Abstract
Mus spicilegus is an Eastern European wild mouse species that has previously been reported to harbor an unusual infectious ecotropic murine leukemia virus (MLV) and proviral envelope genes of a novel MLV subgroup. In the present study, M. spicilegus neonates were inoculated with Moloney ecotropic MLV (MoMLV). All 17 inoculated mice produced infectious ecotropic virus after 8 to 14 weeks, and two unusual phenotypes distinguished the isolates from MoMLV. First, most of the M. spicilegus isolates grew to equal titers on M. dunni and SC-1 cells, although MoMLV does not efficiently infect M. dunni cells. The deduced amino acid sequence of a representative clone differed from MoMLV by insertion of two serine residues within the VRA of SUenv. Modification of a molecular clone of MoMLV by the addition of these serines produced a virus that grows to high titer in M. dunni cells, establishing a role for these two serine residues in host range. A second unusual phenotype was found in only one of the M. spicilegus isolates, Spl574. Spl574 produces large syncytia of multinucleated giant cells in M. dunni cells, but its replication is restricted in other mouse cell lines. Sequencing and mutagenesis demonstrated that syncytium formation could be attributed to a single amino acid substitution within VRA, S82F. Thus, viruses with altered growth properties are selected during growth in M. spicilegus. The mutations associated with the host range and syncytium-inducing variants map to a key region of VRA known to govern interactions with the cell surface receptor, suggesting that the associated phenotypes may result from altered interactions with the unusual ecotropic virus mCAT1 receptor carried by M. dunni.
Studies on virus-host interactions that affect disease induction by the murine leukemia viruses (MLVs) have made extensive use of the inbred laboratory mouse strains with high levels of endogenous virus expression and a high incidence of disease such as AKR and BXH2. Such studies have been highly productive, but use of these laboratory strains provides only limited insight into host response to infection. This is because the common laboratory strains were derived from a small number of progenitors and therefore do not reflect the full range of genetic diversity within Mus (3). Also, the high-virus-titer strains typically used for such studies were bred for their susceptibility to infection and disease and therefore lack the genetic resistance factors that protected their wild mouse progenitors. For example, none of the common inbred strains carry the ecotropic MLV resistance gene Fv4 (10). Also, the major laboratory mouse alleles of the resistance gene, Fv1, Fv1n and Fv1b, have not been identified among wild mouse species (12). For these reasons, we believe that it is worthwhile to examine virus susceptibility and the disease process in wild mouse species. Many of these species have experienced long-term exposure to natural MLV infections, have become adapted to endemic infection, and have evolved protective mechanisms that prevent virus-induced mutation and disease. In this report, we describe studies initiated to examine the susceptibility of one wild mouse species, Mus spicilegus, to exogenous infection.
M. spicilegus (formerly M. hortulanus) inhabits Central and Eastern Europe and Asiatic Russia, overlapping with M. macedonicus and M. mus musculus (16). M. spicilegus is similar to these two species morphologically and genetically, but not behaviorally, and there is no evidence that interbreeding occurs. M. spicilegus is unusual in its monogamous mating system and in its habit of constructing large mounds of earth and seeds. Several previous studies have examined M. spicilegus for endogenous and infectious MLVs. M. spicilegus differs from the laboratory strains and other European species in that it does not contain endogenous viral env genes related to MCF and xenotropic MLVs (13). Tomonaga and Coffin (20) more recently demonstrated that these mice carry proviral env sequences of a novel subgroup that is equidistant from ecotropic and nonecotropic viruses and that likely represents an ancestral form of MLV. In addition to its unusual proviral content, M. spicilegus also differs from the other European mice in that it is the only one from which infectious virus has been isolated (21). This isolate is a novel ecotropic virus that was transmitted as an infectious agent in randomly bred laboratory colonies but was lost from these mice sometime after 1985 (22).
In the present study, we examined the susceptibility of M. spicilegus to exogenous MLV infection. For this purpose, Moloney MLV (MoMLV) was chosen because of its broad host range and pathogenic properties. Results of this study show that infectious ecotropic virus can be recovered from MoMLV-inoculated M. spicilegus but that these isolates differ phenotypically from MoMLV. Unlike MoMLV, all of the isolates grow to high titer in M. dunni cells, and one highly unusual isolate is capable of inducing large syncytia in M. dunni. Using site-specific mutagenesis, we identified a small number of amino acid changes within a segment of the SUenv VRA region responsible for these phenotypes.
MATERIALS AND METHODS
Mouse cells and viruses.
M. spicilegus mice were originally obtained from a randomly bred colony first established in 1981 by R. Sage and subsequently maintained by M. Potter (National Cancer Institute, Bethesda, Md.). The founders of this colony were originally trapped in Halbturn, Austria. This line has been maintained by random breeding in our laboratory for more than 10 years.
Neonatal M. spicilegus mice were inoculated intraperitoneally with ecotropic MoMLV originally obtained from J. Hartley (National Institute of Allergy and Infectious Diseases [NIAID], Bethesda, Md.). Inoculated mice were sacrificed 8 to 14 weeks after inoculation, and cell suspensions of spleen and thymus cells were assayed for infectious ecotropic virus. These cells were used to infect cultured SC-1 (8), M. dunni (14), and mink lung cells plated at 2 × 105 cells per 60-mm dish with Polybrene (4 μg/ml; Aldrich, Milwaukee, Wis.). Cells were passaged and examined for infectious virus by testing for reverse transcriptase (RT) or by the XC test (19). For the XC test, SC-1 plates were irradiated and overlaid with 106 XC cells/plate. Plates were stained 3 days later and examined for plaques of syncytia. For the RT assay, samples of culture fluid were centrifuged for 5 min at 860 × g to remove cells. RT activity in the supernatants was then measured as described by Wilson and Eiden (23).
Additional cell lines used to examine the host range of recovered viruses included NIH 3T3 and A9. Virus stocks of AKV MLV, NB-tropic Friend MLV (FrMLV), and Moloney HIX MCF virus were obtained from J. Hartley (NIAID).
Southern blotting.
High-molecular-weight DNA was isolated from virus-infected cultures by standard protocols. DNAs were digested with restriction enzymes, electrophoresed on 0.4% agarose gels, transferred to nylon membranes (Hybond N+; Amersham, Piscataway, N.J.), and hybridized with radiolabeled probe. As probe, a 216-bp segment of the MoMLV env gene was amplified by PCR with use of the following as primers: 5′-GGACAAGATCCAGGGCTTACA-3′ (forward primer) and 5′-TACTAAGTTTAGCAGCCTATT-3′ (reverse primer).
Cloning and sequencing.
Hirt DNA was prepared from M. dunni cells infected with virus isolates Spl603-3 and Spl574 and was used as PCR substrate (9). The PCR (20-μl final volume) contained 0.1 μg of the template, 2.5 U of AmpliTaq Gold DNA polymerase (PE Applied Biosystems, Foster City, Calif.), and 20 pmol of primers.
The primers to amplify the viral env genes were as follows: Unienv, 5′-GGATACACGCCGCTCACGTA-3′ (MoMLV env universal forward primer); UniLTR, 5′-CGGGCGACTCAGTCTATCGG-3′ (MoMLV long terminal repeat universal reverse primer); VRA1, 5′-GACGGTATGGGCAACTTCTG-3′ (MoMLV SUenv specific forward primer); VRA2, 5′-GCCCCAATAGGCACAGTAGA-3′ (MoMLV SUenv specific reverse primer); 3′env, 5′-TGTCCGAAGTGACCGGAC-3′ (MoMLV TM specific forward primer); and ULTR, 5′-CATGCCTTGCAAGATGGC-3′ (MoMLV TM specific reverse primer).
Unienv and UniLTR primers were used to amplify the full-length env gene. VRA1 and VRA2 were designed to amplify the VRA region of SU, and 3′env and ULTR amplified the env TM domain (Fig. 1A).
FIG. 1.
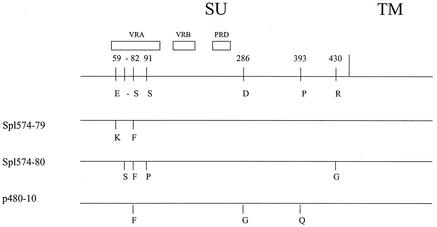
(A) General structure of MoMLV. An open box is used to indicate the position of VRA. The arrows identify the PCR primers and their products. LTR, long terminal repeat. (B) The S82F substitution was introduced into MoMLV by a series of substitutions. The solid vertical lines indicate the positions of the three substitutions in the 2.5-kb env gene PCR product of Spl574 clone p480-10. The arrows indicate the restriction sites for BamHI (B) and HpaI (H). The internal HpaI fragment was first removed and replaced by the comparable segment of MoMLV to produce p480-Mo, and then the internal BamHI segment of p480-Mo was replaced by the S82F-containing BamHI segment of p480-10.
The mCAT1 receptor was amplified from M. spicilegus DNA by PCR with forward (sCAT1: AAACCCCGGACATATTTGCT) and reverse (sCAT2: GGGGGTTCTTGACTTCTTCC) primers derived from the NIH 3T3 ecotropic receptor sequence.
The PCRs were carried out in a GeneAmp PCR system 9700 machine (PE Applied Biosystems). The reactions were performed for 35 cycles with a 30-s DNA denaturation step at 95°C, a 30-s annealing step, and a 1-min extension step at 72°C. The annealing temperature in the first cycle was 63°C, was subsequently reduced by 1°C each cycle for the next eight cycles, and was then maintained at 55°C for the remaining 27 cycles. The VRA, full-length env, and mCAT1 receptor PCR products were cloned into the pCR2.1-TOPO vector and sequenced. The TM PCR product was sequenced directly.
Mutagenesis.
Two strategies were used for mutagenesis. The addition of two adjacent serine residues into a site within the env VRA region was accomplished by using the Stratagene (La Jolla, Calif.) ExSite PCR-Based Site-Directed Mutagenesis kit. HpaI digestion of the MoMLV clone p63-2 produced a 1.3-kb env fragment containing VRA that corresponds to nucleotides 5818 to 7197 of the viral genome. Plasmid p63-2 contains an integrated MoMLV provirus and was a gift of J. Silver (NIAID) (2). This 1.3-kb fragment was subcloned into the vector pCR2.1-TOPO. Amino acid changes were introduced into the env gene by using the following oligonucleotides: forward primer 5′-AGCAGCGGCAGCAGCCCAGGCTGTTCCAGA-3′ and reverse primer 5′-CCCTGAGCAACAAGGGGGCCCCGG-3′. The mutated fragment was then removed as a 1.3-kb HpaI fragment and was ligated to p63-2, from which the corresponding 1.3-kb fragment had been deleted.
To obtain recombinant MoMLV with the single amino acid substitution S82F, we first prepared a Hirt extract (9) of cells infected with virus isolate Spl574. Primers Unienv and UniLTR were used to amplify a segment containing env and long terminal repeat sequences. These products of about 2.5 kb were cloned into the pCR2.1-TOPO vector and sequenced. The deduced amino acid sequence of one of these clones, p480-10, contained three substitutions, including a single substitution within VRA, S82F (Fig. 1B). To create an MoMLV clone containing only the single VRA substitution, a series of exchanges were made (Fig. 1B). First, clone p480-Mo was constructed by replacing the HpaI fragment of p480-10 with the corresponding fragment of MoMLV plasmid p63-2. Second, a 870-bp BamHI containing only the S82F substitution was excised from p480-10 and used to replace the corresponding fragment within p480-Mo to produce p480-Mo-S82F. Finally, the 1.3-kb HpaI fragment containing the single mutation S82F was removed from p480-Mo-S82F and ligated into p63-2, from which the corresponding fragment had been excised.
All mutants were confirmed by DNA sequencing.
Transfection and infection.
To determine whether these mutations were associated with novel syncytium-inducing and host range phenotypes, mutated proviral DNAs were introduced into NIH 3T3 cells by using the Qiagen (Valencia, Calif.) PolyFect Transfection Kit. After 3 days in culture, supernatants were harvested and assayed for RT. To measure syncytium formation in M. dunni cells, 2 × 104 M. dunni cells in six-well tissue culture plates were infected with MoMLV or mutant virus containing medium in the presence of Polybrene (4 μg/ml). After 2 to 4 days, the cells were examined by light microscopy either directly or after being fixed in methanol and stained with methylene blue. Cells were photographed by using a Nikon TS100 microscope and DXM1200 digital camera.
Pseudotype assay.
LacZ pseudotype virus was generated by transfection of TELCeB6 cells with the ecotropic MoMLV envelope expression vector, pVPack-Eco (wild-type; Stratagene) and pVPack-S82F (mutant). TELCeB6 produces noninfectious viral particles harboring the MFGnlslacZ retroviral vector. Viral supernatants were collected and filtered and used to infect NIH 3T3, M. dunni, and SC-1 cells that had been plated in six-well culture dishes at a density of 1.5 × 105 per well. The cells were infected with 1 ml of virus in the presence of 8 μg of Polybrene/ml for 3 h before 2 ml of fresh medium was added to each well. Two days after infection, cells were fixed with 0.5% glutaraldehyde and stained to reveal the presence of β-galactosidase activity. Infectious titers were expressed as the number of blue CFU per milliliter of virus supernatant.
RESULTS
Virus isolation.
A total of 17 neonates in three M. spicilegus litters were inoculated intraperitoneally with MoMLV. The mice were examined for replicating virus 8 to 14 weeks after inoculation. Single-cell suspensions of spleen and thymus cells were plated on mouse SC-1 cells or M. dunni cells and mink cells. Cultures were examined for evidence of replicating virus by RT or by XC test. None of the 17 inoculated mice yielded virus capable of infecting mink cells. All 17 mice produced virus that could be detected in infected-mouse-cell cultures by XC plaque assay or by RT (data not shown). Virus stocks prepared from 10 of these infected-cell cultures were used to infect SC-1 and M. dunni cells (Table 1). Nine of these 10 stocks produced titers on M. dunni that were at least as high as that on SC-1 cells. Since the original infecting virus, MoMLV, is poorly infectious in M. dunni cells (14), the virus isolated from inoculated M. spicilegus differs from MoMLV.
TABLE 1.
Titers of virus isolates from M. spicilegus inoculated with MoMLV as neonates
| Mouse | Wks postinfection | Log10 virus titer (spleen and thymus derived)a
|
|
|---|---|---|---|
| M. dunni | SC-1 | ||
| 574 | 8 | 5.5 | 3.7 |
| 575 | 8 | NDb | 1.9 |
| 601-2 | 10 | >5.0 | >5.0 |
| 601-3 | 10 | 4.4 | 4.1 |
| 602-2 | 10 | 4.3 | 4.5 |
| 602-3 | 10 | 4.8 | >5.0 |
| 603-3 | 10 | 4.6 | 4.7 |
| 604-1 | 14 | 4.6 | 4.7 |
| 605-1 | 14 | 4.8 | >5.0 |
| 605-2 | 14 | 4.5 | 4.7 |
Measured as number of XC PFU in 0.2 ml of spleen extracts. The titers of MoMLV used for inoculation were 2.5 for M. dunni cells and 5.2 for SC-1 cells.
Not detectable.
Cloning and sequencing.
High-molecular-weight DNA was prepared from M. dunni cells infected by 13 of the 17 M. spicilegus isolates. Southern blotting confirmed the presence of MoMLV-related sequences in all 13 DNAs with the use as probe of a 216-bp segment of the MoMLV env (data not shown). All 13 DNAs contained proviral sequences indistinguishable from MoMLV following digestion with KpnI and SacI, enzymes that cleave MoMLV on both sides of the sequences corresponding to the probe. One of these DNAs, prepared from cells infected with isolate Spl603-3, was arbitrarily selected for further characterization as representative of isolates that replicated efficiently in M. dunni cells. Primers Unienv and UniLTR were used to amplify the full-length env, and the PCR products were cloned into pCR2.1-TOPO. Three independent clones were sequenced and showed the same single difference in comparison to MoMLV (Fig. 2). All three clones of this mutant, termed MLV-SS, contained an insertion of two adjacent serine residues between amino acids 75 and 76 in the VRA region of the 5′ end of env.
FIG. 2.
Comparison of the deduced amino acid sequences of the VRA regions of MoMLV and five other ecotropic MLVs. HoMLV, M. hortulanus (M. spicilegus) MLV (21, 22). MLV-SS is a Moloney clone into which two serine residues have been added. Dashes indicate no change; asterisks represent deletions.
Comparisons with other sequenced ecotropic viruses showed that virtually all have a two-amino-acid-residue insertion at this same site relative to MoMLV (Fig. 2). For FrMLV and Rauscher MLV, these inserted amino acids are both serine residues, whereas only the first is serine for AKV and neither is serine for the more distantly related wild mouse isolate CasBRM. FrMLV, Rauscher MLV, and AKV ecotropic MLV are all infectious for M. dunni cells (6, 14) (Table 2). Interestingly, the wild mouse virus isolated previously from M. spicilegus, HoMLV, has only one added amino acid residue in this region relative to MoMLV (22). HoMLV is not infectious for M. dunni cells, although it is otherwise ecotropic in host range and env sequence, and likely uses the mCAT1 receptor, since HoMLV infects hamster × mouse somatic cell hybrids carrying only the single mouse chromosome with the mCAT1 gene (21, 22). These results implicate this region of VRA in virus-receptor interactions consistent with previous studies on the structure of the receptor binding site (7). Also, it has been shown that the mCAT1 receptor of M. dunni differs from that of the laboratory strains in sequence as well as phenotype (6). These observations taken together with our data suggest that these deleted residues are responsible for the failure of MoMLV and HoMLV to infect M. dunni cells because of altered receptor binding.
TABLE 2.
Virus titers of MoMLV variants in different mouse cell lines
| Virus | Log10 virus titera
|
||
|---|---|---|---|
| M. dunni | SC-1 | NIH 3T3 | |
| AKV MLV | 4.1 | 4.4 | 4.4 |
| MoMLV | 2.5 | 5.2 | 5.4 |
| Spl574 | 4.9 | 2.5 | 3.5 |
| MLV-SS | 4.1 | 3.3 | 4.1 |
| MoMLV-S82F | 4.8 | 2.0 | 2.5 |
| FrMLV | 4.0 | 4.6 | NTb |
Titers are given as XC PFU in 0.2 ml.
Not tested.
Mutagenesis.
An infectious molecular clone of Moloney, p63-2, was altered by site-specific mutagenesis by the addition of two serine residues between the two glycine residues at positions 75 and 76 in the VRA region of the 5′ end of env. The clone was transfected into NIH 3T3 cells, and virus-containing media were collected. This mutant, MLV-SS, was titered on various mouse cells by using the XC test. MLV-SS replicated to high titers in M. dunni cells and in other mouse cells, although it replicated more efficiently on M. dunni than on SC-1 cells (Table 2). These results indicate that this two-base deletion in MoMLV relative to the other ecotropic viruses is responsible for the inefficient replication of this virus on M. dunni cells and that addition of the two bases is sufficient to correct this defect.
Syncytium-inducing virus.
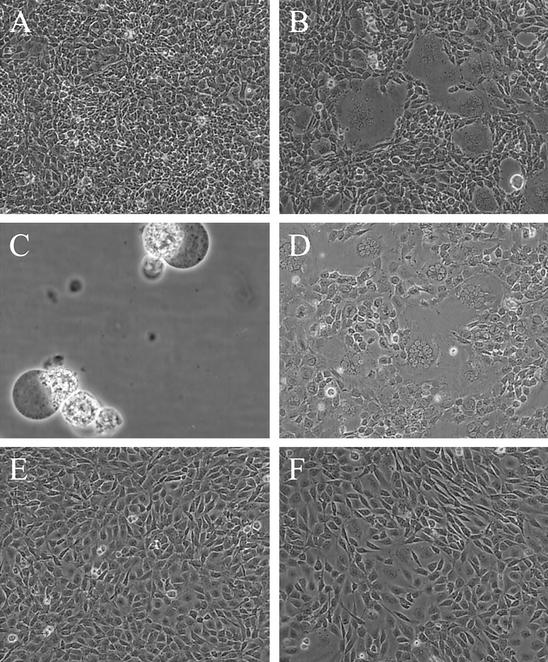
A second novel phenotype was detected among the viruses isolated from the 17 M. spicilegus mice infected with MoMLV. A single isolate, Spl574, produced large multinucleated syncytia on M. dunni cells that appeared 2 days after infection (Fig. 3B). These syncytia often contain more than 15 nuclei, although the size of syncytia in individual cultures is affected by cell density; infection of cells near confluency produces fewer and smaller syncytia. Syncytium induction is a consistent consequence of M. dunni cell infection, and the virus can be titered by the number of plaques of syncytia in M. dunni cultures.
FIG. 3.
(A) Uninfected M. dunni cells. (B) M. dunni cells 2 days after infection with Spl574. (C) Floating cells collected from an M. dunni culture 3 days after infection with Spl574. (D) M. dunni cells 2 days after infection with mutant virus MoMLV-S82F. (E) Uninfected NIH 3T3 cells. (F) NIH 3T3 cells 5 days after infection with Spl574. Objective lens magnification was ×10, except for panel C, for which magnification was ×40.
Infected M. dunni cultures rapidly deteriorate after the appearance of syncytia on the 2nd day after infection. By the next day, virus-infected cultures are characterized by cell lysis generally marked by the ballooning of cells (Fig. 3C) and the accumulation of extracellular debris in the culture fluid.
Syncytium formation was rarely noted following infection of cultures of other established mouse cell lines, including NIH 3T3, SC-1, and A9. Among these cells, only NIH 3T3 cells showed any syncytium induction at all (Fig. 3F). However, on these cells, the number of syncytia was small, the individual syncytia rarely contained more than three nuclei, and syncytia were rarely seen before 5 days after infection. On the other mouse cells, no discernible cytopathic effects could be detected after 2 weeks of observation.
Titration of the Spl574 isolate on the different cells showed that this virus grows efficiently only on M. dunni cells (Table 2). Virus titers determined by the XC test were reduced by 2 logs on NIH 3T3 cells and by 3 logs on SC-1.
Because the XC test relies on syncytium induction and because Spl574 has a novel syncytium phenotype, we also determined virus titer by using retrovirus vectors expressing the normal and mutant env glycoproteins in a single-cycle infection assay that does not rely on syncytium induction. As shown in Table 3, this assay produced titers for MoMLV and S82F that were comparable to the results from the XC test. The S82F pseudotype showed high titers on M. dunni cells compared to MoMLV but was poorly infectious in NIH 3T3 and SC-1 cells. No syncytia were observed in any of the pseudotype-infected cultures, indicating that virus infection is insufficient to cause syncytia, which suggests that formation of syncytia is likely to require viral env glycoprotein expression on the cell surface.
TABLE 3.
Effect of the S82F envelope mutation on viral infectivity
| Target cell | Log10 titer of LacZ pseudotypea
|
|
|---|---|---|
| LacZ (wild type) | LacZ (S82F) | |
| NIH 3T3 | 5.7 | 1.4 |
| M. dunni | 1.5 | 6.2 |
| SC-1 | 5.3 | 1.3 |
Titers are given as number of CFU per milliliter.
To establish the involvement of the ecotropic receptor, mCAT1, in this phenomenon, we established cultures of M. dunni cells chronically infected with either ecotropic AKV MLV or with Moloney HIX MCF virus, a virus that uses the Xpr1 polytropic/xenotropic receptor. These cells were then superinfected with Spl574 and were monitored for syncytia. Syncytia were observed in the MCF virus-infected cells but not in cells infected with AKV MLV (data not shown). These results confirm that the syncytium-inducing virus interacts with the ecotropic receptor mCAT1.
Sequencing and mutagenesis.
Because the ability of MLVs to form syncytia has been attributed to sequence variation at multiple sites in TM and SU (11, 15, 18), we sequenced the entire env gene of Spl574. The TM domain was 100% identical to that of MoMLV: the region encoding the cytoplasmic tail was not truncated, and no nucleotide changes were noted in the fusion domain (data not shown). Within SU, however, there were 6 amino acid changes, 4 of which were within the VRA region (Fig. 4). Five of these differences represented amino acid substitutions, and the fourth was an insertion of a serine residue. Of these changes, however, only one, the S82F substitution, was found in all sequenced clones.
FIG. 4.
Deduced amino acid sequences of the env genes of three clones of the syncytium-inducing virus Spl574. The open boxes indicate the relative positions of the VRA, VRB, and PRD segments within SU. Vertical lines indicate the positions of substitutions or insertions in the three clones, and the numbers represent the amino acid position.
This single amino acid substitution was introduced into the MoMLV clone, p63-2. The resulting virus, MoMLV-S82F, resembled Spl574 in its ability to form syncytia on M. dunni cells (Fig. 3D). This virus also resembled the original isolate in that it grew to lower titers in NIH 3T3 and SC-1 cells (Table 2) and showed marginal syncytium-inducing ability on NIH 3T3 cells (not shown). These results indicate that a single amino acid substitution in VRA is responsible for syncytium formation in M. dunni cells as well as for differences in replication efficiency in other mouse cell lines.
Sequence analysis of M. spicilegus mCAT1.
These results suggest that the virus inoculated into M. spicilegus is exposed to selective pressures that can result in altered growth properties. One possible explanation for this is that M. spicilegus, like M. dunni, may encode an mCAT1 receptor that differs from that of the prototypical NIH 3T3 receptor, and that such differences might select for env variants. The M. dunni receptor has been sequenced, and differences within the third extracellular loop have been associated with reduced susceptibility to MoMLV (5, 6) (Fig. 5).
FIG. 5.
Comparison of the deduced amino acid sequences of the third extracellular regions of the mCAT1 receptor of NIH 3T3 (mCAT1), SC-1 (sCAT1), M. dunni (dCAT1), and M. spicilegus (HCAT1). The two potential N-linked glycosylation sites are underlined. Y235 and E237 are critical for efficient virus infection and binding of MLV envelope protein (1).
In order to test the possibility that the MoMLV variants from M. spicilegus resulted from adaptation to a variant of the mCAT1 receptor, we cloned and sequenced the third extracellular loop of the mCAT1 receptor from this mouse. We also sequenced the corresponding segment of the SC-1 mCAT1 receptor because replication of Spl574 was consistently lower on these cells than on NIH 3T3 cells (Fig. 5). This segment of both the M. spicilegus and SC-1 receptors is identical to that of NIH 3T3. The entire SC-1 mCAT1 gene was then sequenced, but no differences with the NIH 3T3 gene were identified (data not shown). Sequence variation in the virus binding region of these wild mouse receptor genes does not therefore account for the inefficient replication of Spl574 in SC-1 cells, nor does it provide an explanation for the origin of this isolate in M. spicilegus.
DISCUSSION
These results show that MoMLV inoculated into M. spicilegus is subjected to selective pressures that favor replication of viruses with altered amino acid sequences within the VRA domain of SUenv. Virtually all of these isolates, unlike the original MoMLV, are capable of efficiently infecting M. dunni cells, and 1 of the 17 isolates induces syncytia in M. dunni cells. The ability to efficiently infect M. dunni cells was attributed to two different amino acid changes in a small segment of the 5′ region of the env VRA region: insertion of 2 amino acids between G75 and G76 and a single substitution, S82F. Previous studies determined that the segment of VRA containing these changes is involved in the formation of the receptor binding site in the viral receptor binding domain (4, 7), and the present study suggests that either of these amino acid changes specifically enhances interactions with the M. dunni receptor variant.
The relative inability of MoMLV to replicate on M. dunni cells has been attributed to sequence variations within the third extracellular loop of the M. dunni receptor (5, 6). We therefore speculated that the M. spicilegus isolates represented adaptations to a variant M. spicilegus receptor. However, our sequence analysis indicates that not only does the M. spicilegus mCAT1 receptor not resemble the M. dunni variant, but it appears to be identical to the NIH 3T3 mCAT1. Similarly, sequence differences in mCAT1 cannot account for the restricted replication of the S82F mutant in SC-1 cells. Thus, it is not clear why these MoMLV variants were selected in M. spicilegus, but our sequencing results suggest that some mechanism other than receptor sequence variation, such as glycosylation or other posttranslational modification, may be responsible.
The mechanism by which retroviruses cause syncytia is under active investigation and clearly involves receptor interactions. Previous mutagenesis studies on FrMLV have shown that three amino acids are crucial for receptor binding and virus infection: S84 (S82 in MoMLV), D86, and W102 (4). Substitutions at all three of these sites abolished infectivity. Our studies indicate that a specific substitution at one of these sites in MoMLV, S82F, results in a syncytium-inducing virus. Interestingly, a substitution in another of these critical residues, W102G, is responsible for the only other syncytium-inducing MLV variant caused by mutations in SU. This FrMLV-related ecotropic virus isolate, TR1.3, causes syncytia in SC-1 cells (15). The present study provides further evidence of the importance of these residues in receptor interactions and confirms that alteration of this interaction can result in syncytium induction.
The receptor binding site in SUenv is created by two helices and two loops at the top of the receptor binding domain. Amino acid S84 in FrMLV (S82 in MoMLV) is positioned as a single residue between Loop1 and the small helix (4). The two inserted serine residues that restore infectivity of MoMLV in M. dunni are within Loop1. Mutational analysis has shown that substitutions of alanine and glycine at S84 do not alter binding, whereas substitution with isoleucine eliminates infectivity (4). As shown here, substitution with phenylalanine, like isoleucine, substantially reduces infectivity on cells with mCAT1, and this may be related to the fact that both of these substitutions introduce bulky side chains at this position. The observation that M. dunni cells show increased susceptibility to viruses with either the S82F substitution or the serine insertions in Loop1 suggests that these changes produce subtle alterations in the binding site that enhance interaction with the variant M. dunni receptor.
Syncytium induction is relatively rare in MLVs but is common in lentiviruses, where it is associated with pathogenicity. The syncytium-inducing mouse viruses are also pathogenic. Thus, the syncytium-inducing Friend virus variant TR1.3 causes hind limb paralysis in mice, and this disease is associated with the appearance of syncytia in midbrain endothelial cells (15). A recent study described a novel wild mouse retrovirus, M813, that causes syncytia in MuLV-infected cells and, in vivo, produces a peripheral lymphoma associated with large multinucleated cells of macrophage origin (17). In the present study, the inoculated M. spicilegus mice were sacrificed within 14 weeks of infection, at which time there was no gross indication of disease. We have not yet introduced the variant viruses into mice to determine their disease-inducing potential, but the restricted replication of the S82F mutant in laboratory mouse cells suggests that pathogenicity may be restricted to wild mouse species.
Acknowledgments
We acknowledge Alicia Buckler-White for sequencing and the expert technical assistance of M. Charlene Adamson (deceased). We thank Caroline Ball for editorial assistance in the preparation of the manuscript.
REFERENCES
- 1.Albritton, L. M., J. W. Kim, L. Tseng, and J. M. Cunningham. 1993. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J. Virol. 67:2091-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacheler, L., and H. Fan. 1981. Isolation of recombinant DNA clones carrying complete integrated proviruses of Moloney murine leukemia virus. J. Virol. 37:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck, J. A., S. Lloyd, M. Hafezparast, M. Lennon-Pierce, J. T. Eppig, M. F. W. Festing, and E. M. C. Fisher. 2000. Genealogies of mouse inbred strains. Nat. Genet. 24:23-25. [DOI] [PubMed] [Google Scholar]
- 4.Davey, R. A., Y. Zuo, and J. M. Cunningham. 1999. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J. Virol. 73:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiden, M. V., K. Farrell, and C. A. Wilson. 1994. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J. Virol. 68:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eiden, M. V., K. Farrell, J. Warsowe, L. C. Mahan, and C. A. Wilson. 1993. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J. Virol. 67:4056-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fass, D., R. A. Davey, C. A. Hamson, P. S. Kim, J. M. Cunningham, and J. M. Berger. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277:1662-1666. [DOI] [PubMed] [Google Scholar]
- 8.Hartley, J. W., and W. P. Rowe. 1975. Clonal cell lines from a feral mouse embryo which lack host-range restrictions for murine leukemia viruses. Virology 65:128-134. [DOI] [PubMed] [Google Scholar]
- 9.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda, H., F. Laigret, M. A. Martin, and R. Repaske. 1985. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J. Virol. 55:768-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, J. S., and R. Risser. 1993. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J. Virol. 67:67-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak, C. A. 1985. Analysis of wild-derived mice for the Fv-1 and Fv-2 murine leukemia virus restriction loci: a novel wild mouse Fv-1 allele responsible for lack of host range restriction. J. Virol. 55:281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak, C. A., and R. R. O'Neill. 1987. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J. Virol. 61:3082-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lander, M. R., and S. K. Chattopadhyay. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J. Virol. 52:695-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park, B. H., B. Matuschke, E. Lavi, and G. N. Gaulton. 1994. A point mutation in the env gene of a murine leukemia virus induces syncytium formation and neurologic disease. J. Virol. 68:7516-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patris, B., and C. Baudoin. 1998. Female sexual preferences differ in Mus spicilegus and Mus musculus domesticus: the role of familiarization and sexual experience. Anim. Behav. 56:1465-1470. [DOI] [PubMed] [Google Scholar]
- 17.Prassolov, V., D. Ivanov, S. Hein, G. Rutter, C. Munk, J. Lohler, and C. Stocking. 2001. The Mus cervicolor MuLV isolate M813 is highly fusogenic and induces a T-cell lymphoma associated with large multinucleated cells. Virology 290:39-49. [DOI] [PubMed] [Google Scholar]
- 18.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowe, W. P., W. E. Pugh, and J. W. Hartley. 1970. Plaque assay techniques for murine leukemia viruses. Virology 42:1136-1139. [DOI] [PubMed] [Google Scholar]
- 20.Tomonaga, K., and J. M. Coffin. 1999. Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J. Virol. 73:4327-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voytek, P., and C. Kozak. 1988. HoMuLV: a novel pathogenic ecotropic virus isolated from the European mouse, Mus hortulanus. Virology 165:469-475. [DOI] [PubMed] [Google Scholar]
- 22.Voytek, P., and C. A. Kozak. 1989. Nucleotide sequence and mode of transmission of the wild mouse ecotropic virus, HoMuLV. Virology 173:58-67. [DOI] [PubMed] [Google Scholar]
- 23.Wilson, C. A., and M. V. Eiden. 1991. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J. Virol. 65:5975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]