Abstract
Campylobacter jejuni has an N-linked protein glycosylation pathway that is required for efficient cell invasion and chick gastrointestinal colonization by the microbe. In this study, we constructed insertion mutants of 22 putative glycoprotein genes and examined the ability of each to invade the human intestinal epithelial cell line INT-407. Among the mutants tested, one carrying an insertion in Cj1496c was defective for invasion into INT-407 cells; this defect was also observed in an in-frame deletion mutant of Cj1496c (ΔCj1496c). The ΔCj1496c mutant C. jejuni also showed a reduced ability to colonize chick ceca. Site-specific mutagenesis combined with Western blot analysis suggested that the Cj1496c protein is glycosylated at N73 and N169. However, the ΔCj1496c mutant expressing a nonglycosylated form of Cj1496c exhibited levels of invasion and colonization equivalent to those of the parent strain, suggesting that glycans are not directly involved in the function of Cj1496c.
Campylobacter jejuni, a microaerophilic gram-negative bacterium, is the leading cause of bacterial gastroenteritis and food poisoning worldwide, with the number of reported cases currently exceeding 80 per 100,000 people in several developed countries (7). C. jejuni is a common commensal organism of the gastrointestinal tract of many birds and other animals (29). Consequently, C. jejuni infection in humans is often associated with the consumption of contaminated poultry, cross-contamination of other food with raw poultry, and other sources, such as contaminated water and milk products (7, 27). Acute C. jejuni infection causes watery to bloody diarrhea with fever, nausea, and vomiting (4, 35).
Although colonization mechanisms of C. jejuni during infection of the intestinal tract are still unclear, many properties of this organism play important roles, including motility (3), chemotaxis (16), the ability to adhere to and invade intestinal epithelial cells (5), and the production of a cytolethal distending toxin (22). Another trait that plays a role in pathogenicity is an N-linked protein glycosylation system encoded by the pgl locus. Strains with mutations in the pgl locus are diminished for the invasion of intestinal epithelial cells and in the colonization of the gastrointestinal tracts of chicks and mice (16, 19, 20, 33).
A substantial number of proteins in C. jejuni are glycosylated, but the role glycosylation plays in their functions and why the pgl system is required for many aspects of host association are unclear. To analyze the basis for why pgl mutation leads to deficiency in host association, we are taking a systematic approach of generating null mutations in each gene known to encode a glycosylated protein in C. jejuni, as determined by a lectin binding study that reported the identity of many of them (40). In this study, we report results of constructing and testing 22 different mutants with lesions in known glycoprotein-encoding genes. One of these, Cj1496c, is required for the attachment and/or invasion to INT-407 intestinal epithelial cells and the colonization of the chick gastrointestinal tract, although glycosylation is not critical for this role.
MATERIALS AND METHODS
Bacterial strains and media.
C. jejuni 81-176 and its derivatives used in the present study are listed in Table 1. C. jejuni was routinely grown on Mueller-Hinton (MH) agar under microaerophilic conditions at 37°C as previously described (15). When necessary, media were supplemented with the following antibiotics: chloramphenicol (15 μg ml−1), streptomycin (2 mg ml−1), trimethoprim (10 μg ml−1), and cefoperazone (30 μg ml−1). All C. jejuni strains were stored in MH broth with 20% glycerol at −80°C. Escherichia coli strains were grown in LB broth or agar. The following antibiotics were used for E. coli strains when necessary: ampicillin (100 μg ml−1), chloramphenicol (15 μg ml−1), and tetracycline (12.5 μg ml−1). All E. coli strains were stored at −80°C in LB broth with 20% glycerol.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 DlacU169 (f80lacZDM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 13 |
| DH5α/pRK212.1 | Contains conjugative plasmid for the conjugation of plasmid DNA into Campylobacter | 6 |
| C. jejuni | ||
| 81-176 | Clinical isolate | 21 |
| DRH212 | 81-176 rpsLSm | 15 |
| TK062 | DRH212 pglB::skywalker | This study |
| TK075 | DRH212 Cj1565c::skywalker | This study |
| TK108 | DRH212 Cj1496c::skywalker | This study |
| TK211 | TK062 ΔpglB | This study |
| Tk219 | TK108 ΔCj1496c | This study |
| TK325 | DRH212 Cj1496c deleted from nucleotides 133 to 508 and interrupted with SmaI site | This study |
| TK364 | DRH212 Cj1496c (N73N169) chromosomal mutant | This study |
| Plasmids | ||
| pFD1 | Contains gene for the Himar1 transposase and the Himar1 minitransposon | 31 |
| pDRH265 | pUC19 with 1.4 kb cat-rpsL cloned into SmaI site | 15 |
| pX-wing | pFD1 derivative containing the skywalker transposon | This study |
| PECO102 | pRY112 derivative with cat promoter in XhoI-BamHI site | 38 |
| pTK258 | pECO102 with Cj1496c coding sequence | This study |
| pTK265 | pECO102 with Cj1496c (N73N169) | This study |
| pTK312 | pECO102 with Cj1496c-flag | This study |
| pTK358 | pECO102 with Cj1496c (N73)-flag | This study |
| pTK359 | pECO102 with Cj1496c (N169)-flag | This study |
| pTK360 | pECO102 with Cj1496c (N73N169)-flag | This study |
Construction of insertion and deletion mutants.
Defined chromosomal insertion mutants were constructed in streptomycin-resistant 81-176 (DRH212) as described previously by Hendrixson et al. (15). The genes to be disrupted were amplified with approximately 500 bp upstream and downstream of the coding sequence by PCR with primers based on the sequence of C. jejuni NCTC 11168 (30). The PCR products were cloned into pUC19 or pGEM-T vector. The resulting plasmids were then digested with restriction enzymes that cut only once in the coding sequence of the gene to be deleted. When necessary, 5′ or 3′ overhangs were filled in with T4 DNA polymerase to make a blunt end. The genes were then interrupted by the ligation of a SmaI cat-rpsL cassette from pDRH265 into the digested site. For the disruption of genes lacking convenient restriction sites, transposon mutagenesis was carried out. The cat-rpsL fragment was amplified by PCR with primers containing 5′ MluI sites and ligated into MluI-digested pFD1, creating pX-wing harboring the transposon skywalker; the in vitro transposition procedure was described previously (15). Constructed suicide plasmids were electroporated into DRH212. Transformants were selected by growth on chloramphenicol, and the interrupted locus was verified by PCR analysis. To create in-frame deletions in Cj1496c and pglB, ΔCj1496c-F and ΔCj1496c-R and ΔpglB-F and ΔpglB-R, respectively. were synthesized. The sequences of these primers are shown in Table 2. The PCR was carried out using these primers and Pfu DNA polymerase. Plasmids containing a 2.0-kb fragment harboring Cj1496c and a 3.0-kb fragment harboring pglB were used as template. Following thermal cycling, template DNA was eliminated by DpnI digestion and DH5α was transformed by amplified fragment. The resulting suicide plasmids were electroporated into the appropriate insertionally inactivated mutant. Transformants were selected on 2 mg ml−1 of streptomycin and screened for the loss of the cat-rpsL cassette by screening on MH agar with 15 μg ml−1 of chloramphenicol. The deletion of the targeted genes was verified by PCR analysis.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| ΔCj1496c-F | TTTAATCACCTAATTACTAGCATTATTATCCATCTTTTTCTCCTTTGAAAAAACGACTGA |
| ΔCj1496c-R | TCAGTCGTTTTTTCAAAGGAGAAAAAGATGGATAATAATGCTAGTAATTAGGTGATTAAA |
| ΔpglB-F | CTCTAGAATTAATCACCAAGTCCGCAAACAAAACTAAATAAGGG |
| ΔpglB-R | CCCTTATTTAGTTTTGTTTGCGGACTTGGTGATTAATTCTAGAG |
| N73Q-F | AAAGAGGCTGAGGTTCAAGCAACTTTGGCAAAAATTG |
| N73Q-R | CAATTTTTGCCAAAGTTGCTTGAACCTCAGCCCTCTTT |
| N169Q-F | GCTTTTGAAAAATTTAGATAATCAAGCTAGTAATTAGGTGATTAA |
| N169Q-R | TTAATCACCTAATTACTAGCTTGATTATCTAAATTTTTCAAAAGC |
| ΔCj1496c::smaI-F | CATTATTTTAATCACCTAATTACTAGCCCGGGTCTTGCTTCATCAAATTCTCTAG |
| ΔCj1496c::smaI-R | CTAGAGAATTTGATGAAGCAAGACCCGGGCTAGTAATTAGGTGATTAAAATAATG |
Complementation of mutants.
The coding sequences from the second codon to the stop codon of Cj1496c were amplified by PCR. A FLAG tag sequence (5′-GATTATAAAGATGATGATGATAAA-3′) was introduced by PCR to create a gene encoding a fusion protein with the FLAG epitope at the C-terminal end. The amplified fragments were cloned into pGEM, and the resulting plasmids were digested with BglI and XhoI. The fragments were then purified and cloned into pEco102. Plasmids for complementation were introduced into E. coli DH5α/pRK212.1. Conjugations were performed as described by Guerry et al. (12).
Site-directed mutagenesis.
Introduction of point mutations in Cj1496c was performed by a site-directed mutagenesis method using a combination of Pfu DNA polymerase and DpnI as described above. The primer pairs used for this purpose were N73Q-F and N73Q-R and N169Q-F and N169Q-R, whose sequences are shown in Table 2.
Construction of a Cj1496c chromosomal mutant.
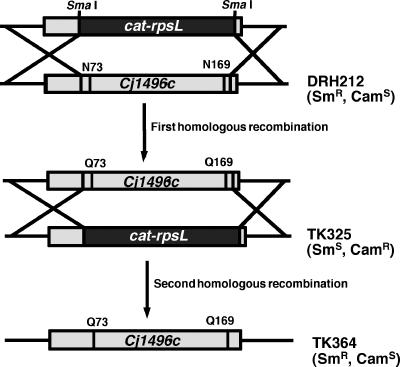
A diagram of the method for constructing a Cj1496c mutant in which the two glycosylation sequons were altered is shown in Fig. 1. Fusions of upstream and downstream DNA fragments of Cj1496c interrupted with SmaI site were created by the site-directed mutagenesis method as described above using ΔCj1496c::smaI-F and ΔCj1496c::smaI-R. The resulting plasmid was digested with SmaI and ligated with the cat-rpsL cassette. This plasmid was electroporated into DRH212, creating TK325. Then a pGEM clone with a Cj1496c allele encoding a nonglycosylated form was electroporated into TK325 and selected on MH plates containing 2 mg ml−1 of streptomycin. The proper replacement of the cat-rpsL allele by the mutant Cj1496c allele lacking the glycosylation sequon was confirmed by sequencing.
FIG. 1.
Diagram of the method for constructing a chromosomal mutant of Cj1496c (N73Q and N169Q) in C. jejuni.
Chick colonization assays.
White leghorn strain delta chicken eggs were supplied by a local farm and maintained in an egg incubator (Sportsman Incubator Model 1202; Georgia Quail Farms) for 21 days at 37.8°C, with appropriate humidity and rotation of eggs according to the manufacturer's instructions until the chicks hatched. For testing the cecal colonization capacity of C. jejuni 81-176 derivatives, each strain was streaked on MH agar and grown at 37°C under microaerophilic conditions for 24 h. Twelve to 36 h after hatching, chicks were divided into groups of five to seven and infected orally with 100 μl of each inoculum. Dilutions were plated on MH agar to determine the number of bacteria in each inoculum. Each group of chicks was housed separately in brooders and given water and food ad libitum. Chicks were sacrificed at day 7 postinfection, and ceca were collected, weighed, and resuspended in phosphate-buffered saline (PBS) to a final concentration of 0.1 g of cecal content per ml. Tenfold serial dilutions of each sample were made and plated on MH agar containing 10 μg ml−1 trimethoprim and 30 μg ml−1 cefoperazone to select for the growth of C. jejuni. Plates were incubated for 48 h at 37°C under microaerophilic conditions. The colonization capacity of C. jejuni in ceca from each chick was reported as the number of CFU per gram of cecal contents.
Motility assays.
Motility phenotypes of strains were tested in MH motility media containing 0.4% agar as previously described (15).
SDS-PAGE and immunoblot analysis.
Whole-cell and protein samples were diluted 1:1 with 2× sodium dodecyl sulfate (SDS) sample buffer and heated to 100°C for 5 min. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) using either 10 or 15% polyacrylamide gels. Separated proteins were transferred to a nitrocellulose membrane and probed with anti-FLAG M2 monoclonal antibody-peroxidase conjugate at 1:1,000 and anti-major outer membrane protein (MOMP) rabbit polyclonal antibody (generously supplied by Q. Zhang, Iowa State University) at 1:1,000, followed by goat anti-rabbit horseradish peroxidase-conjugated secondary antibody at 1:10,000. Antiserum against MOMP was used as control for the cellular fractionation. Proteins were detected with Western lightning chemiluminescence reagent plus.
Adherence and invasion assay.
INT-407 cells were seeded into 24-well plates at semiconfluency (∼1 × 105) approximately 16 h prior to infection. Bacteria were grown in MH biphasic medium. One milliliter of culture medium containing 2 × 107 bacteria was added to each well (multiplicity of infection, 200). Infected monolayers were incubated for 2 h at 37°C in 5% CO2-95% air atmosphere to allow invasion to occur. For time course analyses, the invasion period varied from 30 to 120 min. Following the invasion period, wells for assaying adhesion and invasion (total cell-associated bacteria) were washed three times with Dulbecco's modified Eagle's medium (DMEM) and lysed with 0.1% Triton X-100 in phosphate-buffered saline for 15 min at room temperature. At this time, wells for assaying invasion were washed three times with DMEM and incubated for another 2 h in fresh tissue culture medium containing gentamicin (100 μg ml−1) to kill extracellular bacteria. After the gentamicin kill period, the infected monolayers were washed three times with DMEM and lysed as described above. The number of viable bacteria released from the cells was assessed after serial 10-fold dilutions of the lysates on MH plates.
Chemotaxis assay.
To examine chemotactic behavior of C. jejuni strains, a chemical-in-plug method was carried out (18). Briefly, a 20-ml volume of 2% agar containing PBS and the test chemical was pipetted into petri plates and allowed to solidify at room temperature overnight. Then agar was cut into 6-mm-diameter plugs with a Pasteur pipette. The test chemicals used in this study were 0.1 M l-fucose, l-asparate, l-cystein, l-glutamate, l-serine, pyruvate, succinate, fumarate, citrate, l-malate, and α-ketoglutarate. Bacteria were grown in MH biphasic medium and washed once with PBS. The bacterial pellet was resuspended in 0.4% agar containing PBS and adjusted to an optical density at 600 nm of 0.1. Then 20 ml of this bacterial suspension was added to each of several petri plates. Hard-agar plugs containing a test chemical were placed with sterile toothpicks in the soft agar. The chemotactic behavior was observed after 3 h of incubation under microaerophilic conditions at 37°C.
Cell fractionation.
Cell fractionation was carried out as previously described for C. jejuni (26). Cells were grown in MH biphasic medium in a microaerophilic atmosphere at 37°C. A total of 200 ml of cell suspension was centrifuged (10,000 × g, 10 min at 4°C), and the resulting pellet was resuspended in 10 ml 20% (wt/vol) sucrose and 30 mM Tris/HCl (pH 7.4) at room temperature. EDTA was added to a final concentration of 1 mM, and the suspension was poured into a 100-ml conical flask and stirred at 180 rpm for 10 min at room temperature. The suspension was then centrifuged (10,000 × g for 10 min at 4°C), and the pellet was resuspended in ice-cold 0.5 mM MgCl2 to a volume of 10 ml and stirred gently for 10 min on ice. The suspension was then centrifuged again (10,000 × g for 10 min at 4°C), and the supernatant was collected as the periplasmic fraction. The pellet containing spheroplasts was resuspended in 10 ml 10 mM HEPES (pH 7.3). After one cycle of freeze and thaw, the suspension was sonicated to release cell contents. Cell debris was pelleted at 10,000 × g for 10 min and membranes at 100,000 × g for 60 min at 4°C. The supernatant and pellets from this step were used as sources of cytoplasmic and cell membrane proteins, respectively.
Enzyme assay.
Malate dehydrogenase activity was assayed at a wavelength of 340 nm at room temperature. Rates were obtained by adding 10 μl 14.3 mM NADH to a cuvette containing the following: 500 μl 0.2 M potassium phosphate buffer (pH 7.5) containing 0.2 M KH2PO4 and K2HPO4 at a ratio of 16:84, 16.7 μl 20 mM oxaloacetic acid, 33.3 μl of cell fraction, and 440 μl of H2O. The specific activity of malate dehydrogenase was calculated using an absorption coefficient of NADH of 6.3 mM−1 cm−1 at 340 nm. A unit of malate dehydrogenase activity is defined as 1 μmol of NADH oxidized per min per mg of protein.
RESULTS
Deletion of Cj1496c results in reduced invasion frequency.
Campylobacter jejuni pgl mutants show reduced abilities to adhere to and/or invade intestinal epithelial cells (16, 19, 20, 33). As mutations in the pgl locus can affect many proteins at the same time, we hypothesized that some glycoproteins may be involved in cell invasion by C. jejuni and others may not be. To test this hypothesis, we chose a strategy of inactivating each gene encoding a previously reported glycoprotein. The sequence determinant (sequon) of glycosylation includes the sequence NXS/T, and N-linked glycosylation is presumed to take place within the inner membrane and periplasm (28). Thus, we selected genes that encode both a signal peptide and the putative NXS/T sequon as targets of mutagenesis. Twenty-two insertion mutants were constructed as described in Materials and Methods and tested in cell invasion assays using the human intestinal epithelial line INT-407. Genes tested in this study are shown in Table 3.
TABLE 3.
Glycoprotein genes tested in this study
| Cj gene | Annotation |
|---|---|
| Cj0143c | Periplasmic solute binding protein for ABC transport system |
| Cj0175c | Putative iron uptake ABC transport system periplasmic iron binding protein |
| Cj0200c | Probable periplasmic protein |
| Cj0238 | Probable integral membrane protein |
| Cj0289c | Major antigenic peptide (PEB3) |
| Cj0376 | Probable periplasmic protein |
| Cj0420 | Probable periplasmic protein |
| Cj0511 | Probable secreted proteinase |
| Cj0734c | Histidine binding protein precursor (HisJ) |
| Cj0843c | Putative secreted transglycosylase |
| Cj0906c | Probable periplasmic protein |
| Cj0944c | Probable periplasmic protein |
| Cj0998c | Probable periplasmic protein |
| Cj1018c | Branched-chain amino acid ABC transport system periplasmic binding protein |
| Cj1032 | Probable membrane fusion component of efflux system |
| Cj1214c | Hypothetical protein |
| Cj1380 | Probable periplasmic protein |
| Cj1444c | Putative capsule polysaccharide export system periplasmic protein (KpsD) |
| Cj1496c | Probable periplasmic protein |
| Cj1565c | Paralyzed flagellum protein (PflA) |
| Cj1643 | Putative periplasmic protein |
| Cj1670c | Probable periplasmic protein (CgpA) |
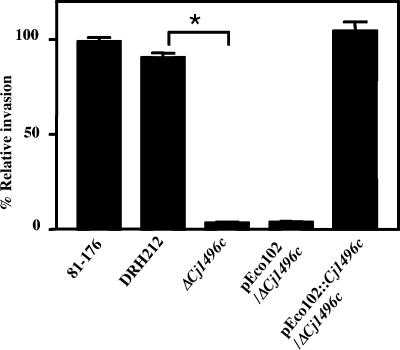
Among the strains tested, those with mutations in Cj1496c and pflA, which was previously reported as a cell invasion determinant, invaded with an efficiency at least 10-fold lower than that of the wild type (data not shown). We selected Cj1496c for further characterization. To confirm that the invasion defect did not result from a polar effect of insertion with the cat-rspL cassette, a strain with an in-frame deletion allele of Cj1496c was constructed. This mutant (ΔCj1496c) showed an approximately 20-fold-reduced ability to invade cells compared to that of 81-176 or its Smr derivative DRH212 (Fig. 2). This mutant phenotype could be completely complemented by the expression of Cj1496c from the cat gene promoter on pEco102. These results demonstrated that Cj1496c influences the cell invasion of C. jejuni.
FIG. 2.
INT-407 cell invasion assay. Cells were infected with the wild type and mutants for 2 h, and invasion values were calculated from the number of bacteria that survived 2 h of incubation in the presence of gentamicin. Values are given relative to the invasion of the wild-type strain 81-176, which was set at 100%. The experiment was performed in triplicate and repeated on at least three separate occasions. The error bars represent standard deviations of triplicate wells. Statistical significance was assessed with an unpaired Student t test. *, P < 0.01.
Reduced colonization of chick ceca by the ΔCj1496c mutant.
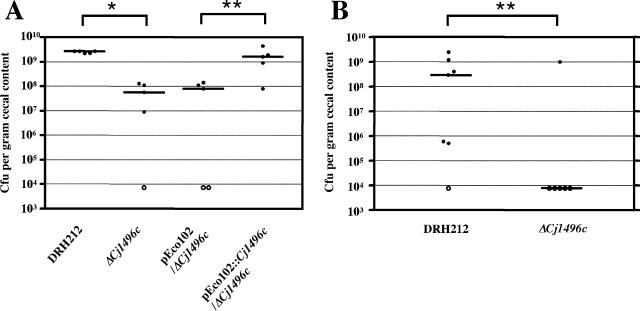
To determine whether Cj1496c has any role in gastrointestinal colonization, we carried out chick colonization experiments. Approximately 12 to 36 h after hatching, chicks were orally infected with 104 C. jejuni and, at 7 days postinfection, C. jejuni in the ceca were enumerated. All chicks in the group inoculated with parental strain DRH212 were colonized, and bacterial loads in the ceca were greater than 109 CFU per gram of cecal content (Fig. 3A). By contrast, the ΔCj1496c mutant exhibited significantly reduced chick colonization levels compared to DRH212. The ΔCj1496c mutant complemented with pEco102::Cj1496c colonized to levels similar to those of DRH212 (Fig. 3A).
FIG. 3.
Colonization of 1-day-old chicks. One-day-old chicks were inoculated with 1 × 104 (A) and 1 × 103 (B) CFU bacteria. The number of bacteria present in the ceca at 7 days postinfection is reported as the number of CFU per gram cecal content. Each closed circle represents the number of bacteria obtained from an individual chick. Open circles represent chicks containing bacterial loads below the limit of detection (<104 CFU per gram cecal content). The median of bacterial concentration recovered is represented by a bar. Statistical significance was assessed with the Mann-Whitney U test. *, P < 0.01; **, P < 0.05.
The burden of C. jejuni in each chick of the group infected with the ΔCj1496c mutant tended to be variable. We assumed that this variation in bacterial burden may reflect a role for the gene product in initially establishing colonization. To test this hypothesis, we inoculated chicks with a reduced number of bacteria (103 bacteria per chick). At this reduced infectious dose, DRH212 was still recovered from six out of seven chicks, whereas the ΔCj1496c mutant was recovered from only one out of six chicks. Taking the data from chick infection experiments using both the higher and lower doses, we conclude that ΔCj1496c establishes colonization poorly compared with the wild type but can reach high levels in the chick gastrointestinal tract once it colonizes (Fig. 3B).
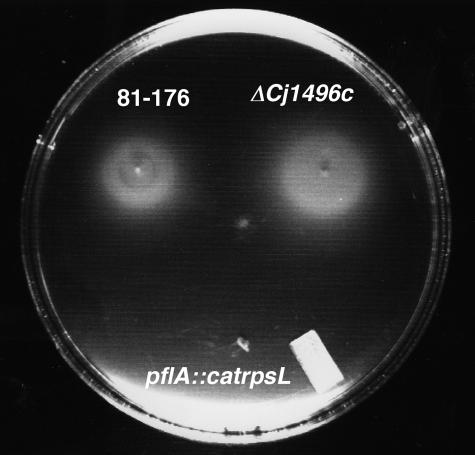
ΔCj1496c mutant swarms faster than the wild type but has the same profile of chemotactic responses as that of the parent strain.
Flagellar motility is positively correlated in C. jejuni with both cell invasion and colonization (9, 11, 16, 38). To determine whether the deficiency in cell invasion observed with the ΔCj1496c mutant is associated with a motility defect, we examined its behavior compared with that of the wild-type strain DRH212 on MH semisolid agar. The mutant displayed a slightly enhanced zone of spreading on a motility plate compared to DRH212 (Fig. 4). The average diameters ± standard deviations (n = 8) of motility rings on semisolid agar plating for 18 h of incubation at 37°C in DRH212 and ΔCj1496c were 9.3 ± 0.4 and 10.7 ± 0.7 mm (P < 0.01), respectively. This is not related to different growth rates of the mutant and the wild type, as they are equivalent (data not shown).
FIG. 4.
Motility phenotypes of DRH212 and ΔCj1496c and pflA::catrpsL mutants in MH motility medium.
To examine whether mutation in Cj1496c affects chemotaxis, we characterized the chemotactic behavior of the ΔCj1496c mutant in the presence of various chemicals reported previously to be chemoattractants (18). Chemotaxis was tested by a chemical-in-plug method as described in Materials and Methods, and chemoattraction was scored as the accumulation of cells forming within the soft agar around the hard-agar plug containing the chemical under investigation. The accumulation of bacterial cells indicating chemoattraction was observed around the plugs containing l-asparate, l-glutamate, l-serine, pyruvate, succinate, l-malate, and α-ketoglutarate but not around plugs containing PBS, l-cystein, l-fucose, or citrate. There was no difference in responses observed with the ΔCj1496c mutant and wild-type bacteria (data not shown).
Kinetic analysis of cell invasion and effect of centrifugation.
To further characterize the association of wild-type and Cj1496c mutant bacteria with host cells, both total cell-associated and intracellular (gentamicin-resistant) bacteria were counted 30, 60, and 120 min after infection. The numbers of each class for both DRH212 and ΔCj1496c steadily increased over the time of this experiment (Table 4), but the percentage of recoverable CFU of the ΔCj1496c mutant was less than 10% of that of the wild type at each time point. Differential survival inside cells was unlikely to be responsible for this observation because reduced numbers of cell-associated mutant bacteria were observed even just 30 min after incubation, and the level of reduction in the numbers of internalized bacteria did not change during the experiment. These results suggest that there may be an adherence defect of the ΔCj1496c mutant that ultimately alters its invasion into INT-407 cells.
TABLE 4.
Kinetic analyses of cell invasion
| Strain (inocula) | No. of cell-associated (internalized) bacteria ata:
|
||
|---|---|---|---|
| 30 min | 60 min | 120 min | |
| DRH212 (1.4 × 107) | 3.4 × 105 ± 0.6 × 105 (3.3 × 104 ± 0.3 × 104) | 4.1 × 105 ± 0.1 × 105 (1.5 × 105 ± 0.2 × 105) | 4.5 × 105 ± 0.8 × 105 (3.5 × 105 ± 0.3 × 105) |
| ΔCj1496c (1.9 × 107) | 1.9 × 104 ± 0.1 × 104 (2.6 × 103 ± 0.3 × 103) | 2.2 × 104 ± 0.2 × 104 (5.5 × 103 ± 0.3 × 103) | 3.2 × 104 ± 0.3 × 104 (1.2 × 104 ± 0.3 × 104) |
The number of cell-associated and internalized bacteria was determined at each time point outlined in Materials and Methods. Data are means ± standard deviations.
Because the ΔCj1496c mutant showed altered motility, we hypothesized that defective cell contact caused by altered motility might contribute to reducing the number of intracellular mutant bacteria. To test this hypothesis, we compared the effect of centrifugation after inoculating the epithelial cells with bacteria. The invasion frequency of the ΔCj1496c mutant did not recover to wild-type levels with centrifugation, although slight increases in bacterial number were observed with both mutant and wild-type strains after centrifugation (Table 5).
TABLE 5.
Effect of centrifugation on cell invasiona
| Strain (inocula) | No. of cell-associated (inernalized) bacteria without centrifugation | % of wild-type bacteria that are cell associated (invaded) | No. of cell-associated (internalized) bacteria with centrifugationb | % of wild-type bacteria that are cell associated (invaded) |
|---|---|---|---|---|
| DRH212 (2.3 × 107) | 6.4 × 105 ± 0.1 × 105 (2.9 × 105 ± 0.1 × 105) | 9.7 × 105 ± 0.1 × 105 (3.1 × 105 ± 0.3 × 105) | ||
| ΔCj1496c (2.5 × 107) | 4.1 × 104 ± 0.1 × 104 (1.2 × 104 ± 0.3 × 104) | 5.7 (3.7) | 1.1 × 105 ± 0.2 × 105 (2.5 × 104 ± 0.3 × 104) | 10.0 (7.7) |
The number of cell-associated and internalized bacteria was determined as outlined in Materials and Methods. Data are means ± standard deviations.
After the addition of bacteria onto wells containing INT-407 cell monolayer, a 24-well plate was centrifuged at 600×g for 10 min to promote bacterium-host cell contact.
Cj1496c is located in the periplasm.
To determine the subcellular location of Cj1496c, we constructed a plasmid expressing Cj1496c tagged with a FLAG epitope at its C terminus. The ΔCj1496c mutant carrying this plasmid exhibited levels of invasion similar to those of the wild type, suggesting that Cj1496c-FLAG is functional (data not shown) and indicating that the tagged protein is behaving like the wild type.
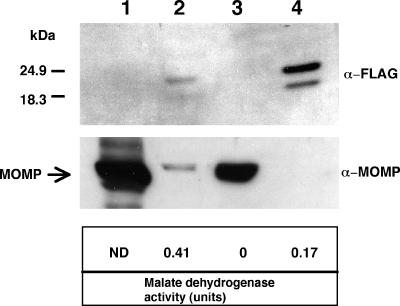
Protein samples from all fractions were analyzed by immunoblotting using anti-FLAG monoclonal antibody. Two bands with different molecular masses were detected specifically in the periplasmic fraction of the ΔCj1496c mutant carrying pEco102::Cj1496c-flag but not in other fractions or whole-cell lysate of ΔCj1496c alone (Fig. 5). Malate dehydrogenase activity was higher in the cytoplasmic fraction than in the periplasmic fraction. The detection of malate dehydrogenase activity in the periplasmic fraction likely reflects the lysis of some spheroplasts or leakage of material from the spheroplasts and not a periplasmic localization of this component. Given the complete lack of observable Cj1496c-FLAG in fractions containing the majority of malate dehydrogenase and MOMP, we conclude that the protein is localized to the periplasmic space in C. jejuni.
FIG. 5.
Intracellular localization of Cj1496c-FLAG. Subcellular fractions of ΔCj1496c expressing Cj1496c-FLAG were separated by SDS-PAGE and, after transfer to nitrocellulose membrane, immunodetected with anti (α)-FLAG monoclonal antibody, with the antiserum raised against recombinant FlgR and with antiserum raised against MOMP. Lane 1, whole-cell lysate of ΔCj1496c; lane 2, cytoplasmic fraction of ΔCj1496c containing pEco102::Cj1496c-flag (TK322); lane 3, membrane fraction of TK322; lane 4, periplasmic fraction of TK322. Malate dehydrogenase activity was measured as marker enzyme of cytoplasmic fraction. ND, not determined.
Cj1496c is glycosylated at two sites.
The N-X-S/T sequon is an essential component of N-linked protein glycosylation in C. jejuni (28). Assuming that this is the only target for N-linked glycosylation, Cj1496c has two sites of potential glycosylation, at positions 73-N-A-T-75 and 169-N-A-S-171. To assess whether these sites are glycosylated, we replaced the asparagine residues at positions 73 and 169 by glutamine residues using site-directed mutagenesis.
We expressed these mutant alleles in both C. jejuni and E. coli; the latter species has no reported system of protein glycosylation (34). In E. coli transformed with pEco102::Cj1496c-flag, two species with different molecular masses were detected. These are consistent with the sizes of the premature form of Cj1496c-FLAG (20.6 kDa) and the mature form lacking a signal peptide (18.6 kDa) (Fig. 6). When expressed in C. jejuni, two species were also observed but each ran with a larger apparent molecular weight than the proteins detected in E. coli. Mutagenesis of the asparagine residue at either position 73 or 169 resulted in a single band bearing the same molecular mass as that of the smaller band found in a strain expressing wild-type proteins. We hypothesized that the single species observed when the N73 and N169 mutants were expressed in C. jejuni represents a form of Cj1496c-FLAG that is glycosylated at only one site. Supporting this hypothesis, a double-mutant allele, in which both asparagine residues were changed, encoded a protein that migrated faster in C. jejuni than the proteins with only a single asparagine altered. The double-mutant protein migrated similarly to how the wild-type protein migrated after expression in the glycosylation-deficient pglB mutant. The migration of the double-mutant protein is also similar to the migration of the putative mature form of wild-type Cj1496c-FLAG after expression in E. coli, which is not expected to glycosylate proteins. These results suggest that Cj1496c can be glycosylated in C. jejuni at two sites, N73 and N169.
FIG. 6.
Cj1496c contains two N-linked glycosylation sites. Whole-cell lysates were separated by SDS-PAGE and, after transfer to nitrocellulose membrane, immunodetected with anti-FLAG monoclonal antibody.
ΔCj1496c mutant expressing nonglycosylated Cj1496c can invade cell and colonize chick ceca at the wild-type level.
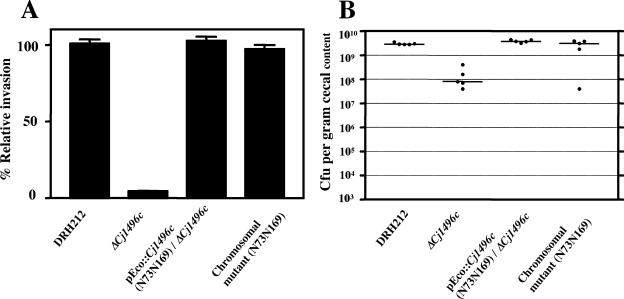
To assess whether glycan modification on Cj1496c plays any role in its function, we carried out an invasion assay and chick colonization experiment with strains expressing the glycosylation sequon mutants described above. The ΔCj1496c mutant expressing pEco102::Cj1496c (N73Q and N169Q) invaded epithelial cells and colonized chick ceca to the same levels as those of the wild-type (Fig. 7). These results imply that glycan modification of Cj1496c has little or no influence on its function.
FIG. 7.
Cell invasion (A) and colonization (B) of mutant expressing nonglycosylated Cj1496c from plasmid and chromosome. (A) Results of an INT-407 cell invasion assay. Values are given relative to the invasion of the parent strain DRH212, which was set at 100%. The experiment was performed in triplicate and repeated on at least three separate occasions. Error bars indicate standard deviations. (B) Colonization of 1-day-old chicks. One-day-old chicks were inoculated with 1 × 104 CFU bacteria. The number of bacteria present in the ceca at 7 days postinfection is reported as the number of CFU per gram cecal content. Each closed circle represents the number of bacteria obtained from an individual chick. The median of bacterial concentration recovered is represented by a bar.
In eukaryotes, glycans play a pivotal role in protein folding and quality control (14). We speculated that the overexpression of Cj1496c by plasmid expression might mask the effect of glycans on its folding or stability. To test this possibility, we constructed a strain in which the wild-type Cj1496c gene on the chromosome was replaced with a mutant allele encoding the nonglycosylated form Cj1496c N73N169. This mutant also invaded INT-407 cells and colonized chick ceca at the wild-type level (Fig. 7).
DISCUSSION
In this study, we identified Cj1496c as a gene whose product, a glycoprotein, influences two key traits of pathogenic C. jejuni. By INT-407 cell invasion assay, we demonstrated that a strain carrying a deletion allele of Cj1496c (ΔCj1496c) showed a reduced ability to adhere to and/or invade cells. We also found that the ΔCj1496c mutant is defective in a chick model of colonization. Our lab and others have observed variability in the outcome of chick infections with C. jejuni strain 81-176, caused by apparent bottlenecks in the infection that eliminate even wild-type organisms from the animals (10, 16). This is most easily demonstrated with mixed infections, where multiple colonization-competent strains are used as inocula. The consistency of poor colonization by the ΔCj1496c mutant, particularly at lower doses of inoculation, along with the consistent high-level colonization of the wild type, leads us to conclude that the Cj1496c gene product is indeed required for efficient chick colonization.
The results of our cell fractionation experiments suggest that Cj1496c is located in the periplasmic space, which is also supported by the observation that FLAG-tagged Cj1496c was protected from protease K digestion of intact cells (data not shown). Therefore, we propose that Cj1496c is indirectly involved in adhesion and/or invasion processes via other bacterial factors which directly react with host molecules rather than through direct interaction with the host cell. As a periplasmic protein, how Cj1496c influences phenotypes, such as chick colonization, adherence, or invasion, is not apparent. One hypothesis to account for the chick colonization defect is that the mutant has a reduced ability to adhere to the intestinal epithelium. Although C. jejuni is readily detected in the crypt lumen without being attached to crypt microvilli in the intestines of chicks (2), adhesins are believed to be involved in the colonization of chicks. For example, strains lacking the fibronectin binding protein cadF colonize chicks poorly (41) but CadF is an outer membrane protein whose binding to the extracellular matrix easily explains its role in adherence.
Indirect mechanisms by which Cj1496c might influence C. jejuni-host interactions include a role as a periplasmic component of relevant signaling or transport activity. Two proteins recently shown to contribute to chick colonization are the methyl-accepting chemotaxis proteins Cj0019 and Cj0262c (16). Methyl-accepting chemotaxis proteins are membrane-spanning signal transduction proteins, and therefore their roles in colonization, like that of the periplasmic Cj1496c, are likely indirect. That strains with mutations in either Cj0019 or Cj00262c are defective for chick colonization suggests that chemotaxis is an important colonization determinant of C. jejuni. Our present data revealed that the Cj1496c mutant is unaffected in its chemotaxis response to several attractants, but we cannot rule out chemotaxis defects because there may be attractants or repellents in vivo that the wild-type responds to but the mutant does not.
Reiterative PSI-BLAST searching showed that Cj1496c has similarity with an intracellular domain of the magnesium/cobalt transporter MgtE. Preliminary data suggest that Cj1496c is not critical for transporting this cation; whereas mutation in Cj1496c did not abolish growth on a low Mg2+-containing medium, mutation in another C. jejuni open reading frame, Cj0726c, which is homologous to bacterial magnesium (and cobalt) transporters, did lead to growth impairment on such a medium (data not shown). PSI-BLAST searching also showed that Cj1496c has similarity with genes found in flagellar gene operons in a number of bacterial species, but no characterization of these genes has yet been reported.
The ΔCj1496c mutant exhibits a hyperswarming phenotype compared with the parent strain DRH212. In E. coli, counterclockwise rotation of flagella results in the bacterium swimming smoothly (running) in a mostly straight line, whereas clockwise rotation causes tumbling (24). A cytoplasmic response regulator, CheY, can bind to the motor in its phosphorylated state and reverse the direction of flagella rotation from counterclockwise (running) to clockwise (tumbling). Swarm size is positively correlated with the tumble frequency, although nonstop tumbling leads to decreased swarm size (37). Therefore, a hypothesis that might explain the hyperswarming phenotype is that the deletion of Cj1496c leads to elevated levels of phosphorylated CheY, thereby increasing the tumbling frequency. The deletion of cheY in C. jejuni results in hyperadherent and hyperinvasive phenotypes (9, 39), while a diploid cheY strain displayed nonadherent, noninvasive, and hypermotile phenotypes (39) similar to what we observed with the ΔCj1496c mutant. The mechanisms involved in this cheY-diploid phenotype have not been elaborated, but perhaps the hyperswarming phenotype of the diploid cheY strain is the result of increased phosphorylated CheY.
An examination of other examples of coordinate regulation of flagellar functions and virulence-associated genes (1, 8, 25, 32) might be instructive for our findings. For example, in Vibrio cholerae, some “hyperswarming” mutants express little or no cholera toxin or toxin-coregulated pilus, two key virulence factors of that pathogen (8). By contrast, some nonmotile mutants of V. cholerae express higher-than-wild-type levels of these factors. These results imply that virulence and flagellar motility may be controlled opposite to one another in V. cholerae and are reminiscent of what we observe with C. jejuni, where the hyperswarming Cj1496c mutant has a reduced capacity for colonization.
Our results suggest that Cj1496c can be glycosylated at two sites. However, based on the results of invasion and colonization experiments, we discerned no influence of glycan modification on the function of Cj1496c. In Mycobacterium tuberculosis, the alteration of sugar attachment sites on a 19-kDa lipoprotein, one of the O-linked glycoproteins in that species, resulted in the generation of a series of smaller forms of the protein as a result of proteolysis (17). In C. jejuni 81-176, the VirB10 protein, part of a plasmid-encoded type IV secretion system, is glycosylated at two sites and this protein is not detected in a pgl mutant, presumably as a result of proteolysis (23). In contrast, our data suggested that the glycosylation of Cj1496c does not have a major influence on the stability of this protein. The absence of apparent function for glycosylation has previously been reported for glycoproteins in eukaryotic cells as well (14).
Acknowledgments
We thank David Hendrixson for his generous assistance with the chick colonization assay and with the use of many of the molecular tools used in this study. We are grateful to members of the DiRita lab, Friedman lab, and Krukonis lab for helpful discussions and comments. We also thank Shinji Takai (Kitasato University).
This work was supported by USDA grant 2002/3520/111672 to V.J.D.
Editor: J. T. Barbieri
REFERENCES
- 1.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. [DOI] [PubMed] [Google Scholar]
- 2.Beery, J. T., M. B. Hugdahl, and M. P. Doyle. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 54:2365-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 4.Butzler, J. P., and M. B. Skirrow. 1979. Campylobacter enteritis. Clin. Gastroenterol. 8:737-765. [PubMed] [Google Scholar]
- 5.Everest, P. H., H. Goossens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37:319-325. [DOI] [PubMed] [Google Scholar]
- 6.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infection in the United States and other industrialized nations, p. 121-139. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed., ASM Press, Washington, D.C.
- 8.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golden, N. J., and D. W. K. Acheson. 2002. Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect. Immun. 70:1761-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant, A. J., C. Coward, M. A. Jones, C. A. Woodall, P. A. Barrow, and D. J. Maskell. 2005. Signature-tagged transposon mutagenesis studies demonstrate the dynamic nature of cecal colonization of 2-week-old chickens by Campylobacter jejuni. Appl. Environ. Microbiol. 71:8031-8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant, C. C., M. E. Konkel, W. Cieplak, Jr., and L. S. Tompkins. 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 61:1764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerry, P., R. Yao, R. A. Alm, D. H. Burr, and T. J. Trust. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 235:474-481. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 14.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 291:2364-2369. [DOI] [PubMed] [Google Scholar]
- 15.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 16.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 17.Herrmann, J. L., P. O'Gaora, A. Gallagher, J. E. Thole, and D. B. Young. 1996. Bacterial glycoproteins: a link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. EMBO J. 15:3547-3554. [PMC free article] [PubMed] [Google Scholar]
- 18.Hugdahl, M. B., J. T. Beery, and M. P. Doyle. 1988. Chemotactic behavior of Campylobacter jejuni. Infect. Immun. 56:1560-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, M. A., K. L. Marston, C. A. Woodall, D. J. Maskell, D. Linton, A. V. Karlyshev, N. Dorrell, B. W. Wren, and P. A. Barrow. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72:3769-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlyshev, A. V., P. Everest, D. Linton, S. Cawthraw, D. G. Newell, and B. W. Wren. 2004. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology 150:1957-1964. [DOI] [PubMed] [Google Scholar]
- 21.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 22.Lara-Tejero, M., and J. E. Galan. 2000. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290:354-357. [DOI] [PubMed] [Google Scholar]
- 23.Larsen, J. C., C. Szymanski, and P. Guerry. 2004. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J. Bacteriol. 186:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lux, R., and W. Shi. 2004. Chemotaxis-guided movements in bacteria. Crit. Rev. Oral Biol. Med. 15:207-220. [DOI] [PubMed] [Google Scholar]
- 25.Millikan, D. S., and E. G. Ruby. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 68:2519-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers, J. D., and D. J. Kelly. 2005. A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology 151:233-242. [DOI] [PubMed] [Google Scholar]
- 27.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nita-Lazar, M., M. Wacker, B. Schegg, S. Amber, and M. Aebi. 2005. The N-X-S/T consensus sequence is required but not sufficient for bacterial N-linked protein glycosylation. Glycobiology 15:361-367. [DOI] [PubMed] [Google Scholar]
- 29.Park, S. F. 2002. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int. J. Food Microbiol. 74:177-188. [DOI] [PubMed] [Google Scholar]
- 30.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davie, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 31.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar, M., and K. Chaudhuri. 2004. Association of adherence and motility in interleukin 8 induction in human intestinal epithelial cells by Vibrio cholerae. Microbes Infect. 6:676-685. [DOI] [PubMed] [Google Scholar]
- 33.Szymanski, C. M., D. H. Burr, and P. Guerry. 2002. Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70:2242-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wacker, M., D. Linton, P. G. Hitchen, M. Nita-Lazar, S. M. Haslam, S. J. North, M. Panico, H. R. Morris, A. Dell, B. W. Wren, and M. Aebi. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790-1793. [DOI] [PubMed] [Google Scholar]
- 35.Walker, R. I., M. B. Caldwell, E. C. Lee, P. Guerry, T. J. Trust, and G. M. Ruiz-Palacios. 1986. Pathophysiology of Campylobacter enteritis. Microbiol. Rev. 50:81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiesner, R. S., D. R. Hendrixson, and V. J. DiRita. 2003. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 185:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe, A. J., and H. C. Berg. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 86:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 39.Yao, R., D. H. Burr, and P. Guerry. 1997. CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol. 23:1021-1031. [DOI] [PubMed] [Google Scholar]
- 40.Young, N. M., J. R. Brisson, J. Kelly, D. C. Watson, L. Tessier, P. H. Lanthier, H. C. Jarrell, N. Cadotte, F. St. Michael, E. Aberg, and C. M. Szymanski. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277:42530-42539. [DOI] [PubMed] [Google Scholar]
- 41.Ziprin, R. L., C. R. Young, L. H. Stanker, M. E. Hume, and M. E. Konkel. 1999. The absence of cecal colonization of chicks by a mutant of Campylobacter jejuni not expressing bacterial fibronectin-binding protein. Avian Dis. 43:586-589. [PubMed] [Google Scholar]