Abstract
Shigella flexneri, the causative agent of shigellosis, is a gram-negative bacterial pathogen that initiates infection by invading cells within the colonic epithelium. Contact with host cell surfaces induces a rapid burst of protein secretion via the Shigella type III secretion system (TTSS). The first proteins secreted are IpaD, IpaB, and IpaC, with IpaB and IpaC being inserted into the host cell membrane to form a pore for translocating late effectors into the target cell cytoplasm. The resulting pathogen-host cross talk results in localized actin polymerization, membrane ruffling, and, ultimately, pathogen entry. IpaD is essential for host cell invasion, but its role in this process is just now coming to light. IpaD is a multifunctional protein that controls the secretion and presentation of IpaB and IpaC at the pathogen-host interface. We show here that antibodies recognizing the surface-exposed N terminus of IpaD neutralize Shigella's ability to promote pore formation in erythrocyte membranes. We further show that MxiH and IpaD colocalize on the bacterial surface. When TTSS needles were sheared from the Shigella surface, IpaD was found at only the needle tips. Consistent with this, IpaD localized to the exposed tips of needles that were still attached to the bacterium. Molecular analyses then showed that the IpaD C terminus is required for this surface localization and function. Furthermore, mutations that prevent IpaD surface localization also eliminate all IpaD-related functions. Thus, this study demonstrates that IpaD localizes to the TTSA needle tip, where it functions to control the secretion and proper insertion of translocators into host cell membranes.
Shigella flexneri is the causative agent of shigellosis, a severe form of bacillary dysentery that is globally responsible for more than 1.1 million deaths each year, especially among young children (www.who.int/vaccine_research/diseases/shigella/en/). S. flexneri is transmitted by the fecal-oral route, typically via contaminated water, and is spread efficiently due to an unusually low 50% infective dose (13). After ingestion, acid-tolerant S. flexneri passes through the stomach to the colonic mucosa, where it is transcytosed across M cells into the underlying gut-associated lymphoid tissues (29). There, the shigellae enter macrophages and efficiently induce apoptosis, leading to the eventual release of the pathogen on the basal side of the colonic epithelium (46). The shigellae then invade these epithelial cells by inducing major cytoskeletal rearrangements (40). The internalized bacteria lyse the resulting vacuolar membrane (33), replicate intracellularly, and move about the cytoplasm via actin-based motility (1), which allows direct cell-to-cell spreading (28).
The Shigella invasive phenotype genetically localizes to a 31-kb region of its large virulence plasmid, which includes the genes for the type III secretion system (TTSS) (7, 34). The mxi-spa operon encodes the type III secretion apparatus (TTSA), and the ipa-ipg operon encodes the type III-secreted protein effectors/translocators IpaA through IpaD and the cytoplasmic IpaB/IpaC chaperone IpgC. The TTSA is composed of a basal body spanning both bacterial membranes and an external needle (4). The TTSA needle, a polymer of MxiH, is 50 nm long and 7 nm in diameter with a central channel that is ∼2.5 nm in diameter (6). After TTSA assembly, secretion is largely arrested until a stimulus such as host cell contact is received. Currently, the mechanism for activation has not been determined, although it is likely that the needle or proteins intimately associated with the needle are involved (16).
Upon receipt of a secretion signal, IpaB, IpaC, and IpaD, which are all essential for bacterial entry, are transported through the TTSA (23). IpaB and IpaC are inserted into the host cell membrane to form a translocon pore through which other effector proteins subsequently pass (2). Additionally, IpaB and IpaC possess direct effector functions that alter host cell physiology, including the induction of apoptosis in macrophages (45) and cytoskeletal rearrangements in epithelial cells (17, 38, 39), respectively. The essential function of IpaD in the invasion process has remained elusive.
IpaD was originally proposed to partner with IpaB to form a plug within the TTSA because the deletion of either protein results in the secretion of large amounts of the remaining TTSS effectors in the absence of a secretion signal (22). Recent findings suggest that IpaD's role is more complex. Deletion mutagenesis was used to show that the N-terminal third of IpaD is partially dispensable for Shigella invasiveness (31). These deletions had a relatively minor impact on invasiveness and secretion control, but they substantially reduced contact hemolysis due, in part, to a slower rate of translocon insertion into erythrocyte membranes (31). Conversely, deletions in the C-terminal two-thirds of IpaD resulted in a complete loss of invasion functions and secretion control. Deletions near the C-terminal end of the protein resulted in massive uninduced secretion of IpaB, IpaC, and IpaD (31). It thus appears that IpaD is needed for secretion control and proper insertion of translocators into host cell membranes.
The closest relative of IpaD is SipD from Salmonella, which is 40% identical and 56% similar to IpaD, with the C terminus being 73% identical and 98% similar (15). Like ipaD null mutants, sipD null mutants are noninvasive and secrete large amounts of the Salmonella translocators SipB and SipC (15). BipD from Burkholderia pseudomallei is also strikingly similar at the C terminus, with a bipD null mutant also being noninvasive (36). Although there are no obvious IpaD homologs in Yersinia spp. and Pseudomonas aeruginosa, LcrV and PcrV, respectively, appear to share some functional homology. Null mutants of lcrV and pcrV are unable to form translocons in host cell membranes (9, 12, 20), while antibodies against LcrV and PcrV have also been shown to prevent translocon insertion (10, 11). It was recently shown that LcrV localizes to the tip of the Yersinia TTSS needle to form a tip complex (25). Previously, Turbyfill et al. demonstrated that IpaD also localizes to the Shigella surface (41). To more precisely define the surface localization of IpaD, we have demonstrated that IpaD is localized at the tip of purified Shigella TTSA needles via an interaction that is dependent upon the IpaD C terminus and, furthermore, we show that anti-IpaD antibodies can prevent Shigella contact-mediated hemolysis.
MATERIALS AND METHODS
Materials.
The ipaA ipgD and ipaD (SF622) null mutants were from P. J. Sansonetti (Institut Pasteur, Paris, France) (27). The mxiH null mutant was generated by A. Allaoui (Brussels, Belgium) (3). Sheep red blood cells were from Colorado Serum Company (Denver, CO). Protein A-Sepharose and TiterMax adjuvant were from Sigma Chemical Co. (St. Louis, MO). Alexa Fluor secondary antibodies were from Molecular Probes (Eugene, OR). Escherichia coli, pET15b, and ligation mix were from Novagen (Madison, WI). Restriction enzymes were from New England Biolabs (Beverly, MA). Oligonucleotide primers were from IDT (Coralville, IA). Other chemicals were reagent grade.
Preparation of ipaD mutants for expression in SF622.
pWPsf4D containing the ipaD gene has been described previously (31). ipaD deletion mutants were made by inverse PCR using pWPsf4D as a template, a primer composed of GAGAGA, a BamHI restriction site, a stop codon, and 18 nucleotides flanking the nucleotides to be deleted (the primer for pWPsf4D4E encoding IpaDΔ329-332 was 5′ GAGAGAGGATCCTCAAAGTTTATCTGTATCTGTACA, that for pWPsf4D3E encoding IpaDΔ330-332 was 5′ GAGAGAGGATCCTCAAAAAAGTTTATCTGTATCTGTA, and that for pWPsf4D2E encoding IpaDΔ331-332 was 5′ GAGAGAGGATCCTCAGAGAAAAAGTTTATCTGTATC) and a primer encoding the 3′ region up to and including the BamHI site of pWPsf4 (5′ GAGAGAGGATCCTCTAGAGTCGA). The PCR product was digested with BamHI, intramolecularly ligated, and transformed into E. coli NovaBlue. The resulting plasmid was electroporated into S. flexneri SF622. Ampicillin selection ensured the presence of the plasmid, while kanamycin resistance and/or Congo red binding indicated the presence of the Shigella virulence plasmid.
Recombinant protein purification.
DΔ1-120/pET15b, which encodes IpaDΔ1-120 was made by ligating the XhoI/BamHI fragment of pWPsf4D1112 into XhoI/BamHI pET15b (31). D23E/pET15b, which encodes IpaDΔ321-332, was made by ligating the NdeI/BamHI fragment from pWPsf4D23E into pET15b (31). D26E/pET15b, which encodes IpaDΔ328-332, was made by ligating the NdeI/BamHI fragment from pWPsf4D26E into pET15b (31). D21E/pET15b, which encodes IpaDΔ281-332, was made by ligating the NdeI/XhoI fragment from pWPsf4D2122 into pET15b (31). Tuner(DE3) E. coli was transformed with the resulting plasmid for high levels of protein expression. Recombinant proteins were purified by nickel chelation chromatography as described previously (21).
Invasion and contact hemolysis.
Contact-mediated hemolysis of sheep erythrocytes has been described previously (30, 31). For the neutralization of Shigella-induced contact-mediated hemolysis, the assay was modified as follows. Bacteria (107 cells in 50 μl of phosphate-buffered saline [PBS]) were combined with 50 μl of immunoglobulin G (IgG) and/or recombinant protein in PBS and allowed to incubate at 37°C for 30 min. This mixture was combined with 50 μl of washed erythrocytes, and the assay was continued. S. flexneri invasion of Henle 407 cells was monitored with a gentamicin-protection assay using an agarose/agar overlay as previously described (30, 31). For the neutralization of invasion, bacteria grown to mid-log phase were harvested by centrifugation and resuspended in PBS. The bacteria were then incubated with 10 μg IgG and/or 20 μg recombinant IpaD for 30 min prior to use in the invasion assays. To reduce exposure risks to laboratory personnel, the ipaA ipgD null mutant was used (27). This strain is deficient in IpaA and IpgD, proteins that are involved in the reorganization of the host cell morphology. This strain has a wild-type phenotype for secretion control, invasion, and hemolysis (27).
Uninduced and induced secretion of TTSS proteins.
Uninduced secretion of the TTSS is determined by measuring protein secretion by bacteria grown overnight in tryptic soy broth (16, 23). Induced secretion is determined by measuring protein secretion after the addition of Congo red, which causes a rapid burst of protein secretion by the Shigella TTSS, thus mimicking the response seen upon host cell contact (16). Both assays were used to determine the phenotype of the IpaD mutants as described previously (16). Briefly, proteins from the clarified overnight growth media were precipitated with 10% trichloroacetic acid, washed, and resuspended in sodium dodecyl sulfate (SDS) sample buffer. Sample buffer was added to a final concentration of 1× to the proteins from the clarified induction media. For both induction states, proteins from equal amounts of bacteria based on A600 were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose, and IpaB, IpaC, and IpaD were detected by using mouse monoclonal antibodies followed by Alexa Fluor 680 goat anti-mouse IgG (Molecular Probes, Eugene, OR). Western blot images were obtained using an Odyssey infrared imager (LI-COR, Lincoln, NE).
Rabbit anti-MxiH antibody production and other antisera.
The preparation of recombinant MxiH has been described previously (8). New Zealand White rabbits were immunized intramuscularly with 50 μg of MxiH four times at 2-week intervals with complete Freund's adjuvant or TiterMax, and incomplete Freund's adjuvant was used on the first and second immunizations. Blood was collected 11 days after the final immunization and tested by enzyme-linked immunosorbent assay against MxiH as previously described (41). Enzyme-linked immunosorbent assay titers ranged from 6,400 to 25,600. Shigella polyclonal and monoclonal antibodies to IpaB, IpaC, and IpaD have been previously described (24, 41).
Purification of IgG.
One milliter of rabbit serum was passed over 0.5 ml of PBS-equilibrated protein A-Sepharose. The resin was washed with PBS, and the IgG was eluted with 0.1 M glycine-HCl, pH 3.0. IgG was dialyzed against PBS and stored at −20°C. Protein concentrations were determined by UV absorbance using an extinction coefficient of 0.724 mg/A280.
Confocal microscopy.
Shigella were grown at 37°C in tryptic soy broth to mid-log phase, collected by centrifugation, and resuspended in 4% formaldehyde in PBS. The bacteria were fixed to the slide, blocked with 1% bovine serum albumin, and treated with the mouse monoclonal anti-IpaD antibody (16F8), followed by Alexa Fluor 488 goat anti-mouse IgG or rabbit anti-MxiH (R73), anti-IpaB (R69), or anti-IpaC (R64) antibody, followed by Alexa Fluor 568 goat anti-rabbit IgG. Imaging was performed using a Zeiss Meta 510 laser scanning confocal microscope, alternately using either the 477-nm laser line and a 505 to 530 band-pass emission filter (Alexa 488) or the 543-nm laser line with a 580-nm long-pass emission filter (Alexa 568). Transmission images were also obtained.
Electron microscopy.
When mxiH is subcloned behind the inducible lac promoter in pRK2 and protein expression is induced in a mxiH null mutant (mxiH/pRK2//SH116), long needles greater than 300 nm are made (16, 37). This strain was grown to mid-log phase in tryptic soy broth containing 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C. Cells were harvested by centrifugation, resuspended in 4% formaldehyde in PBS, collected by centrifugation, and resuspended in cold PBS at 1/30 of the original culture volume. The needles were sheared by passing the cell suspension once through an 18-gauge needle. After removal of the bacterial cells by centrifugation, the supernatant containing the sheared needles was clarified twice by centrifugation at 15,000 × g. The needles were precipitated by the addition of polyethylene glycol 6000 and NaCl to 2% and 0.1 M, respectively. After 20 min on ice, needles were collected by centrifugation at 10,000 × g and resuspended in minimal Tris-buffered saline. The needle sample was diluted, applied to carbon/Formvar-coated copper grids, and treated with anti-IpaD rabbit IgG followed by 5 nm gold-labeled goat anti-rabbit IgG (Nanogold, Yaphank, NY). Negative staining with 2% uranyl acetate permitted the visualization of the purified needles. A similar procedure was used to stain IpaD associated with needles on intact Shigella isolates. In this case, mid-log phase bacteria were harvested and resuspended in PBS at 1/10 the original culture volume before being applied to grids. Grids were imaged using a JEOL 1200-EX II transmission electron microscope at an electron acceleration voltage of 120 KeV.
Far-UV circular dichroism spectroscopy.
Far-UV circular dichroism (CD) spectra were collected using a Jasco J-720 spectropolarimeter equipped with a Peltier temperature controller (Jasco, Inc., Easton, MD). The spectra were acquired using a 0.1-cm-path-length cuvette at 10°C. A resolution of 0.5 nm and a scanning speed of 20 nm/min with a 2-s response time were employed. Spectra presented are averages of three consecutive measurements. Secondary structure content was estimated using the Dichroweb software package (18, 43), which allows the analysis of secondary structure content using the algorithms CONTIN (35), SELCON (32), and CDSSTR (19). The thermal unfolding of the proteins was followed by monitoring the ellipticity at 222 nm over a temperature range of 10 to 90°C with a resolution of 0.5°C and 15°C/h heating rate. Protein concentration was 0.1 mg/ml. CD signals were converted to mean residue molar ellipticities, [θ]R, and the thermal transitions were analyzed using the Jasco spectral manager and Microcal Origin 6.0 software (17).
RESULTS
Neutralization of contact-mediated hemolysis by anti-IpaD antibodies.
It was recently demonstrated by Goure et al. that anti-PcrV antibodies protected erythrocytes from hemolysis by P. aeruginosa by preventing membrane insertion of the TTSS translocon (11). Contact-mediated hemolysis is also the assay for assessing IpaB/IpaC translocon insertion into erythrocytes by Shigella (2, 31). To determine whether anti-IpaD antibodies could also block erythrocyte lysis, S. flexneri was incubated with anti-IpaD rabbit IgG and used in a standard contact-mediated hemolysis assay (Fig. 1A). At 25 ng/μl, rabbit anti-IpaD IgG (R63) (41) neutralized contact-mediated hemolysis by more than 50%, while 100 ng/μl reduced it by over 75%. In contrast, rabbit anti-IpaC IgG (R64) had no effect (Fig. 1A). The anti-IpaD antisera were raised against recombinant IpaD (rIpaD) and recognize epitopes throughout IpaD, including six surface-exposed epitopes within the N-terminal 120 amino acids (41). In contrast, antibodies raised against a peptide containing residues 280 to 295 from the IpaD C terminus (R23) that did not detect IpaD at the Shigella surface (41) did not neutralize S. flexneri contact hemolysis (Fig. 1A).
FIG. 1.
Neutralization of contact hemolysis by anti-IpaD antibodies. (A) Rabbit anti-IpaD IgG (•), rabbit anti-IpaD280-295 IgG (○), and rabbit anti-IpaC IgG (▾) were incubated with bacteria for 30 min prior to a standard contact-mediated hemolysis assay (31). (B) Lysis was restored by competitive titration of rIpaD (•) and IpaDΔ281-332 (▾) but not rIpaDΔ1-120 (○) into the bacteria/rabbit anti-IpaD IgG (50 ng) mixture. Contact hemolysis is presented as values relative to the hemolysis caused by the Shigella ipaA ipgD null mutant, which has been assigned a value of 100% ± standard deviation (SD) (n = 3). Error bars indicate standard deviations.
The specificity of the anti-IpaD IgG in protecting erythrocytes from S. flexneri-induced lysis was tested by adding rIpaD as a competitor for antibody binding. The addition of as little as 50 ng of rIpaD restored complete erythrocyte lysis (Fig. 1B). Similarly, when the C-terminal deletion mutant rIpaDΔ281-332 was used as competitor, lysis was restored. Conversely, when the N-terminal deletion mutant rIpaDΔ1-120 was tested as an antibody binding competitor, it provided no reversal of the protection from hemolysis offered by the IpaD antibodies, suggesting that the neutralizing antibodies recognize a region within residues 1 to 120. Spectral analysis by circular dichroism demonstrated that rIpaDΔ1-120 retains all of its secondary structure and thermal stability (M. Espina et al., submitted for publication) and, thus, appears to be folded correctly. Therefore, the inability of rIpaDΔ1-120 to competitively inhibit R63-mediated neutralization of hemolysis is not an artifact of protein misfolding. Taken together, the data suggest that the surface-exposed N-terminal region of IpaD is critical for translocon insertion.
Previously, it was shown that deletions within the N-terminal region have a greater effect on hemolysis than on invasion (31). To determine whether the rabbit anti-IpaD IgG (R63) could impact invasion, a standard gentamicin-protection assay was performed. The anti-IpaD IgG neutralized invasion to 57% ± 12% of that of wild-type Shigella, which could be restored (118% ± 11%) by the addition of rIpaD as a competitor for antibody binding. These data suggest that these antibodies can neutralize both invasion and contact hemolysis.
IpaD is found on the Shigella cell surface and localizes to the tip of the TTSA needle.
The localization of IpaD to the Shigella cell surface was initially examined using immunofluorescence confocal microscopy. After labeling with rabbit anti-IpaD antibodies and Alexa Fluor 568-conjugated goat anti-rabbit IgG (Fig. 2A and E), a punctate zone staining pattern was seen on the bacterial surface. A similar pattern for LcrV staining on the Yersinia surface was seen by Fields et al. (9). Rabbit anti-MxiH antibodies were then used to detect surface-exposed Shigella TTSA needles. A similar punctate zone staining pattern was seen for the anti-MxiH antibodies (Fig. 2B and F). Staining by anti-IpaD antibodies was not seen on either an ipaD (Fig. 2C and G) or an mxiH null mutant (Fig. 2D and H). When S. flexneri isolates were examined for surface-exposed IpaB or IpaC, anti-IpaB staining was seen in only about 1% of the bacteria and no anti-IpaC staining was seen (data not shown). Thus, IpaD is present on the bacterial surface prior to host cell contact, while IpaB and IpaC are not. Western blot analysis indicates that both IpaD and IpaB are present in long needles sheared from the surface of Shigella; however, IpaB was not detected as being surface exposed on the uninduced bacteria by any of the antibodies tested (data not shown). In contrast, IpaC was not even found associated with the sheared needles.
FIG. 2.
Presentation of IpaD at the Shigella cell surface. For surface localization of IpaD and MxiH, S. flexneri was treated with anti-IpaD mouse monoclonal antibodies/Alexa Fluor 488-conjugated goat anti-mouse IgG (E) or rabbit anti-MxiH antibodies/Alexa Fluor 568-conjugated goat anti-rabbit IgG (F). When an ipaD null mutant (G) or a mxiH null mutant (H) was treated with anti-IpaD antibody/Alexa Fluor 488-conjugated goat anti-mouse IgG, no IpaD was visualized on the cell surface. Panels A to D are differential interference contrast micrographs of panels E to H, respectively.
Because IpaD and MxiH displayed similar staining patterns on the Shigella surface, bacteria were double labeled with anti-IpaD monoclonal antibodies and rabbit anti-MxiH antibodies. Secondary labeling with Alexa Fluor 488-conjugated goat anti-mouse IgG and Alexa 568 goat anti-rabbit IgG then allowed the visualization of IpaD and MxiH independently on the same bacterium (Fig. 3). When the images were merged, it appeared that IpaD and MxiH were colocalized (Fig. 3D). Although MxiH labeling can occasionally be seen without IpaD labeling (data not shown), no IpaD labeling is seen without concomitant MxiH staining (Fig. 3). These data suggested that surface-localized IpaD is associated with the MxiH needle of the Shigella TTSS prior to secretion induction. Recent findings by Mueller et al. (25) further strengthened the argument that IpaD may be localized to the tip of the Shigella TTSA needle.
FIG. 3.
Colocalization of IpaD and MxiH on the Shigella surface. Double labeling of S. flexneri with anti-IpaD monoclonal antibodies/Alexa Fluor 488-conjugated goat anti-mouse IgG and anti-MxiH rabbit antibodies/Alexa Fluor 568-conjugated goat anti-rabbit IgG. Panel A shows the differential interference contrast image of the bacteria. Panel B (green pseudo-colored image) indicates IpaD labeling. Panel C (red pseudo-colored image) indicates MxiH. Panel D is the merged image. Arrows illustrate that IpaD labeling occurs only where there is also MxiH labeling.
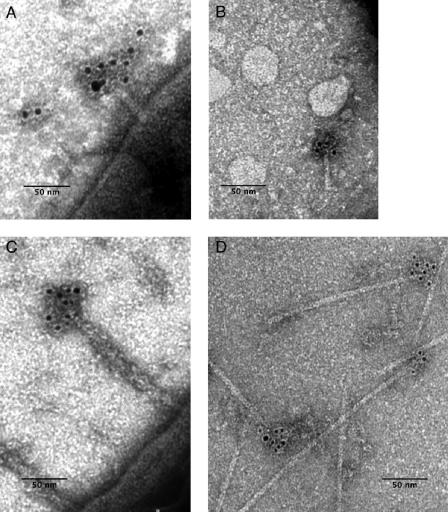
To determine whether IpaD is localized at the TTSA needle tip, immunogold labeling, coupled with transmission electron microscopy, was used. Shigella isolates were harvested and used directly for immunogold labeling of any IpaD associated with the bacterial surface. It was found that immunogold labeling occurred at only the distal tip of MxiH needles that were associated with the bacteria (Fig. 4A and C). To obtain more detailed images (Fig. 4D), needles were sheared from the mxiH null mutant complemented with mxiH/pRK2 (mxiH/pRK2//SH116) (16). Because the mxiH gene in the pRK2 plasmid is under the control of lac promoter, the addition of IPTG is required for protein expression and the restoration of all virulence functions. The needles formed in this strain, however, can be 300 nm or more in length rather than the typical 50-nm-long needles of wild-type strains (Fig. 4D). These long needles are more readily sheared from the Shigella surface for individual examination (16). After formaldehyde treatment, long needles were sheared from the surface of mxiH/pRK2/SH116 and purified. IpaD was seen at the tip of these needles by immunogold labeling (Fig. 4C), while no IpaD staining could be seen for needles prepared from the ipaD null mutant SF622 (data not shown). Gold labeling of IpaD is asymmetric, being observed at only one end of the sheared needles, demonstrating that IpaD resides at only the exposed tip of the MxiH needle. It should be noted that TTSA needles do not easily shear from the Shigella surface unless mxiH is overexpressed to allow long-needle formation. This is presumably due to the base of the needle being protected within the O-polysaccharide layer of the Shigella surface lipopolysaccharide, which has been shown to impact type III secretion (42), and is why Yersinia needles must have a minimum length to make host cell contact (14). Because the base of the MxiH needle is protected from shearing, the polar localization of IpaD on the sheared needles represents the needle tip and not the base. The localization of IpaD on the needle tips is corroborated by the images in Fig. 4A and C. Moreover, no needle fragments had IpaD at both ends and some, presumably broken internal fragments, failed to have IpaD at either end, further indicating that the observed IpaD is located at only the exposed tip of the needle.
FIG. 4.
Transmission electron micrograph of immunogold-labeled IpaD at the tip of negatively stained needles. (A) Shigella isolates were grown to mid-log phase, harvested, and applied to carbon/Formvar-coated copper grids. They were then treated with rabbit anti-IpaD IgG and detected with 5 nm gold-labeled goat anti-rabbit IgG. Uranyl acetate (2%) was used for negative staining. (B) Needles were sheared from the Shigella, applied to carbon/Formvar-coated copper grids and treated as described above for panel A. (C) The Shigella strain mxiH/pRK2/SH116, which makes long needle, was treated as described for panel A. (D) Long needles (≥300 nm) were sheared from mxiH/pRK2/SH116 and treated as described above for panel A. Magnification, ×120,000.
The IpaD C terminus is required for surface localization.
Previous deletion mutagenesis suggested that the IpaD N terminus (residues 1 to 120) is required for efficient translocon insertion into target cell membranes, while the IpaD C terminus (residues 120 to 332) participates in protein folding and TTSS secretion control (31). To correlate IpaD localization at the needle tip with secretion control, Shigella isolates making different IpaD deletion mutants were probed with anti-IpaD rabbit antibodies/Alexa Fluor 568 goat anti-rabbit IgG and assessed for surface-exposed IpaD via immunofluorescence microscopy. The N-terminal deletion mutants IpaDΔ41-80 and IpaDΔ81-120 were largely surface localized with 94 and 88% of the bacteria being stained, respectively. In contrast, only 1% of the bacteria making IpaDΔ161-200 were stained. These results were expected because large deletions (>40 residues) between residues 120 to 320 appear to give rise to an unstable form of IpaD (data not shown) that is unable to complement SF622 (31).
We have previously shown that IpaD's C-terminal tail is critical for invasion functions and that deleting only five residues from its C terminus abolishes invasion, contact hemolysis, and secretion control (31). These IpaD C-terminal deletion mutants also failed to present IpaD on the bacterial surface. Less than 10% of the bacteria making the five-amino-acid deletion (IpaDΔ328-332) had IpaD on their surfaces. Because the C-terminal deletion mutants secrete large amounts of IpaD, these results show that the IpaD surface staining of wild-type bacteria and other mutants is not a result of random cross-linking of secreted IpaD to the needle during the staining process.
When IpaD mutants with C-terminal deletions of two, three, or four amino acids were examined, the proportion of bacteria staining with anti-IpaD antibodies was significantly greater for IpaDΔ331-332 than that for IpaD with three or four residue deletions (Table 1). Correspondingly, the ability of smaller IpaD deletion mutants to complement invasion and contact hemolysis was also enhanced as the deletion became smaller (Table 1). Wild-type invasion levels were seen with IpaDΔ331-332. The control of secretion, both uninduced and induced, strengthened with the addition of each amino acid (Fig. 5).
TABLE 1.
Phenotype of IpaD C-terminal deletion mutants
| IpaD straina | Bacteria with exposed IpaDb | Invasionc | Hemolysisd |
|---|---|---|---|
| SF622 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| IpaD | 97.80 ± 0.51 | 100 ± 10e | 100 ± 14f |
| IpaDΔ328-332 | 0 ± 0.0 | 0.1 ± 0g | 0 ± 0g |
| IpaDΔ329-332 | 10.42 ± 1.73 | 16 ± 7 | 8 ± 3 |
| IpaDΔ330-332 | 47.19 ± 1.93 | 56 ± 21 | 27 ± 10 |
| IpaDΔ331-332 | 77.21 ± 6.94 | 110 ± 10 | 100 ± 15 |
SF622 is the ipaD null mutant. All other strains are SF622 transformed with the plasmid encoding either wild-type IpaD or one of the C-terminal deletion mutants.
Percentage of cells with surface-exposed IpaD as determined by immunofluorescence microscopy. At least 100 bacteria in each of three randomly selected fields were counted (n = 3).
Invasion is relative to IpaD (pWPsf4D/SF622) ± SD (n = 3) and is expressed as a percentage of the control.
Contact-mediated hemolysis is relative to IpaD (pWPsf4D/SF622) ± SD (n = 5) and is expressed as a percentage of the control.
Raw value of invasion of Henle cells by pWPsf4D/SF622 is 207 ± 27 colonies.
Raw value of contact-mediated hemolysis of pwPsf4D/SF622 is 3.5 ± 0.14 (A545).
Value is from reference 31.
FIG. 5.
Analysis of uninduced and induced secretion of IpaD mutants. SF622, the ipaD null mutant, and SF622 transformed with a plasmid expressing either wild-type IpaD or one of the deletion mutants were analyzed for their abilities to secrete IpaB, IpaC, and IpaD. Supernatants from either bacterial cultures grown overnight (A) or bacteria induced to secrete by the addition of Congo red (B) were analyzed by Western blot analysis using mouse monoclonal anti-IpaB, anti-IpaC, and anti-IpaD antibodies with Alexa 680 goat anti-mouse antibodies (Molecular Probes, Eugene, OR). Western blot images were obtained using an Odyssey infrared imager (LI-COR, Lincoln, NE). In panel A, the lanes between SF622 and Δ329-332 were not relevant for this study and were deleted.
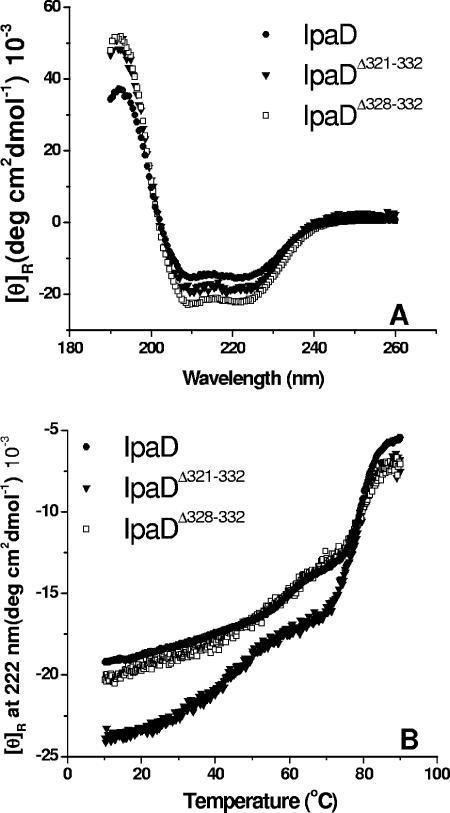
Large deletions within the C-terminal two-thirds of IpaD result in misfolding of the protein, which could easily account for the failure of the protein to localize at the Shigella cell surface (M. Espina et al., submitted for publication). This is not presumed to be the case, however, for small deletions at the immediate C terminus of IpaD. To test for proper protein folding, two of the C-terminal mutant proteins, IpaDΔ321-332 and IpaDΔ328-332, were purified and examined by CD spectroscopy to determine the effect of the deletions or addition on the secondary structure of the mutant proteins (Fig. 6A and Table 2). The CD spectra of IpaD and the short IpaD C-terminal deletion mutants showed similar double minima at 208 nm and 222 nm, suggesting a significant amount of helical structure as predicted by the in silico analysis of IpaD. The secondary structure content was estimated from the far-UV CD spectra using CDSSTR (19) (Table 2). No major changes were observed in the distribution of secondary structure of the IpaD deletion mutants (Table 2). Of the proteins tested, IpaDΔ328-332 showed the greatest change in estimated secondary structure content.
FIG. 6.
Secondary structure analysis of IpaD and IpaD mutants by CD. (A) CD spectra were collected at 10°C at a rate of 20 nm/min employing a resolution of 0.5 nm and are an average of three consecutive measurements. Protein concentrations were 0.1 mg/ml in PBS. The double minima shown are indicative of a highly α-helical protein. (B) Temperature dependence of the molar ellipticity at 222 nm was monitored from 10 to 90°C at 0.5°C intervals at a rate of 15°C/h. Protein concentrations were 0.1 mg/ml in PBS. Although the error bars have been omitted for clarity, the individual data entries have a typical uncertainty of 5% or less (n = 3).
TABLE 2.
Secondary structure estimates for IpaD and IpaD mutants
| % Contenta
|
||||
|---|---|---|---|---|
| Protein | α-Helix | β-Sheet | Turn | Random |
| IpaD | 55b | 10 | 12 | 22.5 |
| IpaDΔ321-332 | 61 | 12 | 8 | 18.5 |
| IpaDΔ328-332 | 66 | 5 | 8 | 20 |
The estimated percent content of each type of secondary structure is provided.
Secondary structure contents are estimated to be ± 3%.
The relative thermal stabilities of the IpaD mutants were then determined by monitoring the changes in the ellipticity at 222 nm as a function of temperature (Fig. 6B). IpaD and IpaDΔ328-332 displayed major thermal transitions at about 60 and 80°C (Fig. 6B). The 12-residue deletion at the IpaD C terminus, IpaDΔ321-332, exhibited a broader change at the onset temperature, suggesting that it might experience greater structural perturbation than the full-length protein (Fig. 6B). These results show that small deletions (no more than five residues) or additions at the IpaD C terminus do not have a major impact on its folding or thermal stability. It is therefore likely that the defect in surface localization for IpaD possessing small C-terminal deletions is not a problem with protein structure. The most likely problem with these IpaD mutants thus appears to be the elimination of a region that is necessary for protein-protein interactions at the needle tip.
IpaD localization at the cell surface of mxiH mutant strains.
Kenjale et al. produced 46 mutations spanning MxiH and demonstrated that these mutations either have no specific effect on Shigella virulence properties or fall into five distinct classes, ranging from severe defects that abolish needle polymerization to mild defects that prevent effector secretion (16). These 46 strains were examined for IpaD surface localization (Table 3). Interestingly, only six of these strains failed to exhibit IpaD on their cell surfaces. None of these mutants are capable of invading cultured cells. Three of these strains cannot polymerize needles due to deletions of three, four, or five residues from the C terminus of MxiH. Additionally, IpaD was not detected on the surfaces of S. flexneri with MxiH mutation W10A or K72A or the double mutation P44A/Q51A. The W10A mutant produces short needles, suggesting a polymerization defect that might be unable to localize IpaD to the surface. It is postulated that W10A would prevent required contacts between MxiH monomers and thus inhibit packing and needle formation (J. E. Deane et al., submitted for publication). K72A is also defective in needle polymerization as well as in the secretion of effectors. Like W10, K72 is predicted to be necessary for contact with an adjacent MxiH monomer within the needle and, therefore, is required for needle polymerization (Deane et al., submitted). Not all mutations that impact needle polymerization affected IpaD surface localization. Y57A, for example, is one of nine MxiH mutations that also cause needle polymerization defects, yet IpaD was found to be surface localized in this strain as well as in the other eight polymerization defective mutants. The P44A/Q51A double mutant, however, is unique among the six strains that cannot surface localize IpaD. This double mutant does not have a needle polymerization defect, but it is noninducible with Congo red (16). P44 and Q51 are postulated to map to the outside of the packed needle rather than being involved in monomer-monomer interactions (Deane et al., submitted). Further studies will need to be initiated to understand the impact that these particular MxiH mutations have on the inability to localize IpaD to the surface. Thus, the mutations in MxiH that completely prevent needle formation also prevent IpaD surface localization. But, of the remaining mutant MxiH phenotypes, the vast majority are still able to maintain IpaD at the tip. Therefore, a direct correlation between needle packing or secretion defects and the inability to localize IpaD to the surface of uninduced bacteria cannot be made.
TABLE 3.
IpaD surface localization on Shigella-expressing mutant forms of MxiHa
| Mutation class | Mutants in class | Mutants not displaying IpaD |
|---|---|---|
| mxiH null mutant | mxiH null | |
| Wild-type-like | MxiH, S2A, V3A, N7A, D8A, T11A, S13A, S14A, L15A, S16A, E17A, V74A | |
| Class I needle polymerization | Δ3, Δ4, Δ5, W10A, Y57A | Δ3, Δ4, Δ5, W10A |
| Class II needle length regulation defects | K69A, I71A, K72A, D73A, D75A, I78A, I79A, Q80A, R83A | K72A |
| Class III constitutive “on” secretion | P44A, Q51A, 4 loop mutants | |
| Class IV constitutive “uninducible” secretion | D73A, P44A/Q51A | P44A/Q51A |
| Class V “effector” secretion | K69A, K72A, R83A | K72A |
| No specific phenotype assigned | T4A, V5A, P6A, L12A, L54A, T67A, V68A, V70A, N81A, F82A |
Surface localization of IpaD was determined by immunofluorescence using rabbit anti-IpaD antibodies. A complete description of these strains can be found in reference 16.
DISCUSSION
Previous studies have clearly identified IpaD as an essential virulence factor for Shigella (23). The precise role that IpaD plays in virulence, however, has remained enigmatic. The data presented here demonstrate that IpaD localizes to the tip of the Shigella TTSA needle prior to the induction of type III secretion and that proper localization of IpaD to the surface is a prerequisite for cellular invasion and is very likely necessary for the proper insertion of the IpaB/IpaC translocon into the host cell membrane. Previously, we have shown that the deletion of a significant portion of the IpaD N terminus (residues 1 to 120) has little impact on invasion but results in lowered hemolysis due to a slower rate of translocon insertion into target cells (31). In contrast, the IpaD C terminus (residues 121 to 332) is essential for IpaD function since large deletions within this region abolish secretion control, invasion, and contact hemolysis (31). The localization of IpaD to the bacterial surface appears to correlate with these observed virulence phenotypes. N terminus deletion mutants were found to be surface localized, while none of the large C-terminal deletion mutants were visible on the bacteria. In fact, deletions greater than two or three residues at the C terminus result in a dramatic decrease in IpaD present on the Shigella surface (Table 1). CD spectroscopy demonstrates that the secondary structures of these deletion mutants have not been significantly altered. Thus, compromised structural integrity is not likely to be responsible for the inability of these proteins to function.
This is the first demonstration that members of the “IpaD protein family,” which includes SipD of Salmonella typhimurium serovar Typhimurium and BipD of Burkholderia pseudomallei, localize to the tip of the TTSA needle. IpaD, SipD, and BipD have substantial sequence homology, especially within their C-terminal halves, suggesting functional conservation for this region (15) (the ClustalW program; data not shown). Like ipaD, the sipD and bipD null mutants are noninvasive, with the sipD-null mutant also demonstrating uncontrolled translocator secretion (15, 36). Recently, LcrV was found to form a complex at the tip of the Yersinia TTSA needle (25). Furthermore, PcrV and AcrV from Pseudomonas aeruginosa and Aeromonas salmonicida, respectively, complemented the lcrV null mutant to form a similar tip complex (25). As with the IpaD homologs, the LcrV orthologs show amino acid similarity, especially at their C termini (26). Unlike the LcrV orthologs, which are interchangeable, however, the IpaD homologs are unable to restore virulence functions to an ipaD null mutant (data not shown) (31). It is not yet known whether the IpaD homologs may be able to localize to the MxiH needle tip but are unable to control secretion.
When LcrV and IpaD are compared, the sequence homology that is seen at the C terminus within either set of homologs is not evident. Nevertheless, because IpaD, LcrV, and several of their homologs all appear to control translocon insertion or, at the very least, translocator secretion, it is intriguing to speculate that IpaD and LcrV homologs will all localize to the needle tip to form the link between the needle and the translocon. Mueller et al. were able to cross-link sheared needles to demonstrate an interaction between LcrV and YscF (25). Due to the differences in the bacterial systems, needles can be efficiently sheared from the Shigella surface only if the mxiH is overexpressed to form long needles (16). The fraction of IpaD in this macromolecular structure now becomes even smaller than that of the wild-type-length needles. Thus, when the needles are separated by SDS-polyacrylamide gel electrophoresis and subjected to Western blot analysis, it is possible to visualize a weak IpaD band. When the needles are cross-linked, however, the MxiH ladder can be visualized but the IpaD cannot, presumably because the small amount of IpaD is distributed among the many MxiH ladder bands. Although we believe that, as with YscF-LcrV, there is a direct interaction between IpaD and MxiH, we have thus far been unable to establish this in vivo, though the data presented in Table 3 hint that this is the case.
These studies also demonstrate that antibodies to IpaD can protect erythrocytes from contact-mediated lysis by S. flexneri, suggesting that they can neutralize IpaD-dependent presentation of IpaB/IpaC translocons to target cell membranes. Previously, the N-terminal 120 amino acids of IpaD were shown by Turbyfill et al. to be accessible to antibodies and, therefore, surface-exposed on the bacterium (41). Because IpaDΔ1-120 was unable to compete with the anti-IpaD IgG to restore erythrocyte lysis, it appears that the neutralizing epitopes may lie within the first 120 residues of IpaD. LcrV and PcrV are also known as protective or V antigens because antibodies against them protect cultured cells and model animals from Y. pestis and P. aeruginosa infections, respectively (10, 44). Similar protective effects were shown for anti-AcrV antibodies (5). The protective nature of the LcrV and PcrV can now be attributed to their prominent location atop the TTSA needles. We here show that this phenomenon may also occur for IpaD, since antibodies that target the IpaD N terminus protect erythrocytes from Shigella-induced lysis.
IpaD and IpaB have been proposed to interact to form a plug that prevents uncontrolled secretion (22). In this study, within the context of uninduced Shigella, IpaD was surface localized, while IpaB could be found associated with the sheared needles by only Western blot analysis (data not shown). IpaC was not seen associated with the uninduced needles (data not shown). Thus, this study demonstrates that IpaD is the first of the three Shigella virulence proteins involved in translocon formation to be secreted to a position at the needle tip, while, based on its location within the needle, IpaB is probably the second. Based on previous work and this study, we present the following working model. Upon the completion of needle polymerization, IpaD enters the needle channel and takes up a position at the tip of the needle. Based on the requirement for the C terminus to maintain IpaD atop the needle and the inability to detect surface-exposed epitopes within this region, the C terminus of IpaD is likely to lie beneath the main body of the protein or perhaps even be buried within the secretion channel itself. The IpaD C terminus may actually interact with MxiH or, alternatively, IpaB located within the channel. Biophysical characterization of IpaD has revealed the presence of an intramolecular coiled coil that is predicted to involve residues 130 to 174 and 270 to 315 (Espina et al., submitted). Thus, this structure would extend away from the needle and position the N-terminal domain back toward the needle. Although we do not have in vivo data demonstrating a direct interaction between IpaD and MxiH, our model suggests that IpaD may maintain its position at the needle tip via such an interaction. Meanwhile, because the N-terminal epitopes have been shown to be surface exposed and antibodies to these epitopes appear to be neutralizing, it is likely that the 120 residues of the N terminus must lie outside the needle-tip complex. Upon the interaction with a physiological signal in the gut, which perhaps is sensed by the structurally dynamic N-terminal domain (Espina et al., submitted), a conformational change occurs within IpaD to allow IpaB, which has been waiting in queue within the needle secretion channel, to take a position at the needle tip with IpaD. Whether this secretion signal is environmental and prior to host cell contact or whether it actually is host cell contact is not yet known. Once host cell contact is made, however, IpaC is released to complete the needle-translocon conduit. Therefore, in an ipaD null mutant, IpaD is not present at the tip of the needle to maintain IpaB within the needle and, subsequently, atop the needle, thereby allowing the cytoplasmic Ipa stores to be released in an uncontrolled fashion (22). More work will need to be done to explain the phenotype of the ipaB null mutant, but the loss of secretion control by these mutants suggests that IpaD also has a role in maintaining the IpaD “off” conformation. ipaB null mutants continue to have IpaD located on the bacterial surface.
Taken together, the results presented here show that IpaD is located at the tip of the Shigella TTSA needle by interactions involving the IpaD C terminus. Because it is the most distal component of the TTSA, IpaD may be involved in sensing host cell contact and triggering the first step in translocon secretion and insertion. From this position, IpaD has a central mechanistic role in Shigella invasiveness and may be a potential target for therapeutic intervention in the prevention of shigellosis.
Acknowledgments
We thank the Picking lab members and M. Olive for critical reading of the manuscript. We thank M. Buechner for technical assistance.
Funding was provided by AI034428 and AI057927 to W.D.P. and by NIH39334 and the KU Center for Research, Inc., for the Microscopy and Analytical Imaging Laboratory.
Editor: J. T. Barbieri
REFERENCES
- 1.Bernardini, M. L., J. Mounier, H. d'Hauteville, M. Coquis-Rondon, and P. J. Sansonetti. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 86:3867-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blocker, A., P. Gounon, E. Larquet, K. Niebuhr, V. Cabiaux, C. Parsot, and P. Sansonetti. 1999. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 147:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blocker, A., N. Jouihri, E. Larquet, P. Gounon, F. Ebel, C. Parsot, P. Sansonetti, and A. Allaoui. 2001. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39:652-663. [DOI] [PubMed] [Google Scholar]
- 4.Blocker, A., K. Komoriya, and S. Aizawa. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. USA 100:3027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burr, S. E., K. Stuber, and J. Frey. 2003. The ADP-ribosylating toxin, AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion pathway. J. Bacteriol. 185:6583-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordes, F. S., K. Komoriya, E. Larquet, S. Yang, E. H. Egelman, A. Blocker, and S. M. Lea. 2003. Helical structure of the needle of the type III secretion system of Shigella flexneri. J. Biol. Chem. 278:17103-17107. [DOI] [PubMed] [Google Scholar]
- 7.Cossart, P., and P. J. Sansonetti. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304:242-248. [DOI] [PubMed] [Google Scholar]
- 8.Darboe, N., R. Kenjale, W. L. Picking, W. D. Picking, and C. R. Middaugh. 2006. Chemical denaturation of MxiH from Shigella and PrgI from Salmonella. Protein Sci. 15:543-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields, K. A., M. L. Nilles, C. Cowan, and S. C. Straley. 1999. Virulence role of V antigen of Yersinia pestis at the bacterial surface. Infect. Immun. 67:5395-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, D. W., A. Vallis, J. P. Wiener-Kronish, A. Roy-Burman, E. G. Spack, B. P. Mullaney, M. Megdoud, J. D. Marks, R. Fritz, and T. Sawa. 2002. Generation and characterization of a protective monoclonal antibody to Pseudomonas aeruginosa PcrV. J. Infect. Dis. 186:64-73. [DOI] [PubMed] [Google Scholar]
- 11.Goure, J., P. Broz, O. Attree, G. R. Cornelis, and I. Attree. 2005. Protective anti-V antibodies inhibit Pseudomonas and Yersinia translocon assembly within host membranes. J. Infect. Dis. 192:218-225. [DOI] [PubMed] [Google Scholar]
- 12.Goure, J., A. Pastor, E. Faudry, J. Chabert, A. Dessen, and I. Attree. 2004. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72:4741-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale, T. L. 1998. Bacillary dysentery, p. 479-493. In L. Collier, A. Balows, M. Sussman, and W. R. Hansler (ed.), Topley and Wilson's microbiology and microbial infections, vol. 3. Arnold, London, United Kingdom. [Google Scholar]
- 14.Journet, L., C. Agrain, P. Broz, and G. R. Cornelis. 2003. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302:1757-1760. [DOI] [PubMed] [Google Scholar]
- 15.Kaniga, K., D. Trollinger, and J. E. Galan. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 177:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenjale, R., J. Wilson, S. F. Zenk, S. Saurya, W. L. Picking, W. D. Picking, and A. Blocker. 2005. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 280:42929-42937. [DOI] [PubMed] [Google Scholar]
- 17.Kueltzo, L. A., J. Osiecki, J. Barker, W. L. Picking, B. Ersoy, W. D. Picking, and C. R. Middaugh. 2003. Structure-function analysis of invasion plasmid antigen C (IpaC) from Shigella flexneri. J. Biol. Chem. 278:2792-2798. [DOI] [PubMed] [Google Scholar]
- 18.Lobley, A., L. Whitmore, and B. A. Wallace. 2002. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18:211-212. [DOI] [PubMed] [Google Scholar]
- 19.Manavalan, P., and W. C. Johnson, Jr. 1987. Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 167:76-85. [DOI] [PubMed] [Google Scholar]
- 20.Marenne, M. N., L. Journet, L. J. Mota, and G. R. Cornelis. 2003. Genetic analysis of the formation of the Ysc-Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, YscF and YopN. Microb. Pathog. 35:243-258. [DOI] [PubMed] [Google Scholar]
- 21.Marquart, M. E., W. L. Picking, and W. D. Picking. 1995. Structural analysis of invasion plasmid antigen D (IpaD) from Shigella flexneri. Biochem. Biophys. Res. Commun. 214:963-970. [DOI] [PubMed] [Google Scholar]
- 22.Menard, R., P. Sansonetti, and C. Parsot. 1994. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 13:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills, J. A., J. M. Buysse, and E. V. Oaks. 1988. Shigella flexneri invasion plasmid antigens B and C: epitope location and characterization with monoclonal antibodies. Infect. Immun. 56:2933-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller, C. A., P. Broz, S. A. Muller, P. Ringler, F. Erne-Brand, I. Sorg, M. Kuhn, A. Engel, and G. R. Cornelis. 2005. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310:674-676. [DOI] [PubMed] [Google Scholar]
- 26.Nanao, M., S. Ricard-Blum, A. M. Di Guilmi, D. Lemaire, D. Lascoux, J. Chabert, I. Attree, and A. Dessen. 2003. Type III secretion proteins PcrV and PcrG from Pseudomonas aeruginosa form a 1:1 complex through high affinity interactions. BMC Microbiol. 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niebuhr, K., S. Giuriato, T. Pedron, D. J. Philpott, F. Gaits, J. Sable, M. P. Sheetz, C. Parsot, P. J. Sansonetti, and B. Payrastre. 2002. Conversion of PtdIns(4,5)P(2) into PtdIns(5)P by the S. flexneri effector IpgD reorganizes host cell morphology. EMBO J. 21:5069-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niebuhr, K., and P. J. Sansonetti. 2000. Invasion of epithelial cells by bacterial pathogens the paradigm of Shigella. Subcell. Biochem. 33:251-287. [DOI] [PubMed] [Google Scholar]
- 29.Parsot, C., and P. J. Sansonetti. 1996. Invasion and the pathogenesis of Shigella infections. Curr. Top. Microbiol. Immunol. 209:25-42. [DOI] [PubMed] [Google Scholar]
- 30.Picking, W. L., L. Coye, J. C. Osiecki, A. Barnoski Serfis, E. Schaper, and W. D. Picking. 2001. Identification of functional regions within invasion plasmid antigen C (IpaC) of Shigella flexneri. Mol. Microbiol. 39:100-111. [DOI] [PubMed] [Google Scholar]
- 31.Picking, W. L., H. Nishioka, P. D. Hearn, M. A. Baxter, A. T. Harrington, A. Blocker, and W. D. Picking. 2005. IpaD of Shigella is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect. Immun. 73:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Provencher, S. W., and J. Glockner. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20:33-37. [DOI] [PubMed] [Google Scholar]
- 33.Sansonetti, P. J., A. Ryter, P. Clerc, A. T. Maurelli, and J. Mounier. 1986. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect. Immun. 51:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasakawa, C., K. Kamata, T. Sakai, S. Makino, M. Yamada, N. Okada, and M. Yoshikawa. 1988. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J. Bacteriol. 170:2480-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sreerema, N., and R. W. Woody. 1993. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209:32-44. [DOI] [PubMed] [Google Scholar]
- 36.Stevens, M. P., A. Haque, T. Atkins, J. Hill, M. W. Wood, A. Easton, M. Nelson, C. Underwood-Fowler, R. W. Titball, G. J. Bancroft, and E. E. Galyov. 2004. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology 150:2669-2676. [DOI] [PubMed] [Google Scholar]
- 37.Tamano, K., S. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran, N., A. B. Serfis, J. C. Osiecki, W. L. Picking, L. Coye, R. Davis, and W. D. Picking. 2000. Interaction of Shigella flexneri IpaC with model membranes correlates with effects on cultured cells. Infect. Immun. 68:3710-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran Van Nhieu, G., E. Caron, A. Hall, and P. J. Sansonetti. 1999. IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J. 18:3249-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran Van Nhieu, G., and P. J. Sansonetti. 1999. Mechanism of Shigella entry into epithelial cells. Curr. Opin. Microbiol. 2:51-55. [DOI] [PubMed] [Google Scholar]
- 41.Turbyfill, K. R., J. A. Mertz, C. P. Mallett, and E. V. Oaks. 1998. Identification of epitope and surface-exposed domains of Shigella flexneri invasion plasmid antigen D (IpaD). Infect. Immun. 66:1999-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M. C. Prevost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313-1317. [DOI] [PubMed] [Google Scholar]
- 43.Whitmore, L., and B. A. Wallace. 2004. DICHROWEB: an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32:W668-W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williamson, E. D., S. M. Eley, A. J. Stagg, M. Green, P. Russell, and R. W. Titball. 2000. A single dose sub-unit vaccine protects against pneumonic plague. Vaccine 19:566-571. [DOI] [PubMed] [Google Scholar]
- 45.Zychlinsky, A., B. Kenny, R. Menard, M. C. Prevost, I. B. Holland, and P. J. Sansonetti. 1994. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol. Microbiol. 11:619-627. [DOI] [PubMed] [Google Scholar]
- 46.Zychlinsky, A., M. C. Prevost, and P. J. Sansonetti. 1992. Shigella flexneri induces apoptosis in infected macrophages. Nature 358:167-169. [DOI] [PubMed] [Google Scholar]