's instructions. The C. jejuni strains carrying insertion mutations in the genes encoding γ-glutamyltranspeptidase (ggt), cytochrome c (cytC), and dimethyl sulfoxide (DMSO) reductase (dmsA) were constructed by inserting a terminatorless aphA3 cassette within the respective genes (amplified by PCR) by standard recombinant DNA methods. The mutated genes were then introduced into C. jejuni 81-176 by natural transformation. All mutant strains were verified by PCR analysis.
Bacterial invasion assay.
The ability of C. jejuni to enter cultured intestinal epithelial cells was evaluated by the standard gentamicin protection assay. Briefly, T84 intestinal epithelial cells grown to 70% confluence in 24-well dishes were infected with C. jejuni grown to mid-logarithmic phase at a multiplicity of infection of 100. Plates were centrifuged at 200 × g for 5 min to maximize bacterium-host cell contact and incubated for 2 h at 37°C in an atmosphere of 5% CO2. Monolayers were then washed three times with Hanks balanced salt solution, followed by incubation with Dulbecco modified Eagle medium containing gentamicin (100 μg ml−1) for 2 h to kill extracellular bacteria. Epithelial cells were then washed three times and lysed in phosphate-buffered saline containing 0.1% deoxycholic acid to release the intracellular bacteria. The intracellular bacteria (i.e., those that survived the gentamicin treatment) were enumerated by plating of serial dilutions on tryptic soy agar plates supplemented with 5% defibrinated sheep blood.
Mouse infections.
The ability of C. jejuni to colonize mice was tested with a recently established mouse model of infection, which will be described in detail elsewhere (R. O. Watson et al., submitted for publication). Briefly, this animal model makes use of a mouse line which lacks the Myd88 adaptor protein and therefore is defective in Toll-like receptor signaling. Absence of Myd88 permits stable colonization by C. jejuni. Equal numbers (∼109 bacterial CFU) of CFU of C. jejuni 81-176 and its isogenic mutants were simultaneously administered orally to 6-week-old Myd88−/− mice. Colonization by the different strains was monitored by enumerating the number of CFU in the feces of the inoculated animals. Equal amounts of homogenized feces were plated on blood agar plates containing Campylobacter-selective supplements (Oxoid SR0167E) with or without kanamycin (50 μg ml−1). At the end of the experiment, mice were sacrificed and the bacterial loads in the intestines were determined by plating on selective plates as described above. Statistical analysis of the results was carried out with the Wilcoxon (Mann-Whitney) rank test.
Nucleotide sequence accession numbers.
The annotated genome sequence has been deposited in the GenBank/EMBL sequence database under accession numbers DQ489494, DQ492685, DQ493918, DQ493919, DQ493920, DQ493921, DQ493922, DQ493923, DQ493924, DQ493925, and DQ493926.
RESULTS AND DISCUSSION
High-throughput sequencing of the C. jejuni 81-176 genome.
Total DNA of C. jejuni 81-176 containing chromosomal, as well as plasmid, DNA was isolated, fragmented, and specifically prepared for sequencing with the Genome Sequencer 20 system (see Materials and Methods). Rather than the generation of a closed, finished genome, our goal was to use high-throughput nucleotide sequencing to identify the unique relevant genomic features of this strain. Two sequencing runs were performed, which produced 60,905,794 bp of sequence assembled in 43 contigs, resulting in 34-fold coverage (Table 1). Adjacent contigs were subsequently linked by additional nucleotide sequencing of PCR-generated DNA fragments with primers specific for the ends of adjacent contigs. Remaining gaps of 56 and 1,589 bp located within the LOS and capsule loci were closed with published sequences (19, 31). We thus obtained an almost closed genome totaling 1,594,651 bp, with only two remaining gaps located within highly repetitive regions, which are difficult to accurately sequence and assemble. One of the gaps, with an estimated size of 32 bp, is located within internal sequences of a repetitive conserved rRNA cluster. The other gap, predicted to be 111 bp, is located within the highly conserved Cj0794 gene of reference strain NCTC 11168.
TABLE 1.
C. jejuni 81-176 genome sequencing raw statistics
| Parameter | Value |
|---|---|
| No. of instrument runs | 2 |
| Total no. of reads | 558,331 |
| Total no. of bases | 60,905,794 |
| No. of reads assembled | 533,496 |
| No. of contigs | 43 |
| Total no. of bases in contigs | 1,668,622 |
| No. of bases covering plasmid DNA | 80,673 |
| No. of bases of pVira | 35,468c |
| No. of bases of pTetb | 45,205d |
| Avg coverage depth (n-fold) | 34 |
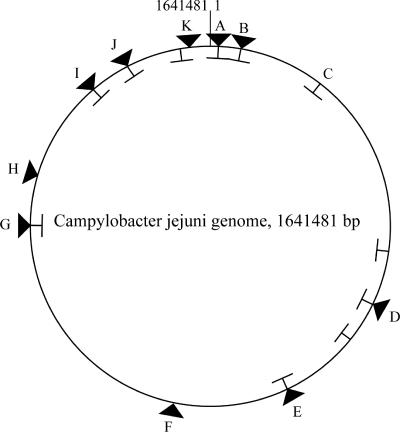
The C. jejuni 81-176 genome is slightly smaller than those of the other two published sequenced strains, NCTC 11168 and RM1221. This reflects the absence of inserted phages (as in C. jejuni RM1221) and the presence of a reduced number of genes involved in the synthesis of capsule and posttranslational modification of flagellar proteins. The genomic structure of C. jejuni 81-176 is syntenic with the genomes of C. jejuni NCTC 11168 and RM1221 (Fig. 1), except where the synteny is disrupted by small duplications or deletions or the insertion of genomic islands of various sizes (Fig. 1; see Fig. S1 in the supplemental material; see also below). C. jejuni 81-176 carries two resident plasmids, pVir and pTet, whose nucleotide sequences have been previously reported (5, 7). As expected, the nucleotide sequences of these two plasmids were virtually identical to those previously published.
FIG. 1.
Differences in synteny between C. jejuni NCTC 11168 and C. jejuni 81-176. A schematic comparison of the genomes of C. jejuni NCTC 11168 and 81-176 indicating the positions of insertions (triangles) and deletions (T symbols) in 81-176 is shown. Detail maps of the loci marked with capital letters are shown in other figures (A, see Fig. S2 in the supplemental material; B, see Fig. 3; C, see Fig. 7; D, see Fig. 5A; E, see Fig. S3 in the supplemental material; F, see Fig. 4; G, see Fig. S4 in the supplemental material; H, see Fig. S5 in the supplemental material; I, see Fig. S6 in the supplemental material; J, see Fig. 2; K, see Fig. 6A).
C. jejuni 81-176 genes that are absent from or are pseudogenes in reference strains NCTC 11168 and RM1221.
Comparison of the nucleotide sequence of C. jejuni 81-176 with those of reference strains NCTC 11168 (50) and RM1221 (16) revealed the presence of 37 genes that are absent from both reference strains and are located in 11 regions throughout the chromosome (Fig. 1; see Table S1 in the supplemental material). On the basis of the nature of the inserted genetic material and/or the predicted function of the encoded proteins, these genes can be grouped into six general categories.
(i) Electron transport, energy metabolism, and respiratory pathways.
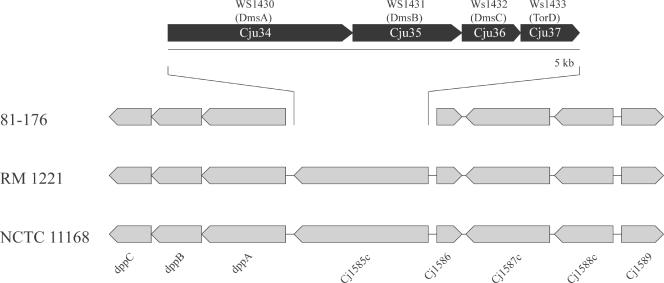
C. jejuni is an obligate microaerophile with a remarkably versatile respiratory chain which allows the utilization of a large variety of electron donors and acceptors. Under low-oxygen conditions, C. jejuni can use a variety of alternative electron acceptors, including fumarate, nitrate, nitrite, trimethylamine-N-oxide, DMSO, and sulfite (45, 53, 60). Such versatility is thought to be important for the ability of this pathogen to colonize the low-oxygen environment of the intestinal tract. C. jejuni 81-176 encodes respiratory functions additional to those encoded by reference strain NCTC 11168 or RM1221 which may contribute to its unique ability to efficiently colonize the intestines of human or animal hosts. For example, instead of the putative oxidoreductase encoded by Cj1585c in reference strain NCTC 11168, C. jejuni 81-176 possesses a gene cluster that encodes predicted proteins with significant sequence similarity to DmsA, DmsB, DmsC, and DmsD/TorD, which are components of a putative anaerobic DMSO reductase of Wolinella succinogenes (Fig. 2). It has been shown in Escherichia coli that DmsA and DmsB form a catalytic dimer that is anchored to the membrane by DmsC (59). The DmsA/DmsB complex is targeted to the cytoplasmic membrane by the twin-arginine translocase (TAT) system (8). Indeed, C. jejuni 81-176 encodes the putative components of the TAT system, TatA (Cj0576c), TatB (Cj0579c), and TatC (Cj0578c). Furthermore, immediately adjacent to the dms locus, C. jejuni 81-176 encodes a putative homolog of E. coli TorD (Fig. 2). This cytoplasmic protein binds tightly to the Tat signal peptide of DmsA, and therefore it has been proposed to function as a chaperone that escorts the DmsA protein to the TAT translocon (26, 27). Taken together, these observations indicate that C. jejuni 81-176 has an additional DMSO reductase system that may be important for respiration under oxygen-restricted conditions.
FIG. 2.
Gene arrangement of the dmsA locus in C. jejuni 81-176. In the genome of C. jejuni 81-176, the putative oxidoreductase gene Cj1585c is replaced by a cluster of four genes whose products show very significant sequence similarity to the components of a DMSO reductase complex encoded by an operon of W. succinogenes.
C. jejuni NCTC 11168 encodes a molybdoprotein (Cj0264c) that serves as its sole active trimethylamine-N-oxide/DMSO reductase (60). This protein is missing in C. jejuni RM1221 but present in C. jejuni 81-176. Although there is an apparent redundancy in C. jejuni 81-176 between the DmsA homolog Cju34 and its Cj0264c homolog, the amino acid sequences of these two proteins with putatively similar functions are quite divergent (27% identity and 42% similarity). It is unclear whether this sequence diversity reflects differences between their functions.
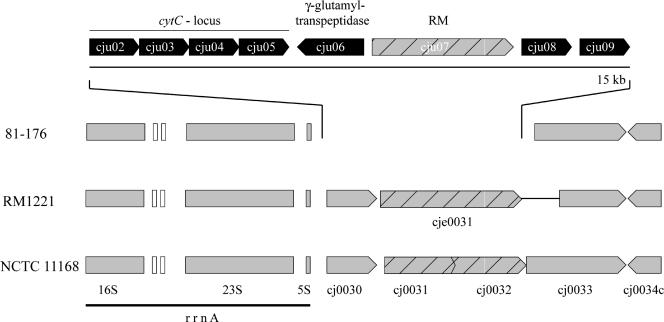
C. jejuni 81-176 encodes all of the different cytochromes c and cytochrome c oxidoreductases present in reference strains NCTC 11168 and RM1221. In addition, however, 81-176 possesses a cluster of genes encoding putative proteins with amino acid sequence similarity to cytochrome c and a number of cytochrome c biogenesis proteins (Fig. 3). These proteins are closely related to corresponding gene products of Campylobacter lari, with sequence identities between 53% and 80%. Although the specificity of this putative electron transfer pathway is not known, this finding is another example of the expanded respiratory pathways in this C. jejuni strain.
FIG. 3.
Gene arrangement of a novel cytochrome c biosynthesis locus in C. jejuni 81-176. Two genes of NCTC 11168 are replaced in C. jejuni 81-176 by a 15-kb region encoding biosynthesis genes for cytochrome c with the closest homologs found in C. lari. In addition, this region also encodes a γ-glutamyltranspeptidase, a type II restriction-modification enzyme, and two hypothetical proteins with unknown functions. White boxes indicate the positions of tRNA genes.
A number of studies have demonstrated the importance of fumarate as a major electron acceptor when C. jejuni is grown under oxygen-limited conditions (60, 62). Furthermore, a recent report has shown that genes involved in fumarate metabolism are upregulated when C. jejuni is present in the intestinal tracts of chickens (69), suggesting that specific adaptations that could optimize this respiration mode may enhance its ability to colonize this site. Fumarate is transported by a number of C4-dicarboxylate carriers, such as DcuA and DcuB (28). These carriers are present only in anaerobic and facultative anaerobic bacteria, such as C. jejuni. DcuA and DcuB are thought to operate preferentially as uptake carriers, while DcuC is thought to be involved in the efflux of succinate, the end product of fumarate fermentation (28). C. jejuni NCTC 11168 and RM1221 encode homologs of DcuA (Cj0088/CJE0083) and DcuB (Cj0671/CJE0772) but lack a functional DcuC protein since its putative homologs (Cj1389/CJE1580 and Cj1528/CJE1699) are pseudogenes in these reference strains (16, 50). In contrast, C. jejuni 81-176 encodes homologs of all three of these C4-dicarboxylate carriers, including two apparently functional DcuC homologs (see Table S2 in the supplemental material), which are likely to be active under anaerobic conditions, as shown for E. coli. Therefore, it is possible that the presence of a more robust anaerobic respiration system in strain 81-176 requires more efficient efflux systems to maintain an optimal redox balance.
The nucleotide sequences of C. jejuni NCTC 11168 and RM1221 revealed the presence of a relatively large number of pseudogenes (16, 50). Some of these pseudogenes are the result of frame shifts, but others exhibited significant degeneration of the ORFs, suggesting that mutations may have accumulated over time. Interestingly, we found several ORFs that appear as pseudogenes in NCTC 11168 or RM1221 but encode apparently functional proteins in C. jejuni 81-176 (see Table S2 in the supplemental material). Genes encoding restriction and modification enzymes, two-component signal transduction proteins, and membrane proteins involved in several transport processes, like iron uptake, anion and cation transport, and sugar and peptide transport, are often pseudogenes in C. jejuni. This variability may explain some of the diversity in several metabolic traits that is often observed among different C. jejuni isolates (16, 50). For example, C. jejuni 81-176 possesses a gene for a glycerol-3-phosphate transporter (GlpT), although this gene appears disrupted in C. jejuni RM1221. Glycerol-3-phosphate is a central intermediate in glycolysis and in phospholipid metabolism (34). The enzyme glycerol-3-phosphate dehydrogenase (GpsA; Cj1196c/CJE1330) of C. jejuni converts glycerol-3-phosphate to glyceraldehyde-3-phosphate, which can enter the glycolysis pathway. Given the fact that C. jejuni is unable to utilize glucose because of the lack of phosphofructokinase (66), the ability to take up glycerol-3-phosphate may confer significant metabolic advantages on this strain.
(ii) Potassium uptake.
The genes kdpA, kdpB, and kdpC, which encode a potassium-transporting ATPase (3), are apparently functional in C. jejuni 81-176 although they are pseudogenes in C. jejuni NCTC 11168 and RM1221. Besides this potassium transport system, all three C. jejuni strains encode the KtrA/KtrB potassium uptake system, a high-affinity uptake system described for Vibrio alginolyticus (47, 64). The kdp system is induced under low-potassium conditions and repressed under high potassium concentrations. The coordinated activity of these systems may contribute to the establishment of a balanced intracellular potassium concentration, which is important for maintaining constant turgor pressure within the cell. In E. coli, turgor pressure regulates the expression of the kdpABC genes via the membrane-bound KdpD regulatory protein (39). Interestingly, the kdpD gene is truncated and presumably nonfunctional in C. jejuni 81-176, suggesting a different regulatory mechanism for the expression of the potassium transport system. It is possible that the presence of an additional functional potassium uptake system may benefit the growth of C. jejuni 81-176 in environments such as that within the phagocytic vacuole, where the potassium concentration may be low (67).
(iii) γ-Glutamyltranspeptidase.
Within the same segment that contains the novel cytochrome c in C. jejuni 81-176 (Fig. 3), there is an ORF also absent from reference strains NCTC 11168 and RM1221 which encodes a protein with very significant amino acid sequence similarity to a γ-glutamyltranspeptidase of Helicobacter pylori. This enzyme has been shown to be required for H. pylori colonization in animal models (11, 42). The mechanisms by which this γ-glutamyltranspeptidase contributes to virulence are not understood, although it has been reported that its addition to cultured gastric cells induces apoptosis (61). Alternatively, γ-glutamyltranspeptidases are part of the antioxidant glutathione pathway and therefore it is possible that they may contribute to counteract oxidative stress. It is noteworthy that in addition to C. jejuni 81-176, we have detected this γ-glutamyltranspeptidase by PCR analysis in 3 out of 15 human isolates analyzed (data not shown), suggesting that this enzyme may contribute to the virulence of a subset of C. jejuni isolates.
(iv) Restriction-modification systems.
A recent study has described a great deal of diversity in the DNA restriction and modification systems encoded by C. jejuni isolates (43). It is believed that this diversity may influence the ability of C. jejuni to horizontally acquire genetic information. C. jejuni 81-176 encodes a number of DNA restriction-modification systems, and consistent with the diversity observed in other strains, some of these systems are absent from the genomes of reference strains NCTC 11168 and RM1221. For example, C jejuni 81-176 encodes a complete additional type I DNA restriction and modification system (see Fig. S3 in the supplemental material). The GC content of this genomic region (28.8%) is different from that of the rest of the C. jejuni chromosome, suggesting that these genes may have been acquired horizontally. In this context, the three subunits of the restriction-modification system encoded in this locus possess the closest sequence similarity to the HsdR, HsdS, and HsdM proteins of Campylobacter upsaliensis (see Table S1 in the supplemental material).
(v) Serine protease.
C. jejuni 81-176 encodes a putative serine protease that belongs to the autotransporter family and is absent from both reference strains. The enzymatic domain of this protein, located at its amino terminus, belongs to the subtilinase family of serine proteases and has the closest significant amino acid sequence similarity to a serine protease from C. upsaliensis (see Fig. S5 in the supplemental material). The autotransporter domain is located at the carboxy terminus, which ends in phenylalanine, a characteristic shared by most of the described autotransporter proteins (23).
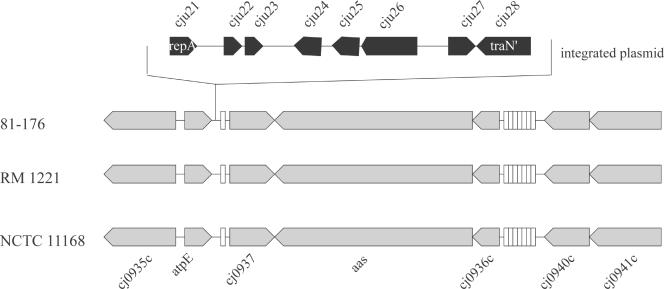
(vi) Integrated plasmid.
C. jejuni 81-176 carries two previously described plasmids, pTet and pVir (7), which have been proposed to contribute to the virulence of this strain (4, 5). The nucleotide sequence analysis of C. jejuni 81-176 did not reveal the presence of any other extrachromosomal element or the presence of large integrated prophages or insertion sequences, as seen in C. jejuni RM1221. However, we identified a 6-kb insertion element located immediately adjacent to a Leu tRNA which exhibits the features of an integrated plasmid (Fig. 4). This element encodes homologs of the TraN and TraG proteins of type IV secretion systems, proteins with homology to CCOA0053 and CCOA0054 of plasmid pCC178 of Campylobacter coli RM2228 (16), and one protein nearly identical to CJE1155 which is encoded by the CJIE3 integrated element of C. jejuni RM1221 (16). The TraN homolog is truncated at its N terminus, and the gene encoding a putative TraG homolog carries a frameshift mutation, indicating that these genes do not encode functional proteins. This integrated element also encodes a homolog of the RepA protein of several C. jejuni plasmids. Another remnant of an extrachromosomal element in C. jejuni 81-176 is represented by a unique ORF (Cju30), located immediately adjacent to the Ser tRNA (see Fig. 7), which encodes a protein with significant amino acid sequence similarity to plasmid-stabilizing protein ParE. Since these integrated elements lack essential functions that would allow their transmission and/or replication and do not encode any other obvious function, they probably represent vestigial elements that do not contribute to the biology of this strain.
FIG. 4.
Remnants of an integrated plasmid in the genome of C. jejuni 81-176. Adjacent to a leucine tRNA gene in the C. jejuni 81-176 genome, there are a group of genes with significant amino acid sequence similarity to proteins known to be important for plasmid replication (Cju21) or transfer (Cju25, Cju26, and Cju28). Furthermore, this region encodes polypeptides with significant amino acid sequence similarity to proteins encoded in plasmids present in C. jejuni RM1221 (Cju22 and Cju23) or C. coli RM2228 (Cju27). The positions of tRNA genes in this region are indicated by open boxes.
FIG. 7.
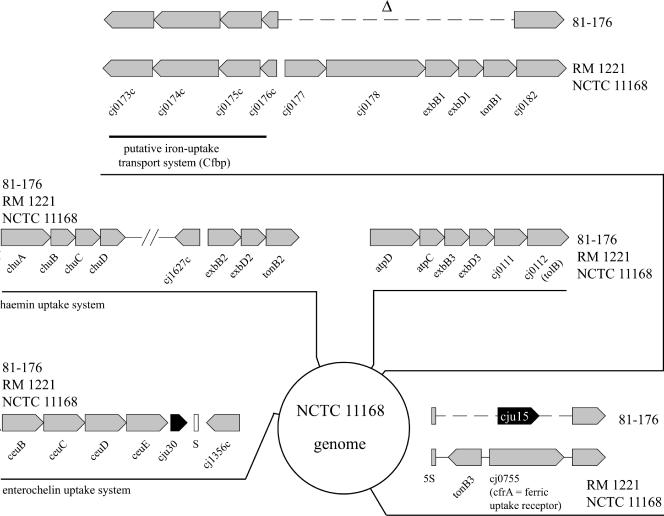
Iron uptake loci in C. jejuni 81-176. The genome location and arrangement of the five identified putative iron uptake systems in C. jejuni NCTC 11168 are shown. Two of these loci exhibit significant degradation in C. jejuni 81-176. A putative iron uptake system (bounded by Cj0173c and Cj0182 in reference strain NCTC 11168) is missing critical components in C. jejuni 81-176. In addition, in C. jejuni 81-176 tonB3 and cfrA, which encodes a ferric uptake receptor, are replaced by a protein of unknown function (Cju15) (see text for details).
Identification of integration hot spots in the C. jejuni genome.
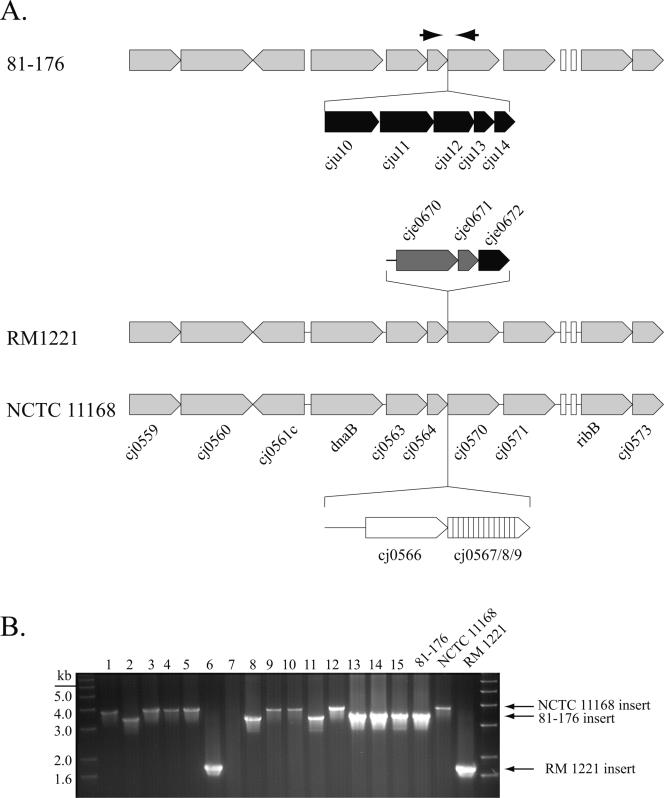
Comparison of the genome sequence of C. jejuni 81-176 with those of reference strains NCTC 11168 and RM1221 revealed the existence of chromosomal loci that appear to be hot spots for the integration of highly diverse genetic material. For example, in C. jejuni 81-176 there are five ORFs encoding putative proteins of unknown function located between the highly conserved homologs of cj0564 and cj0570 of reference strain NCTC 11168 (Fig. 5A). Interestingly, examination of this region in reference strains NCTC 11168 and RM1221 reveals the presence of DNA segments unique for each of these strains that are also bounded by the cj0564 and cj0570 homologs. To further investigate the potential diversity within this locus, we analyzed 15 clinical isolates of C. jejuni by PCR with primers complementary to sequences in the conserved boundaries. Of the 15 isolates analyzed, 6 yielded fragments of the same size as the one present in 81-176, 7 were similar to the fragment of NCTC 11168, and 1 yielded a fragment similar to the one present in RM1221 (Fig. 5B).
FIG. 5.
Hypervariable region in the C. jejuni genome. (A) A genetic locus between ORFs Cj0564 and Cj0570 of reference strain NCTC 11168 shows significant variability across the sequenced strains of C. jejuni. Each of the strains shows a different genomic fragment integrated at this site. (B) PCR amplification of this region with primers complementary to sequences within conserved ORFs Cj0564 and Cj0570 yielded fragments of various sizes, an indication of the diversity of this region. The gel shows fragments amplified from 15 different clinical isolates. Amplification of DNAs from isolates 1, 3, 4, 5, 9, and 10 yielded a band of a size similar to that of C. jejuni NCTC 11168; isolates 2, 8, 11, 13, 14, and 15 yielded a band of a size similar to that of C. jejuni 81-176; and isolate 6 yielded a band of a size similar to that of C. jejuni RM1221. The lack of amplification of a DNA fragment from isolate 7 may be due to sequence diversity within the Cj0564 and Cj0570 ORFs.
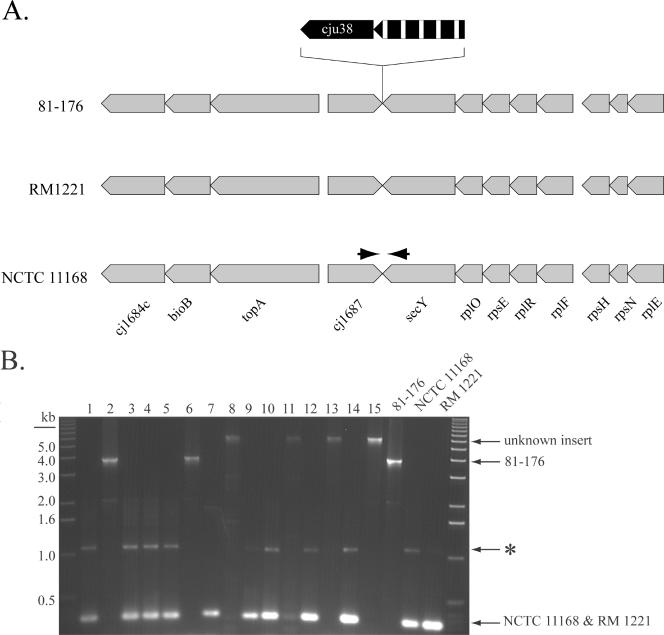
Another example of a region of genetic diversity in strain 81-176 is a locus containing two unique ORFs that are bounded by ORFs identical to cj1687 and cj1688 in reference strain NCTC 11168 (Fig. 6A). One of the ORFs is an apparent pseudogene which would encode a permease of the major facilitator superfamily. The other ORF encodes a putative peptidase with amino acid sequence similarity to a putative dipeptidyl-peptidase of Trypanosoma cruzi and Leishmania major (see Table S1 in the supplemental material). Neither C. jejuni NCTC 11168 nor RM1221 has an insertion at this locus. However, PCR amplification analysis of 15 C. jejuni clinical isolates with primers complementary to the conserved boundaries of this region confirmed that this is a hot spot for integration of new genetic material. Of the 15 isolates analyzed, 2 yielded a fragment identical in size to that of strain 81-176 whereas 4 isolates yielded a fragment about 2.5 kb larger, suggesting the presence of additional genetic material (Fig. 6B). Sequencing into the flanking region of these larger fragments confirmed that new genetic material is integrated between cj1687 and cj1688. In both strains tested (ATCC 49943 and strain 240.0872), we identified genes encoding proteins with homology to putative ATP-binding proteins of ABC transporters and phosphodiesterases which have no similarity to known C. jejuni proteins (data not shown). The remaining nine isolates yielded a fragment of a size that suggests the absence of an insert at this site (Fig. 6B). Taken together, these results indicate that these sites represent loci of high variability among C. jejuni isolates. Although it is not clear why these sites constitute a hot spot for genetic integration, close examination of this region across C. jejuni isolates may help to identify novel genetic material that may confer specific properties on different strains.
FIG. 6.
Identification of an integration hot spot in the C. jejuni genome. (A) C. jejuni 81-176 exhibits an insertion of genetic material between homologous ORFs Cj1687 and secY (NCTC 11168 nomenclature) not seen in sequenced reference strains NCTC 11168 and RM1221. The 3.5-kb insert contains a gene encoding a peptidase (cju38) and a pseudogene of a putative permease of the major facilitator superfamily (hatched arrow). (B) PCR amplification of this region with primers complementary to sequences within Cj1687 and secY yielded fragments of various sizes, an indication of the presence of novel genetic material in this region (see text for more details). The gel shows fragments amplified from 15 different clinical isolates. Amplification of DNAs from isolates 2 and 6 yielded a band similar in size to that of C. jejuni 81-176, while isolates 1, 3, 4, 5, 7, 9, 10, 12, and 14 yielded a band similar in size to that of C. jejuni NCTC 11168 and RM1221, indicating the absence of inserted genetic material in these strains. Amplification from isolates 8, 11, 13, and 15 yielded a band of a size different from those of any of the sequenced strains, suggesting the presence of novel genetic material (see text for more details). *, nonspecific PCR product. The strain numbers correspond to those in Fig. 5.
Hypervariable regions.
Analysis of the two available genome sequences of C. jejuni NCTC 11168 and RM1221 (16, 50), as well as a comparative analysis of several C. jejuni strains with DNA microarrays (13, 35, 36), led to the identification of several hypervariable plasticity regions (51). The most striking variability occurs in gene clusters encoding proteins involved in the biosynthesis of surface structures such as LOS, flagellin, and capsule, as well as their posttranslational modification through glycosylation. The variability is represented not only by sequence divergence among the predicted gene products but also by the presence or absence of specific genes within these loci. As expected, such diversity was also detected in 81-176 in regions involved in the synthesis and modification of these surface structures and confirmed previous observations from more limited nucleotide sequence analysis of some of these loci (19, 31, 63).
Another hypervariable plasticity region of C. jejuni (bounded by cj0295 and cj0306c of reference strain NCTC 11168) covers loci essential for molybdenum transport and pantothenate and biotin synthesis (51). The modABCD genes of C. jejuni, which encode a molybdenum transport apparatus, were reported to be missing in C. jejuni 81-176 in a microarray-based comparison of different strains of C. jejuni. However, a recent study detected a potential homolog of modD in this strain, although no homologs of the other components of the molybdenum transport system were found (54). In contrast to these observations, we found that all of the genes encoding a molybdenum transport system are present in C. jejuni 81-176. Interestingly, there is significant diversity in this gene cluster across the different Campylobacter sp. strains. Although the C. jejuni 81-176 modABCD cluster is nearly identical to the equivalent gene clusters in C. jejuni RM1221 and Campylobacter coli RM2228, it is significantly divergent from the modABCD genes in C. jejuni reference strain NCTC 11168. The plasticity of this region is also reflected by the observation that C. jejuni 81-176 is missing two genes in this region that are conserved in reference strains NCTC 11168 and RM1221, i.e., cj0295, encoding a putative acetyltransferase of the GNAT family, and Cj0299, whose product is similar to a penicillin-binding protein and contains a transpeptidase domain. It is unclear what the selection pressure is to maintain this variability across C. jejuni strains.
The porin protein Omp50 and the flagellar protein FlgE2 are two examples of localized variability across strains within a protein. In this case, the variability is localized to discrete regions of the predicted protein while other regions of the same protein are highly conserved (12, 38). This type of variability cannot usually be detected by standard microarray analysis unless many probes are used to query each gene. The Omp50 protein of C. jejuni 81-176 is very closely related to corresponding protein CJE1304 of C. jejuni RM1221 (94% amino acid sequence identity) but much more divergent from the equivalent protein from NCTC 11168. In contrast, the C. jejuni 81-176 FlgE2 protein shows only 82% and 78% amino acid sequence identity to the FlgE2 homologues in RM1221 and NCTC 11168, respectively. This divergence explains why the gene for FlgE2 was not detected in C. jejuni 81-176 in a microarray analysis and was therefore reported to be missing from this strain.
Diversity of iron uptake systems in C. jejuni.
Acquisition of iron is an important virulence factor for pathogens to cope with the severe iron limitation that occurs in their host environment (10). No siderophore has so far been identified in C. jejuni (15, 52), which is in agreement with the absence of putative genes encoding these factors in the genome sequences of C. jejuni NCTC 11168 and RM1221 and the genome sequence of C. jejuni 81-176 reported here. However, it has been reported that C. jejuni makes use of exogenous siderophores produced by other bacteria or the host (e.g., ferrichrome, enterochelin, and heme compounds) (15, 52). Several putative uptake systems for these siderophores have been identified in the genomes of C. jejuni, and some of them have been functionally characterized (65).
Similar to reference strains NCTC 11168 and RM1221, C. jejuni 81-176 encodes a complete hemin/hemoglobin uptake system (ChuABCD; Cj1614 to Cj1617) and an enterochelin transport system (CeuBCDE; Cj1352 to Cj1355) (Fig. 7). In addition, and as previously reported for other strains, we could not detect the presence of a gene encoding a CeuA homolog in C. jejuni 81-176. The N- and C-terminal regions of the siderophore receptor ChuA are well conserved across C. jejuni strains. However, three internal regions of this protein exhibit considerable variability across strains (data not shown). With the algorithm PRED-TMBB (http://bioinformatics.biol.uoa.gr/PRED-TMBB/), these variable regions are predicted to be exposed loops between the membrane-embedded β sheets. It is not known whether this diversity may influence the ability of these proteins to interact with the siderophore or if this variability in the surface-exposed domains is the result of selective pressure by the immune system.
C. jejuni 81-176 also encodes the CfpB ferric-uptake system (Cj0173c to Cj0176c), which is broadly conserved across strains. However, an ORF capable of encoding a hemin receptor, which is located immediately adjacent to the cfpB genes in the two sequenced reference strains, is absent from C. jejuni 81-176 (Fig. 7). The predicted ferrichrome uptake system encoded by the cfhUABD locus in some strains of C. jejuni (18) is also missing from 81-176 (Fig. 7). Also absent from C. jejuni 81-176 are the TonB-dependent ferric uptake receptor CfrA (Cj0755/CJE0847) and another putative siderophore receptor, Cj0178. This finding is surprising since both systems have been shown to be important for colonization by C. jejuni NCTC 11168 in a chicken infection model (49). Since C. jejuni 81-176 has been shown to efficiently colonize chickens, it must be able to use alternative iron uptake systems to acquire iron in the chicken intestine. It should be noted that the substrate specificity of the CfrA homolog in C. coli is not known since its absence has no effect on the utilization of hemin, enterochelin, or ferrichrome as an iron source (21).
In addition to a reduced spectrum of iron receptor proteins, C. jejuni 81-176 is also missing essential components of the ExbB-ExbD-TonB energy-transducing system that are essential for the function of some iron uptake systems. The genes exbB1, exbD1, tonB1 (Cj0179-0181), and tonB3 (Cj0754c), which are highly conserved in C. jejuni NCTC 11168 and RM1221, are absent in C. jejuni 81-176 (Fig. 7).
The diversity in the iron metabolism genes in C. jejuni is also illustrated by the variability in a number of genes or pseudogenes that encode putative iron-binding proteins. For example, Cj0045c, Cj0072c, Cj0241c, and Cj1224 of reference strain NCTC 11168 encode putative iron-binding proteins with weak similarities to members of the eukaryotic hemerythrin family. The genes Cj0045c and Cj1224 and their homologs presumably encode functional proteins in the three sequenced strains of C. jejuni, while cj0072 and its homologs are apparently pseudogenes in all of these C. jejuni strains. In contrast, Cj0241c is a pseudogene in C. jejuni RM1221 but its homologs in NCTC 11168 and 81-176 seem to encode functional proteins. Another pseudogene in C. jejuni NCTC 11168 (Cj0444) and RM1221 (cJE0496), which encodes a putative TonB-dependent siderophore receptor, seems to encode a functional protein in C. jejuni 81-176. Taken together, these observations indicate a great deal of diversity in the iron uptake systems of different strains of C. jejuni. Whether this diversity contributes specific pathogenic properties on different strains of C. jejuni is not known.
Genes of C. jejuni NCTC 11168 that are absent from C. jejuni 81-176.
Comparison of C. jejuni strains has revealed significant differences in gene content among different isolates. In fact, some microarray-based comparisons have reported up to 20% difference in gene content between certain isolates (13), although this is probably an overestimate as microarray comparisons often cannot distinguish between genes that are absent and genes that significantly deviate from the query. Besides variations in gene content in the hypervariable regions encoding LOS and capsule biosynthesis, as well as glycosylation loci, our analysis revealed the absence of 51 genes in C. jejuni 81-176 compared to reference strain NCTC 11168 (see Table S3 in the supplemental material). Gene deletions encompass either single ORFs or entire clusters of genes. For example, a 9-kb genomic segment (Cj0480 to Cj0490) encoding a putative transcription regulator, an l-fucose permease, and a tartrate transporter is missing from C. jejuni 81-176 although it is highly conserved in both reference strains. Several of the C. jejuni NCTC 11168 genes that could not been identified in the genome sequence of C. jejuni 81-176 are pseudogenes in the reference strain (e.g., Cj0501, Cj0565, Cj0752, and Cj1528). Other genes of C. jejuni NCTC 11168 missing in C. jejuni 81-176 encode putative periplasmic proteins (Cj0299, Cj0424, Cj0425, Cj1376, Cj1722c, and Cj1723c) or lipoproteins (Cj0177) with unknown functions.
Contribution to virulence of the C. jejuni 81-176-specific genes.
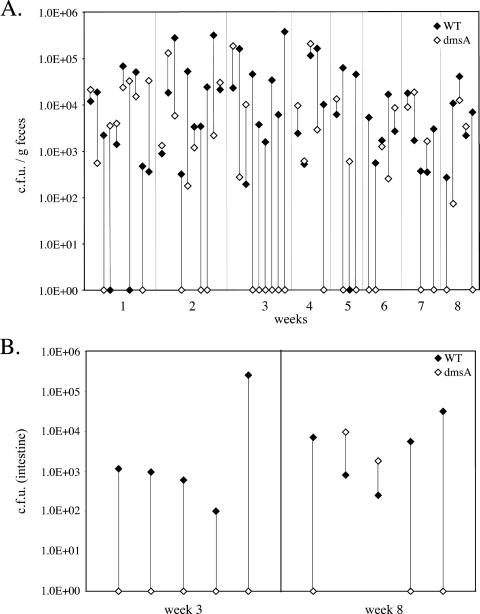
To examine the potential contribution to virulence of genes that appear unique to C. jejuni 81-176, we constructed strains carrying loss-of-function mutations in a subset of these genes and examined their phenotypes in several virulence assays. We first examined the contribution to virulence of a group of genes potentially involved in respiration under low-oxygen conditions, which are most likely encountered during gut colonization. More specifically, we constructed isogenic strains of C. jejuni 81-176 carrying loss-of-function insertion mutations in dmsA, which encodes a putative DMSO reductase, and cytC, which encodes a component of a cytochrome c oxidase (see above). Neither of the mutant strains showed any defect in the ability to grow in BHI liquid medium at 37°C under an atmosphere of 10% CO2 or under anaerobic conditions (data not shown) compared to the wild type. In addition, the mutants were able to enter and survive within intestinal epithelial cells in a manner indistinguishable from that of the wild type (data not shown). When tested in a mouse model of infection, the dmsA mutants showed a significant colonization defect (Fig. 8). Mixed-infection experiments in which mice were administered equal doses of wild-type C. jejuni 81-176 and the isogenic dmsA mutant strain showed that both strains equally colonize the intestine during the first week of infection. However, after 3 weeks of infection, the numbers of CFU of the dmsA mutant strain in the feces of most mice were significantly reduced (P = 0.0092) in comparison to those of the wild type (Fig. 8A) and it could not be cultured from the intestinal tissues of 8 of the 10 infected mice (Fig. 8B). In contrast, the C. jejuni cytC mutant strain showed no detectable colonization defect (P = 0.34 at week 6) in equivalent mixed-infection experiments (data not shown). These results indicate that at least some of the C. jejuni 81-176-specific genes that may enhance its ability to respire under oxygen-limiting conditions can contribute to the ability of this strain to colonize the host and potentially cause disease.
FIG. 8.
Ability of the C. jejuni 81-176 dmsA mutant to colonize mice after mixed oral infection. C. jejuni 81-176 and its dmsA isogenic mutant were simultaneously administered orally to Myd88−/− mice. The numbers of CFU of the two strains in the feces (A) and intestines (B) of the infected animals were determined at different times after infection, as indicated. The numbers of CFU of the wild type (WT) and the dmsA mutant in each mouse are indicated by linked closed and open diamonds, respectively.
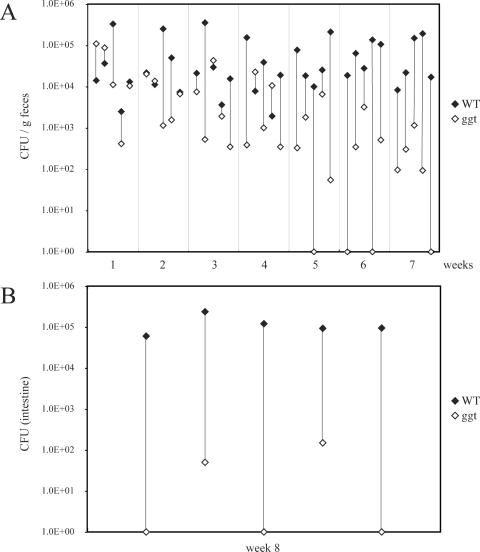
We also tested the potential role of the γ-glutamyltranspeptidase in the pathogenesis of C. jejuni 81-176. An isogenic strain carrying a loss-of-function mutation in ggt was constructed and tested for the ability to grow under microaerophilic and anaerobic conditions, as well as for the ability to enter and survive within intestinal epithelial cells. The C. jejuni ggt::kan mutant strain showed no growth defect and was able to enter and survive within cultured intestinal epithelial cells in a manner indistinguishable from that of the wild type (data not shown). However, when tested for the ability to colonize mice, the ggt mutant showed a very significant defect (Fig. 9). When administered simultaneously with the wild type via the oral route in mixed-infection experiments, both strains were isolated at roughly equal levels from the feces of infected animals during the first week of infection (Fig. 9A). However, over time, larger numbers of wild-type bacteria were isolated from the feces of the infected animals and by week 7, very few numbers of the ggt mutant bacteria were detected in the feces of the inoculated mice (significance of the difference, P = 0.004). The mutant was also detected in much smaller numbers in tissues of animals 8 weeks after infection and was not detected at all in three of the five inoculated animals even though the wild-type strain was recovered from all of the infected animals (significance of the difference, P = 0.004) (Fig. 9B). These results indicate that, similarly to what has been observed for its H. pylori homolog (11, 42), the γ-glutamyltranspeptidase contributes to the virulence of C. jejuni 81-176. Taken together, these results demonstrate that at least some of the genes that are unique to C. jejuni 81-176 contribute to its virulence.
FIG. 9.
Ability of the C. jejuni 81-176 ggt mutant to colonize mice after mixed oral infection. C. jejuni 81-176 and its ggt isogenic mutant were simultaneously administered orally to Myd88−/− mice. The numbers of CFU of the two strains in the feces (A) and intestines (B) of the infected animals were determined at different times after infection, as indicated. The numbers of CFU of the wild type (WT) and the ggt mutant in each mouse are indicated by linked closed and open diamonds, respectively.
Concluding remarks.
Rapid genome comparisons of C. jejuni isolates have been routinely carried out by DNA array (13, 32, 35, 36, 51, 54, 55) and subtractive hybridization (1) approaches. Although powerful, these approaches exhibit some limitations, such as the inability to easily detect deletions or divergent sequences. In addition, only a portion of new, strain-specific genes are often found by using these approaches (1, 54, 55). With the development of new high-throughput sequencing technologies, rapid whole-genome sequencing and comparison of entire bacterial genomes are becoming routine and economically feasible (41, 57). By using a PicoTiterPlate-based approach, we have identified unique genetic attributes of a highly pathogenic strain of C. jejuni. We have identified a number of C jejuni 81-176 genes that may contribute to its unique pathogenic properties. Notably, we have identified several genes that encode electron transport components, as well as alternative respiratory pathways that may confer advantages under oxygen-limiting conditions, such as those encountered within the intestinal tract. We have, also identified genes encoding other metabolic traits and potential virulence factors that may contribute to the virulence of this C. jejuni strain. Given the unique properties of this strain, it is expected that the information presented here will help in the understanding of the general mechanisms of C. jejuni pathogenesis. These studies demonstrate the utility of high-throughput sequencing for the identification of unique genetic attributes of highly variable pathogenic bacteria.
Supplementary Material
Acknowledgments
We thank Maria Lara-Tejero for providing the Myd88−/− mice and members of the Galán laboratory for critical review of the manuscript.
D.H. was supported by an EMBO long-term fellowship. This work was supported by a grant from the Ellison Medical Foundation to J.E.G., who is an Ellison Medical Foundation senior scholar in infectious diseases.
Editor: J. B. Bliska
Footnotes
Supplemental material for this article may by found at http://iai.asm.org/.
REFERENCES
- 1.Ahmed, I. H., G. Manning, T. M. Wassenaar, S. Cawthraw, and D. G. Newell. 2002. Identification of genetic differences between two Campylobacter jejuni strains with different colonization potentials. Microbiology 148:1203-1212. [DOI] [PubMed] [Google Scholar]
- 2.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 3.Altendorf, K., A. Siebers, and W. Epstein. 1992. The KDP ATPase of Escherichia coli. Ann. N. Y. Acad. Sci. 671:228-243. [DOI] [PubMed] [Google Scholar]
- 4.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacon, D. J., R. A. Alm, L. Hu, T. E. Hickey, C. P. Ewing, R. A. Batchelor, T. J. Trust, and P. Guerry. 2002. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect. Immun. 70:6242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769-777. [DOI] [PubMed] [Google Scholar]
- 7.Batchelor, R. A., B. M. Pearson, L. M. Friis, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150:3507-3517. [DOI] [PubMed] [Google Scholar]
- 8.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 9.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 10.Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104-109.10203757 [Google Scholar]
- 11.Chevalier, C., J. M. Thiberge, R. L. Ferrero, and A. Labigne. 1999. Essential role of Helicobacter pylori γ-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol. Microbiol. 31:1359-1372. [DOI] [PubMed] [Google Scholar]
- 12.Dedieu, L., J. M. Pages, and J. M. Bolla. 2004. Use of the omp50 gene for identification of Campylobacter species by PCR. J. Clin. Microbiol. 42:2301-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everest, P. H., H. Goossens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37:319-325. [DOI] [PubMed] [Google Scholar]
- 15.Field, L. H., V. L. Headley, S. M. Payne, and L. J. Berry. 1986. Influence of iron on growth, morphology, outer membrane protein composition, and synthesis of siderophores in Campylobacter jejuni. Infect. Immun. 54:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLOS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry, B. N., S. Feng, Y. Y. Chen, D. G. Newell, P. J. Coloe, and V. Korolik. 2000. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 68:2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galindo, M., W. Day, B. Raphael, and L. Joens. 2001. Cloning and characterization of a Campylobacter jejuni iron-uptake operon. Curr. Microbiol. 42:139-143. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert, M., M. F. Karwaski, S. Bernatchez, N. M. Young, E. Taboada, J. Michniewicz, A. M. Cunningham, and W. W. Wakarchuk. 2002. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277:327-337. [DOI] [PubMed] [Google Scholar]
- 20.Guerry, P., C. P. Ewing, T. E. Hickey, M. M. Prendergast, and A. P. Moran. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68:6656-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerry, P., J. Perez-Casal, R. Yao, A. McVeigh, and T. J. Trust. 1997. A genetic locus involved in iron utilization unique to some Campylobacter strains. J. Bacteriol. 179:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerry, P., C. M. Szymanski, M. M. Prendergast, T. E. Hickey, C. P. Ewing, D. L. Pattarini, and A. P. Moran. 2002. Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 24.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 25.Hu, L., and D. J. Kopecko. 1999. Campylobacter jejuni 81-176 associates with microtubules and dynein during invasion of human intestinal cells. Infect. Immun. 67:4171-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilbert, M., V. Mejean, M. T. Giudici-Orticoni, J. P. Samama, and C. Iobbi-Nivol. 2003. Involvement of a mate chaperone (TorD) in the maturation pathway of molybdoenzyme TorA. J. Biol. Chem. 278:28787-28792. [DOI] [PubMed] [Google Scholar]
- 27.Jack, R. L., G. Buchanan, A. Dubini, K. Hatzixanthis, T. Palmer, and F. Sargent. 2004. Coordinating assembly and export of complex bacterial proteins. EMBO J. 23:3962-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kroger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 29.Jones, M. A., K. L. Marston, C. A. Woodall, D. J. Maskell, D. Linton, A. V. Karlyshev, N. Dorrell, B. W. Wren, and P. A. Barrow. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72:3769-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanipes, M. I., L. C. Holder, A. T. Corcoran, A. P. Moran, and P. Guerry. 2004. A deep-rough mutant of Campylobacter jejuni 81-176 is noninvasive for intestinal epithelial cells. Infect. Immun. 72:2452-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlyshev, A. V., O. L. Champion, C. Churcher, J. R. Brisson, H. C. Jarrell, M. Gilbert, D. Brochu, F. St Michael, J. Li, W. W. Wakarchuk, I. Goodhead, M. Sanders, K. Stevens, B. White, J. Parkhill, B. W. Wren, and C. M. Szymanski. 2005. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 55:90-103. [DOI] [PubMed] [Google Scholar]
- 32.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3(11):0065.1-0065.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 34.Lemieux, M. J., Y. Huang, and D. N. Wang. 2004. Glycerol-3-phosphate transporter of Escherichia coli: structure, function and regulation. Res. Microbiol. 155:623-629. [DOI] [PubMed] [Google Scholar]
- 35.Leonard, E. E., II, T. Takata, M. J. Blaser, S. Falkow, L. S. Tompkins, and E. C. Gaynor. 2003. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates. J. Infect. Dis. 187:691-694. [DOI] [PubMed] [Google Scholar]
- 36.Leonard, E. E., II, L. S. Tompkins, S. Falkow, and I. Nachamkin. 2004. Comparison of Campylobacter jejuni isolates implicated in Guillain-Barre syndrome and strains that cause enteritis by a DNA microarray. Infect. Immun. 72:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Logan, S. M., J. F. Kelly, P. Thibault, C. P. Ewing, and P. Guerry. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587-597. [DOI] [PubMed] [Google Scholar]
- 38.Luneberg, E., E. Glenn-Calvo, M. Hartmann, W. Bar, and M. Frosch. 1998. The central, surface-exposed region of the flagellar hook protein FlgE of Campylobacter jejuni shows hypervariability among strains. J. Bacteriol. 180:3711-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malli, R., and W. Epstein. 1998. Expression of the Kdp ATPase is consistent with regulation by turgor pressure. J. Bacteriol. 180:5102-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGovern, K. J., T. G. Blanchard, J. A. Gutierrez, S. J. Czinn, S. Krakowka, and P. Youngman. 2001. γ-Glutamyltransferase is a Helicobacter pylori virulence factor but is not essential for colonization. Infect. Immun. 69:4168-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, W. G., B. M. Pearson, J. M. Wells, C. T. Parker, V. V. Kapitonov, and R. E. Mandrell. 2005. Diversity within the Campylobacter jejuni type I restriction-modification loci. Microbiology 151:337-351. [DOI] [PubMed] [Google Scholar]
- 44.Morooka, T., A. Umeda, and K. Amako. 1985. Motility as an intestinal colonization factor for Campylobacter jejuni. J. Gen. Microbiol. 131:1973-1980. [DOI] [PubMed] [Google Scholar]
- 45.Myers, J. D., and D. J. Kelly. 2005. A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology 151:233-242. [DOI] [PubMed] [Google Scholar]
- 46.Nachamkin, I. 2002. Chronic effects of Campylobacter infection. Microbes Infect. 4:399-403. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura, T., R. Yuda, T. Unemoto, and E. P. Bakker. 1998. KtrAB, a new type of bacterial K+-uptake system from Vibrio alginolyticus. J. Bacteriol. 180:3491-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oelschlaeger, T. A., P. Guerry, and D. J. Kopecko. 1993. Unusual microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc. Natl. Acad. Sci. USA 90:6884-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 51.Pearson, B. M., C. Pin, J. Wright, K. I'Anson, T. Humphrey, and J. M. Wells. 2003. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554:224-230. [DOI] [PubMed] [Google Scholar]
- 52.Pickett, C. L., T. Auffenberg, E. C. Pesci, V. L. Sheen, and S. S. Jusuf. 1992. Iron acquisition and hemolysin production by Campylobacter jejuni. Infect. Immun. 60:3872-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pittman, M. S., and D. J. Kelly. 2005. Electron transport through nitrate and nitrite reductases in Campylobacter jejuni. Biochem. Soc. Trans. 33:190-192. [DOI] [PubMed] [Google Scholar]
- 54.Poly, F., D. Threadgill, and A. Stintzi. 2005. Genomic diversity in Campylobacter jejuni: identification of C. jejuni 81-176-specific genes. J. Clin. Microbiol. 43:2330-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poly, F., D. Threadgill, and A. Stintzi. 2004. Identification of Campylobacter jejuni ATCC 43431-specific genes by whole microbial genome comparisons. J. Bacteriol. 186:4781-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prendergast, M. M., D. R. Tribble, S. Baqar, D. A. Scott, J. A. Ferris, R. I. Walker, and A. P. Moran. 2004. In vivo phase variation and serologic response to lipooligosaccharide of Campylobacter jejuni in experimental human infection. Infect. Immun. 72:916-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogers, Y. H., and J. C. Venter. 2005. Genomics: massively parallel sequencing. Nature 437:326-327. [DOI] [PubMed] [Google Scholar]
- 58.Russell, R. G., M. J. Blaser, J. I. Sarmiento, and J. Fox. 1989. Experimental Campylobacter jejuni infection in Macaca nemestrina. Infect. Immun. 57:1438-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambasivarao, D., and J. H. Weiner. 1991. Dimethyl sulfoxide reductase of Escherichia coli: an investigation of function and assembly by use of in vivo complementation. J. Bacteriol. 173:5935-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sellars, M. J., S. J. Hall, and D. J. Kelly. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shibayama, K., K. Kamachi, N. Nagata, T. Yagi, T. Nada, Y. Doi, N. Shibata, K. Yokoyama, K. Yamane, H. Kato, Y. Iinuma, and Y. Arakawa. 2003. A novel apoptosis-inducing protein from Helicobacter pylori. Mol. Microbiol. 47:443-451. [DOI] [PubMed] [Google Scholar]
- 62.Smith, M. A., G. L. Mendz, M. A. Jorgensen, and S. L. Hazell. 1999. Fumarate metabolism and the microaerophily of Campylobacter species. Int. J. Biochem. Cell Biol. 31:961-975. [DOI] [PubMed] [Google Scholar]
- 63.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 64.Tholema, N., M. Vor der Bruggen, P. Maser, T. Nakamura, J. I. Schroeder, H. Kobayashi, N. Uozumi, and E. P. Bakker. 2005. All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. J. Biol. Chem. 280:41146-41154. [DOI] [PubMed] [Google Scholar]
- 65.van Vliet, A. H., J. M. Ketley, S. F. Park, and C. W. Penn. 2002. The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26:173-186. [DOI] [PubMed] [Google Scholar]
- 66.Velayudhan, J., and D. J. Kelly. 2002. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: an essential role for phosphoenolpyruvate carboxykinase. Microbiology 148:685-694. [DOI] [PubMed] [Google Scholar]
- 67.Wagner, D., J. Maser, B. Lai, Z. Cai, C. E. Barry III, K. Honer Zu Bentrup, D. G. Russell, and L. E. Bermudez. 2005. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J. Immunol. 174:1491-1500. [DOI] [PubMed] [Google Scholar]
- 68.Wassenaar, T. M., and M. J. Blaser. 1999. Pathophysiology of Campylobacter jejuni infections of humans. Microbes Infect. 1:1023-1033. [DOI] [PubMed] [Google Scholar]
- 69.Woodall, C. A., M. A. Jones, P. A. Barrow, J. Hinds, G. L. Marsden, D. J. Kelly, N. Dorrell, B. W. Wren, and D. J. Maskell. 2005. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect. Immun. 73:5278-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao, R., D. H. Burr, and P. Guerry. 1997. CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol. 23:1021-1031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.