Abstract
Human-pathogenic bacteria that are able to cause persistent infections must have developed mechanisms to resist the immune defense system. Lysozyme, a cell wall-lytic enzyme, is one of the first defense compounds induced in serum and tissues after the onset of infection. Recently, we showed that Staphylococcus aureus is resistant to lysozyme by O acetylating its peptidoglycan (PG) by O-acetyltransferase (OatA). We asked the question of which staphylococcal species PG is O acetylated. We applied various methods, such as genome analysis, PCR, Southern blotting, lysozyme sensitivity assay, and verification of O acetylation of PG by high-performance liquid chromatography (HPLC) analysis. PCR analysis using S. aureus-derived oatA primers and Southern blotting did not yield reliable results with other staphylococcal species. Therefore, we used the HPLC-based assay to directly detect PG O acetylation. Our studies revealed that the muramic acid was O acetylated only in pathogenic, lysozyme-resistant staphylococci (e.g., S. aureus, S. epidermidis, S. lugdunensis, and others). All nonpathogenic species were lysozyme sensitive. They can be divided into sensitive species (e.g., S. carnosus, S. gallinarum, and S. xylosus) and hypersensitive species (e.g., S. equorum, S. lentus, and S. arlettae). In all lysozyme-sensitive species, the analyzed PG was de-O-acetylated. When we transformed the oatA gene from lysozyme-resistant S. aureus into S. carnosus, the corresponding transformants also became lysozyme resistant.
Staphylococci are commonly widespread as natural commensals of humans and warm-blooded animals. The largest populations are usually found in regions of the skin, such as mucosal surfaces surrounding openings in the body surface (e.g., Staphylococcus aureus colonizes anterior nares in >30% of the human population) (1, 33). Currently, 36 species and 17 subspecies are recognized in the genus Staphylococcus, and about one-half of these are indigenous to humans (22). Staphylococci are also opportunistic and adaptable pathogens with the ability to infect, invade, persist, and replicate in any human tissue, including skin, bone, visceral organs, or vasculature (19, 33). Formerly, coagulase-negative staphylococci were considered to be saprophytic or mildly pathogenic for humans. However, several species of coagulase-negative staphylococci, such as S. epidermidis (20, 24, 26), S. saprophyticus (32), S. haemolyticus (3), S. lugdunensis (49, 52), and S. warneri (53), have now been shown to be opportunistic human pathogens.
Infections caused by coagulase-negative staphylococci are commonly associated with implanted devices, and the ability to form a biofilm promotes infection (20, 21). Some nonpathogenic staphylococci are food-borne bacteria. Two examples are S. equorum (autochthonous strain of the red smear cheese surface) (10, 37) and S. arlettae (autochthonous strain growing on dried, salted cod) (50); they have never been isolated from human skin or in connection with infections. S. xylosus (5, 35, 48), S. pulvereri (5), S. succinus (43), S. pasteuri (43), and S. equorum (5, 35) were found to be dominant on traditional fermented meat and on the surface of ripened cheese but have also been isolated from goat milk (48). For more than 50 years, nonpathogenic staphylococci, such as S. carnosus (8, 45, 46, 54), S. equorum (10, 42, 45), S. sciuri (7, 44), S. xylosus (25, 36, 45), and S. pulvereri (8), have been used as starter cultures in fermentation and biopreservation of food (meat, cheese, and probiotics). Since pathogenic staphylococci colonize the host over a longer period and cause chronic infections and are exposed to all tissue bactericides that kill or inhibit the growth of microbes, they must have developed mechanisms that evade lysozyme defense. It is very remarkable that more than 80 years ago, Alexander Fleming discovered lysozyme and proposed that nonpathogenic microorganisms fail to cause disease, because they are very sensitive to lysozyme-mediated killing (17). Lysozyme is a component of granules of neutrophils and the major secretory product of macrophages, found in mammalian secretions and tissues; and undoubtedly one of the principal important enzymes of our innate immune system (16, 27). Although lysozyme preferentially hydrolyzes the β-1,4-glycosidic linkages between N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) (6), it does not recognize peptidoglycan (PG) modified with O-acetyl groups that enables bacteria such as S. aureus and other pathogenic staphylococci to overcome the innate defense system. S. aureus acetylates its cell wall at the C-6 position of N-acetylmuramic acid producing the 2,6-N,O-diacetylmuramic acid derivative (4). This modification acts as a steric hindrance and inhibits the binding of the lysozyme to the polysaccharide substrate. We have previously provided evidence that the O acetylation of the PG correlates with the observed high lysozyme resistance by S. aureus and that it is mediated by a PG-specific, membrane-bound O-acetyltransferase (OatA) (4). In this study, we show that OatA is widespread only among pathogenic staphylococci, which is typical for a virulence factor. Moreover, we show that nonpathogenic staphylococci are lysozyme sensitive and possess no O-acetylated PG.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains were used in the lysozyme diffusion-based assay and in the high-performance liquid chromatography (HPLC)-based assay. The staphylococcal strains used were as follows: S. arlettae DSM20672, S. aureus SA113 (ATCC 35556), S. aureus SA113 ΔoatA::km, S. auricularis ATCC 33753, S. capitis subsp. capitis ATCC 27840, S. caprae DSM20608, S. carnosus TM300, S. chromogenes DSM20454, S. cohnii subsp. cohnii CCM2734, S. condimenti DSM11684, S. delphini DSM20771, S. epidermidis ATCC 14990, S. equorum subsp. equorum DSM20674, S. equorum subsp. linens DSM15097, S. gallinarum DSM20610, S. haemolyticus CCM2737, S. hominis subsp. hominis DSM20328, S. hyicus subsp. hyicus NCTC103350, S. intermedius CCM2734, S. kloosii DSM20676, S. lugdunensis ATCC 43809, S. muscae DSM7068, S. pasteuri ATCC 51129, S. piscifermentans SKO3, S. pulvereri DSM9930, S. saccharolyticus DSM20359, S. saprophyticus DSM20229, S. sciuri subsp. lentus DSM20352, S. schleiferi subsp. schleiferi DSM4807, S. simulans subsp. simulans ATCC 27848, S. succinus subsp. casei DSM15096, S. succinus subsp. succinus DSM14617, S. vitulinus DSM15615, S. warneri DSM20316, and S. xylosus DSM20266. Macrococcus caseolyticus DSM20597 and Micrococcus luteus ATCC 9341 were also used.
Bacteria were routinely grown under standard conditions in B medium (BM) (1% tryptone [Gibco BRL Life Technologies GmbH, Eggenstein, Germany], 0.5% yeast extract [Gibco BRL], 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose) or tryptic soy broth (Difco). Media were supplemented when appropriate with tetracycline (12.5 μg/ml) unless noted otherwise. The pTX15 vector (staphylococcal plasmid with xylose-inducible promoter) was used for cloning and expression of the oatA gene in S. carnosus (40).
Construction of plasmids, homologous recombination, and complementation.
Standard methods and vectors were used for PCR amplification, cloning, sequencing, in vitro recombination, and Southern blotting. DNA from S. aureus SA113 was amplified by PCR with Deep Vent polymerase (New England Biolabs) according to the instructions of the supplier. Primers were obtained from MWG-Biotech (Ebersberg, Germany). The PCR product was cloned into the polylinker (BamHI-EcoRI) of the pTX15 vector (40), upstream of the promoter region. Restriction enzymes were purchased from Gibco BRL, Boehringer, or New England Biolabs GmbH (Schwalbach, Germany). Plasmid DNA was introduced into S. carnosus by protoplast transformation. Plasmid DNA and chromosomal DNA were sequenced using a LI-COR DNA sequencer Long Reader (Lincoln Corporation, Inc., Lincoln, Neb.). Computer sequence analysis was performed with MacDNASIS Pro (Hitachi Software Engineering, San Bruno, Calif.).
Lysozyme agar diffusion-based assay.
The growth inhibition that is caused by lysozyme was tested on tryptic soy agar (TSA) plates. Overnight cultures of the bacteria were diluted in TSA soft agar to 0.5 × 106 CFU per ml and poured on TSA plates. In the agar, 0.5-mm-diameter wells were cut out, and in each well, 4 mg of lysozyme (concentration of 200 mg/ml, suspended in water, sterile filtered) was added. After overnight incubation at 37°C, the growth inhibition was measured.
Highly purified cell wall preparation.
Staphylococci were inoculated in 1,000 ml of liquid BM and incubated at 37°C until the optical density at 578 nm (OD578) reached 0.4 to 0.6. Cultures were swirled in ice-alcohol baths and then centrifuged at 6,000 rpm for 20 min at 4°C. The cells were washed once with ice-cold 100 mM Tris-HCl (pH 6.8) and resuspended in 15 ml of the same buffer. The cells were added dropwise to 45 ml of boiling 5% sodium dodecyl sulfate (SDS) buffered with 100 mM Tris-HCl (pH 6.8) and boiled for a further 30 min. After the solution was cooled to room temperature, the SDS-insoluble material was collected by centrifugation at 10,000 rpm for 15 min at 20°C. The pellet was washed five times with warm water until no more SDS could be detected by the method of Hayashi (23). Cells were resuspended in 1 to 2 ml of water and broken with glass beads (150 to 210 microns) in a Fast Prep FP120 machine. Nonbroken cells were removed by low-speed centrifugation at 1,000 rpm for 5 min. The collected broken cell walls were centrifuged at 15,000 rpm for 30 min at 20°C. The pellet was resuspended in 100 mM Tris-HCl (pH 7.5) with the addition of 20 mM MgCl2, DNase, and RNase. DNase and RNase (Sigma Chemical Co., St. Louis, Mo.) were added at a concentration of 10 μg/ml and 50 μg/ml, respectively, and the mixture was incubated for 2 h at 37°C. The peptidoglycan-associated proteins were removed by overnight incubation at 37°C with 50 μg/ml of trypsin in the presence of 20 mM CaCl2. The SDS-insoluble material was reextracted with boiling 1% SDS in 100 mM Tris-HCl (pH 7) for 20 min. The material was collected and washed with water by centrifugation four times as described above. The PG pellet was treated with 8 mM LiCl for 15 min at 41°C, collected by centrifugation at 15,000 rpm for 15 min at 20°C, and resuspended in 100 mM EDTA (pH 7.0) for 15 min at 41°C. The pellet was washed two times with water, lyophilized, and stored at 4°C.
Analysis of O-acetyl groups of peptidoglycan by HPLC.
The range of O acetylation of the isolated peptidoglycan from lysozyme-resistant and lysozyme-sensitive staphylococci was analyzed. PG (20 mg in 1 ml) was incubated for 3 h with 80 mM NaOH at 37°C with shaking to hydrolyze the O-acetyl groups. For a negative control, PG of S. aureus SA113 was incubated for 3 h with 80 mM phosphate-buffered saline (pH 6.4) at 37°C with shaking (11, 15). The quantitation of released acetic acid was measured by HPLC, with an organic acid column HPX-87H (Bio-Rad) under the same conditions as for the acetic acid standard (Bio-Rad) and according to the instructions of the suppliers. The samples of the filtered supernatant (80 μl) were injected into the column and eluted with 0.008 M sulfuric acid at a flow rate of 0.6 ml min−1 and at 35°C.
RESULTS
Lysozyme resistance among staphylococci.
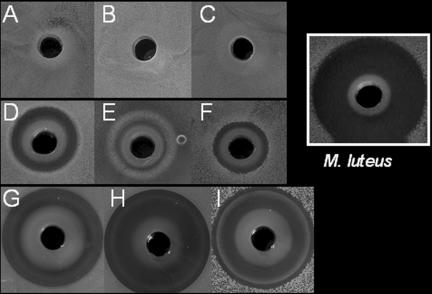
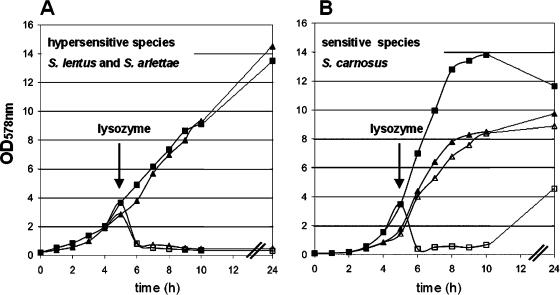
A lysozyme sensitivity plate assay revealed that not all species of the Staphylococcaceae family are lysozyme resistant. Susceptibility of 34 staphylococcal species against lysozyme was tested in the agar diffusion-based assay. We observed that representative pathogenic species were all resistant to lysozyme, while representative nonpathogenic species were sensitive to lysozyme. The lysozyme-sensitive staphylococci can be grouped into sensitive and hypersensitive species: for example, S. carnosus, S. xylosus, and S. gallinarum were sensitive species, whereas S. arlettae, S. equorum, and S. lentus were hypersensitive (Fig. 1 and Table 1). These hypersensitive species were as sensitive against lysozyme as Micrococcus luteus was, which we always use as a control strain (Fig. 1). For a further investigation of lysozyme resistance, characteristic members of each group were chosen to determine the effect of lysozyme on growth in the liquid culture. In a cell lysis assay, lysozyme (final concentration of 300 μg/ml) was added at the exponential growth phase, and its effect on cell density (cell lysis) was monitored. While the optical density of S. aureus was completely unaffected by the presence of lysozyme (data not shown), it decreases rapidly in S. lentus, S. arlettae (Fig. 2A), and S. carnosus (Fig. 2B) cultures, indicating continuous cell lysis. Hypersensitive species lysed almost immediately after the addition of lysozyme, and the cell density measured after 24 h stayed at an OD578 of 0.3. With the S. carnosus culture, the addition of lysozyme also led to rapid cell lysis, but the cell density measured after 24 h increased again to an OD578 of 4.8. In the absence of lysozyme, no cell lysis was observed with any of the strains.
FIG. 1.
Agar diffusion-based assay with 4 mg of lysozyme. (A) S. lugdunensis, (B) S. epidermidis, (C) S. haemolyticus, (D) S. carnosus, (E) S. gallinarum, (F) S. xylosus, (G) S. lentus, (H) S. equorum, and (I) S. arlettae. Growth inhibition caused by lysozyme was tested on TSA plates. Overnight cultures were diluted in TSA soft agar to 0.5 × 106 CFU/ml−1 and poured onto TSA plates. Lysozyme was applied into wells (diameter, 5 mm). Plates were incubated overnight at 37°C. Lysozyme formed white, characteristic precipitation zone. Micrococcus luteus is shown as an example of a lysozyme-hypersensitive bacterium.
TABLE 1.
Distribution of lysozyme resistance and O acetylation of peptidoglycan in pathogenic and nonpathogenic staphylococcal species
| Species | Lysozyme resistancea | Acetate releaseb |
|---|---|---|
| Pathogenic species | ||
| S. aureus ATCC 35556 | R | + |
| S. auricularis ATCC 33753 | R | ND |
| S. capitis ATCC 27840 | R | ND |
| S. caprae DSM20608 | R | ND |
| S. chromogenes DSM20454 | R | ND |
| S. delphini DSM20771 | R | ND |
| S. epidermidis ATCC 14990 | R | + |
| S. haemolyticus CCM2737 | R | + |
| S. hominis subsp. hominis DSM20328 | R | ND |
| S. hyicus subsp. hyicus NCTC103350 | R | + |
| S. intermedius CCM2734 | R | ND |
| S. lugdunensis ATCC 43809 | R | + |
| S. muscae DSM7068 | R | ND |
| S. pasteuri ATCC 51129 | R | ND |
| S. saccharolyticus DSM20359 | R | + |
| S. saprophyticus DSM20229 | R | + |
| S. warneri DSM20316 | R | ND |
| Macrococcus caseolyticus DSM20597c | R | + |
| Nonpathogenic species | ||
| S. arlettae DSM20672 | S | − |
| S. carnosus TM300 | S | − |
| S. cohnii subsp. cohnii CCM2734 | S | ND |
| S. condimenti DSM11684 | S | ND |
| S. equorum subsp. equorum DSM20674 | S | − |
| S. equorum subsp. linens DSM15097 | S | ND |
| S. gallinarum DSM20610 | S | ND |
| S. kloosii DSM20676 | S | ND |
| S. lentus DSM20352 | S | − |
| S. piscifermentans SKO3 | S | ND |
| S. pulvereri DSM9930 | S | ND |
| S. sciuri subsp. lentus DSM20352 | S | ND |
| S. schleiferi subsp. schleiferi DSM4807 | S | ND |
| S. simulans subsp. simulans ATCC 27848 | S | ND |
| S. succinus subsp. casei DSM15096 | S | ND |
| S. succinus subsp. succinus DSM14617 | S | ND |
| S. vitulinus DSM15615 | S | ND |
| S. xylosus DSM20266 | S | ND |
| Micrococcus luteus ATCC 9341 | S | ND |
Lysozyme resistance (R) or sensitivity (S) was determined by the agar diffusion assay.
Acetate release was determined by treating purified PG with alkaline, and the released acetic acid was determined by HPLC. Symbols: +, O acetylation; −, de-O-acetylation. ND, not determined.
Macrococcus caseolyticus can be isolated from abscesses of lambs and until recently was classified as Staphylococcus caseolyticus.
FIG. 2.
Growth curve and lysozyme-induced cell lysis. Overnight cultures were inoculated into fresh BM broth. After 5 h of incubation, lysozyme was added at a concentration of 300 μg/ml (indicated by an arrow). Incubation continued for another 19 h. (A) S. lentus alone ▪ and with lysozyme □ and S. arlettae alone ▴ and with lysozyme ▵ are shown. (B) S. carnosus alone ▪ and with lysozyme □ and S. carnosus carrying pTX15-oatA in the absence ▴ and presence of lysozyme ▵ are shown.
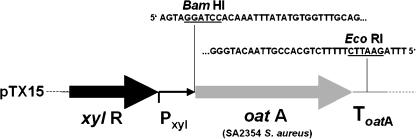
We have shown previously that OatA of S. aureus catalyzes the O acetylation of the PG, thus rendering it lysozyme resistant. In this study, we expressed oatA in S. carnosus via the xylose-inducible promoter of pTX15 (Fig. 3). The tetracycline-resistant transformants of S. carnosus were also lysozyme resistant, as shown in the liquid culture-based assay (Fig. 2B). The cell density of S. carnosus(pTX15-oatA) culture was monitored without and with the addition of lysozyme (final concentration of 300 μg/ml). After the addition of lysozyme, the cell density decreased, indicating cell lysis. S. carnosus(pTX15-oatA) grows poorly compared to the wild type. One explanation is that OatA is a rather hydrophobic protein with 10 predicted transmembrane domains. Moreover, overexpression by induction with xylose may cause membrane jamming, which is known to be associated with toxicity and eventual cell death (2). Nevertheless, by expressing oatA, S. carnosus became lysozyme resistant (Fig. 2B).
FIG. 3.
Fragment of pTX15 encoding xylose-inducible oatA. Sequences up- and downstream of the oatA gene are also shown. ToatA, terminator region.
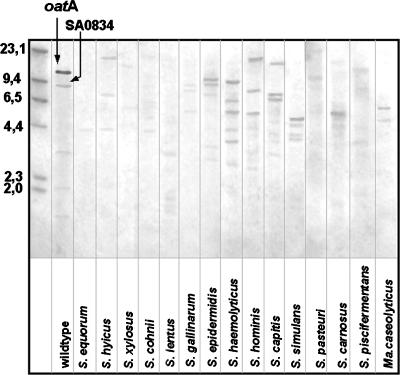
In order to search for oatA homologues among several Staphylococcus species, we used PCR (with primers derived from S. aureus-specific oatA) and Southern blotting method. No specific PCR products were detected in S. equorum, S. arlettae, S. lentus, or S. carnosus. However, in S. epidermidis, S. lugdunensis, S. haemolyticus, S. saprophyticus, S. hyicus, and S. warneri, specific products for oatA were detected. Additionally, our results confirmed that there are paralogues of oatA in the staphylococcal genomes. Sequencing and comparing the S. aureus OatA-derived protein sequence with those from other species (30) revealed homologues from S. epidermidis ATCC 12228 (55), S. epidermidis RP62A (18), S. saprophyticus ATCC 15305 (31), and S. haemolyticus JCSC1435 (47) with 64%, 64%, 56%, and 55% identity, respectively. Consistent with this result, PG from S. epidermidis, S. haemolyticus, and S. saprophyticus was found to be O acetylated. Southern blotting of EcoRI-digested chromosomal DNA from several Staphylococcus species probed with S. aureus oatA at various stringencies revealed no clear results. The homology of oatA at the DNA sequence level between different staphylococci species is low (45% to 30%), and staphylococcal genomes differ markedly in EcoRI restriction pattern, which makes it particularly difficult to obtain oatA-specific signals (Fig. 4). Furthermore, even when signals are strong and seem to be specific, it is difficult to distinguish the oatA gene from its paralogues.
FIG. 4.
Southern blot showing oatA homologues detected in staphylococcal species. EcoRI-digested chromosomal DNA was probed with oatA from S. aureus SA113 (wild type). Molecular sizes (in kilobases) of HindIII-digested phage λ DNA are shown to the left of the blot. The oatA signal and SA0834, the paralogue of oatA (56% identity with oatA at the DNA sequence level), are indicated by arrows. Ma. caseolyticus, Macrococcus caseolyticus.
Comparative analysis of peptidoglycan from different staphylococci species.
It is commonly known that staphylococcal PG possesses a pentapeptide bridge, which is cleavable by lysostaphin if it is composed essentially of pentaglycine such as that of S. aureus and S. carnosus for example. The majority of staphylococci have a pentapeptide bridge that contains one or two serine residues instead of glycine (28, 29), which makes them more resistant against lysostaphin. This is the reason why it was difficult to obtain monomeric muropeptides by lysostaphin/cellosyl digestion of PG with these species, a method that was useful with S. aureus and S. carnosus (4).
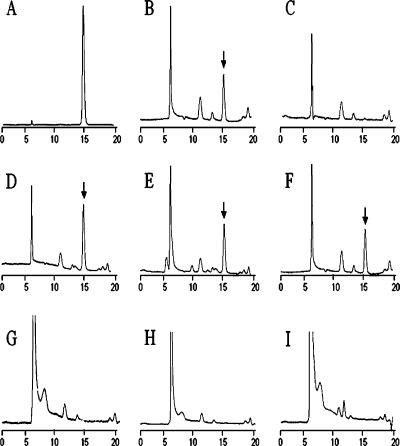
For determining whether the C-6 position of N-acetylmuramic acid was O acetylated, we purified PG from 10 species, carried out alkaline hydrolysis (80 mM NaOH) of the O-acetyl groups, and determined the released acetic acid by HPLC-muropeptide analysis as described previously (4). Pathogenic, lysozyme-resistant species produced significant acetate and exhibited a strong acetate peak in contrast to nonpathogenic, lysozyme-sensitive staphylococci (Table 1 and Fig. 5). The peptidoglycan of Macrococcus caseolyticus, isolated from abscesses of lambs, which until recently was classified as Staphylococcus caseolyticus (22), is also found to be O acetylated (Table 1). The O-acetylated peptidoglycan of wild-type S. aureus served as a positive control (Fig. 5B), and the completely de-O-acetylated peptidoglycan of the oatA deletion mutant (4) served as a negative control (Fig. 5C).
FIG. 5.
HPLC-based detection of O-linked acetate from the peptidoglycan of staphylococci. The acetate peak (retention time, 15.2 min) is indicated by a black arrow. (A) Acetate (40 μmol/ml), (B) S. aureus SA113, (C) oatA deletion mutant, (D) S. epidermidis, (E) S. lugdunensis, (F) S. hyicus, (G) S. carnosus, (H) S. equorum, and (I) S. arlettae.
DISCUSSION
For a long time, it was believed that all staphylococci are lysozyme resistant. Here we show that this is not the case. The O acetylation of the PG was found only in pathogenic staphylococci and in Macrococcus caseolyticus, which is also a member of the Staphylococcaceae family (22, 29). The resistance to lysozyme of S. epidermidis, an opportunistic pathogen, may contribute to its ability to colonize the skin and mucous membranes and its role in chronic, implant-associated infections (20, 24, 26). S. lugdunensis, which also possesses highly O-acetylated peptidoglycan, previously was isolated only from healthy human skin but is now regarded as an opportunistic pathogen implicated in endocarditis, septicemia, chronic osteoarthritis, catheter-associated infections, and infections of soft tissues, bone, and peritoneal fluid (49, 52). S. hyicus, a lysozyme-resistant, opportunistic pathogen of animals, has been involved in infectious exudative epidermitis and septic polyarthritis of pigs, skin lesions in cattle and horses, and osteomyelitis in poultry and cattle (13, 41). This species is also rarely isolated from healthy human skin (18). S. saprophyticus, S. haemolyticus, and Macrococcus caseolyticus have less O-acetylated peptidoglycan than the species described above. S. saprophyticus is frequently found in inguinal and perineal regions of the human body and may serve as the source for inoculation of the urinary tract, leading to urinary tract infections, especially in young women and very rarely in men. This opportunistic pathogen has occasionally been isolated from wound infections and septicemia (32). S. haemolyticus has been isolated frequently from serious human infections, especially from native valve endocarditis but also from septicemia, peritonitis, and urinary tract infections and it is occasionally associated with wound infections (3). Macrococcus caseolyticus was isolated from cattle, sheep, and goats and may be found in their milk and meat products. In only one case, this species was associated with infection as an etiologic agent of the abscesses of slaughtered lambs (38). Lysozyme-resistant S. warneri, S. hominis, and S. capitis constitute the predominant human skin microflora together with S. epidermidis (18), and the O acetylation of the cell wall helps them to survive on the host. Under favorable conditions, however, they can overcome the innate immune system and may cause infection. S. hominis, which predominates on the dry, glabrous skin of arms, legs, and trunk, has been involved in endophthalmitis, rhinitis, endocarditis, peritonitis, septicemia, and arthritis (14, 34). S. warneri, rarely associated with infection, has been occasionally noted as an etiologic agent of osteomyelitis, valve endocarditis, bacteremia, and urinary tract infections (9, 53). S. capitis has been associated with bacteremia and infective endocarditis (12, 51).
Nonpathogenic staphylococci involved in food fermentation, e.g., S. carnosus, S. xylosus, S. equorum, S. arlettae, S. condimenti, and S. piscifermentans, are lysozyme sensitive and possess de-O-acetylated peptidoglycan. For a long time, S. carnosus, S. xylosus, and S. equorum have been used as starter cultures for the production of raw fermented sausages and hams (45). S. piscifermentans, S. condimenti, and S. carnosus are important species in the production of fish sauce, and the cheese industry employs S. succinus subsp. casei, S. equorum, and S. xylosus as starter cultures in red smear cheese production. The staphylococci isolated from heavily salted meat, fish, or cheese have unusual properties that have never been observed for other staphylococci. S. equorum and S. arlettae can tolerate 15% NaCl and display clear growth even at 4°C; however, S. equorum grows slowly even under optimal growth conditions (25 to 32°C) (50). S. carnosus and S. xylosus are also much more halotolerant than other staphylococci (45). Moreover, none of the known virulence factors described for S. aureus, such as hyaluronidase, DNase, enterotoxins (staphylococcal enterotoxins A to E), toxic shock syndrome toxin 1, exfoliative toxins (exfoliative toxins A and B), coagulase, lectinase, protein A, elastase, hemolysins, capsules, and PG-specific O-acetyltransferase (this study) was found in S. equorum and in S. arlettae (39). These species are also sensitive against all antibiotics that are fully ineffective in treatment of S. aureus infections (39). The lysozyme-hypersensitive species S. lentus, which is commonly isolated from the skin and udders of goats and sheep, and S. arlettae, which is infrequently isolated from the skin of goats and poultry, have not been found associated with human or animal infections.
It is becoming apparent that staphylococci have developed a variety of strategies during evolution to survive as human commensals and paradoxically also as human pathogens. OatA is a factor that enables staphylococci to live on human skin and mucous membranes rich in lysozyme, but it is also a virulence factor, which can be useful during staphylococcal invasion, persistence, and infection. OatA is the major factor responsible for lysozyme resistance in pathogenic staphylococci. The finding of lysozyme-hypersensitive species suggests that other factors beside oatA contribute to lysozyme resistance. There are two additional factors that may play an important role in lysozyme resistance: the percentage of wall teichoic acids bound to the C-6 group of the N-acetylmuramic acid and the degree of cross-linking in the murein.
Acknowledgments
We thank Ralf Rosenstein for providing sequence information on the S. carnosus genome.
This work was supported by the DFG Graduate College “Infection Biology” and Forschergruppe (FOR 449/1).
Editor: J. L. Flynn
REFERENCES
- 1.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 2.Baneyx, F., and M. Mujacic. 2004. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 22:1399-1408. [DOI] [PubMed] [Google Scholar]
- 3.Bartoszewicz, M., J. Nowicka, and A. Przondo-Mordarska. 2003. Selected features determine pathogenicity of Staphylococcus haemolyticus. Med. Dosw. Mikrobiol. 55:225-229. [PubMed] [Google Scholar]
- 4.Bera, A., S. Herbert, A. Jakob, W. Vollmer, and F. Gotz. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55:778-787. [DOI] [PubMed] [Google Scholar]
- 5.Blaiotta, G., C. Pennacchia, F. Villani, A. Ricciardi, R. Tofalo, and E. Parente. 2004. Diversity and dynamics of communities of coagulase-negative staphylococci in traditional fermented sausages. J. Appl. Microbiol. 97:271-284. [DOI] [PubMed] [Google Scholar]
- 6.Blake, C. C., L. N. Johnson, G. A. Mair, A. C. North, D. C. Phillips, and V. R. Sarma. 1967. Crystallographic studies of the activity of hen egg-white lysozyme. Proc. R. Soc. Lond. B 167:378-388. [DOI] [PubMed] [Google Scholar]
- 7.Bockelmann, W., T. Hoppe Seyler, U. Krusch, W. Hoffmann, and K. J. Heller. 1997. The microflora of Tilsit cheese. Part 2. Development of a surface smear starter culture. Nahrung 41:213-218. [Google Scholar]
- 8.Bover-Cid, S., M. Izquierdo-Pulido, and M. C. Vidal-Carou. 2000. Mixed starter cultures to control biogenic amine production in dry fermented sausages. J. Food Prot. 63:1556-1562. [DOI] [PubMed] [Google Scholar]
- 9.Buttery, J. P., M. Easton, S. R. Pearson, and G. G. Hogg. 1997. Pediatric bacteremia due to Staphylococcus warneri: microbiological, epidemiological, and clinical features. J. Clin. Microbiol. 35:2174-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carnio, M. C., T. Stachelhaus, K. P. Francis, and S. Scherer. 2001. Pyridinyl polythiazole class peptide antibiotic micrococcin P1, secreted by foodborne Staphylococcus equorum WS2733, is biosynthesized nonribosomally. Eur. J. Biochem. 268:6390-6401. [DOI] [PubMed] [Google Scholar]
- 11.Clarke, A. J. 1993. Extent of peptidoglycan O acetylation in the tribe Proteeae. J. Bacteriol. 175:4550-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cone, L. A., E. M. Sontz, J. W. Wilson, and S. N. Mitruka. 2005. Staphylococcus capitis endocarditis due to a transvenous endocardial pacemaker infection: case report and review of Staphylococcus capitis endocarditis. Int. J. Infect. Dis. 9:335-339. [DOI] [PubMed] [Google Scholar]
- 13.Devriese, L. A., and J. Derycke. 1979. Staphylococcus hyicus in cattle. Res. Vet. Sci. 26:356-358. [PubMed] [Google Scholar]
- 14.Dudkiewicz, B., and E. Szewczyk. 1993. Etiology of bacterial endocarditis in materials from Cardiology and Cardiac Surgery Clinics of the Lodz Academy. Med. Dosw. Mikrobiol. 45:357-359. [PubMed] [Google Scholar]
- 15.Dupont, C., and A. J. Clarke. 1991. Evidence for N→O acetyl migration as the mechanism for O acetylation of peptidoglycan in Proteus mirabilis. J. Bacteriol. 173:4318-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahlgren, A., S. Hammarstrom, A. Danielsson, and M. L. Hammarstrom. 2003. Increased expression of antimicrobial peptides and lysozyme in colonic epithelial cells of patients with ulcerative colitis. Clin. Exp. Immunol. 131:90-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming, A. 1922. On a remarkable bacteriolytic element found in tissues and secretions. Proc. R. Soc. B 93:306-317. [Google Scholar]
- 18.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillet, Y., B. Issartel, P. Vanhems, J. C. Fournet, G. Lina, M. Bes, F. Vandenesch, Y. Piemont, N. Brousse, D. Floret, and J. Etienne. 2002. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753-759. [DOI] [PubMed] [Google Scholar]
- 20.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 21.Götz, F., and G. Peters. 2000. Colonization of medical devices by coagulase-negative staphylococci, p. 55-88. In F. A. Waldvogel and A. L. Bisno (ed.), Infections associated with indwelling medical devices, 3rd ed. ASM Press, Washington, D.C.
- 22.Götz, F., T. Bannerman, and K. H. Schleifer. 2004. The genera Staphylococcus and Macrococcus, vol. 3.17. Springer, New York, N.Y.
- 23.Hayashi, K. 1975. A rapid determination of sodium dodecyl sulfate with methylene blue. Anal. Biochem. 67:503-506. [DOI] [PubMed] [Google Scholar]
- 24.Heilmann, C., and G. Peters. 2000. Biology and pathogenicity of Staphylococcus epidermidis. ASM Press, Washington, D.C.
- 25.Hugas, M., and M. Roca. 1997. The selection of Staphylococcus spp. autochthonous strains as starter culture in slightly fermented meat sausages. Eurocarne 54:45-47. (In Spanish.) [Google Scholar]
- 26.Jarvis, W. R., and W. J. Martone. 1992. Predominant pathogens in hospital infections. J. Antimicrob. Chemother. 29(Suppl. A):19-24. [DOI] [PubMed] [Google Scholar]
- 27.Keshav, S., P. Chung, G. Milon, and S. Gordon. 1991. Lysozyme is an inducible marker of macrophage activation in murine tissues as demonstrated by in situ hybridization. J. Exp. Med. 174:1049-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloos, W. E., and K. H. Schleifer. 1986. Genus Staphylococcus, p. 1013-1035. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 29.Kloos, W. E., K.-H. Schleifer, and F. Götz. 1992. The genus Staphylococcus. Springer-Verlag, New York, N.Y.
- 30.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 31.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 102:13272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latham, R. H., K. Running, and W. E. Stamm. 1983. Urinary tract infections in young adult women caused by Staphylococcus saprophyticus. JAMA 250:3063-3066. [PubMed] [Google Scholar]
- 33.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 34.McEwan, N. A., G. Kalna, and D. Mellor. 2005. A comparison of adherence by four strains of Staphylococcus intermedius and Staphylococcus hominis to canine corneocytes collected from normal dogs and dogs suffering from atopic dermatitis. Res. Vet. Sci. 78:193-198. [DOI] [PubMed] [Google Scholar]
- 35.Meugnier, H., M. Bes, C. Vernozy-Rozand, C. Mazuy, Y. Brun, J. Freney, and J. Fleurette. 1996. Identification and ribotyping of Staphylococcus xylosus and Staphylococcus equorum strains isolated from goat milk and cheese. Int. J. Food Microbiol. 31:325-331. [DOI] [PubMed] [Google Scholar]
- 36.Morita, H., R. Sakata, Y. Tsukamasa, A. Sakata, and Y. Nagata. 1997. Reddening and bacteriological property of salami without addition of nitrite and nitrate using Staphylococcus carnosus and Staphylococcus xylosus as starter cultures. Anim. Sci. Technol. 68:787-796. [Google Scholar]
- 37.Mounier, J., R. Gelsomino, S. Goerges, M. Vancanneyt, K. Vandemeulebroecke, B. Hoste, S. Scherer, J. Swings, G. F. Fitzgerald, and T. M. Cogan. 2005. Surface microflora of four smear-ripened cheeses. Appl. Environ. Microbiol. 71:6489-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagase, N., A. Sasaki, K. Yamashita, A. Shimizu, Y. Wakita, S. Kitai, and J. Kawano. 2002. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 64:245-250. [DOI] [PubMed] [Google Scholar]
- 39.Novick, R. P. 2000. Pathogenicity factors and their regulation. ASM Press, Washington, D.C.
- 40.Peschel, A., B. Ottenwalder, and F. Gotz. 1996. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 137:279-284. [DOI] [PubMed] [Google Scholar]
- 41.Phillips, W. E., Jr., and W. E. Kloos. 1981. Identification of coagulase-positive Staphylococcus intermedius and Staphylococcus hyicus subsp. hyicus isolates from veterinary clinical specimens. J. Clin. Microbiol. 14:671-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Place, R. B., D. Hiestand, H. R. Gallmann, and M. Teuber. 2003. Staphylococcus equorum subsp. linens, subsp. nov., a starter culture component for surface ripened semi-hard cheeses. Syst. Appl. Microbiol. 26:30-37. [DOI] [PubMed] [Google Scholar]
- 43.Rantsiou, K, R. Urso, L. Iacumin, C. Cantoni, P. Cattaneo, G. Comi, and L. Cocolin. 2004. Culture-dependent and -independent methods to investigate the microbial ecology of Italian fermented sausages. Appl. Environ. Microbiol. 71:1977-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rebecchi, A., S. Crivori, P. G. Sarra, and P. S. Cocconcelli. 1998. Physiological and molecular techniques for the study of bacterial community development in sausage fermentation. J. Appl. Microbiol. 84:1043-1049. [DOI] [PubMed] [Google Scholar]
- 45.Sondergaard, A. K., and L. H. Stahnke. 2002. Growth and aroma production by Staphylococcus xylosus, S. carnosus and S. equorum—a comparative study in model systems. Int. J. Food Microbiol. 75:99-109. [DOI] [PubMed] [Google Scholar]
- 46.Stahnke, M. L. H. 1999. Volatiles produced by Staphylococcus xylosus and Staphylococcus carnosus during growth in sausage minces. Lebensm.-Wiss. Technol. 32:357-364. [Google Scholar]
- 47.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of S. haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valle, J., S. Piriz, R. de la Fuente, and S. Vadillo. 1991. Staphylococci isolated from healthy goats. Zentbl. Vetmed. Reihe B 38:81-89. [DOI] [PubMed] [Google Scholar]
- 49.Van Hoovels, L., P. De Munter, J. Colaert, I. Surmont, E. Van Wijngaerden, W. E. Peetermans, and J. Verhaegen. 2005. Three cases of destructive native valve endocarditis caused by Staphylococcus lugdunensis. Eur. J. Clin. Microbiol. Infect. Dis. 24:149-152. [DOI] [PubMed] [Google Scholar]
- 50.Vilhelmsson, O., H. Hafsteinsson, and J. K. Kristjansson. 1997. Extremely halotolerant bacteria characteristic of fully cured and dried cod. Int. J. Food Microbiol. 36:163-170. [DOI] [PubMed] [Google Scholar]
- 51.Wang, S., C. C. Liu, H. W. Tseng, Y. J. Yang, C. H. Lin, A. H. Huang, and Y. H. Wu. 1999. Staphylococcus capitis bacteremia of very low birth weight premature infants at neonatal intensive care units: clinical significance and antimicrobial susceptibility. J. Microbiol. Immunol. Infect. 32:26-32. [PubMed] [Google Scholar]
- 52.Weightman, N. C., K. E. Allerton, and J. France. 2000. Bone and prosthetic joint infection with Staphylococcus lugdunensis. J. Infect. 40:98-99. [DOI] [PubMed] [Google Scholar]
- 53.Wood, C. A., D. L. Sewell, and L. J. Strausbaugh. 1989. Vertebral osteomyelitis and native valve endocarditis caused by Staphylococcus warneri. Diagn. Microbiol. Infect. Dis. 12:261-263. [DOI] [PubMed] [Google Scholar]
- 54.Zalacain, I., M. J. Zapelena, M. P. Pena, I. Astiasaran, and J. Bello. 1997. Lipid fractions of dry fermented sausages change when starter culture and/or Aspergillus lipase are added. J. Food Sci. 62:1076-1079. [Google Scholar]
- 55.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577-1593. [DOI] [PubMed] [Google Scholar]