Abstract
The Tf1 element of Schizosaccharomyces pombe is a long terminal repeat-containing retrotransposon that encodes functional protease, reverse transcriptase, and integrase proteins. Although these proteins are known to be necessary for protein processing, reverse transcription, and integration, respectively, the function of the protein thought to be Gag has not been determined. We present here the first electron microscopy of Tf1 particles. We tested whether the putative Gag of Tf1 was required for particle formation, packaging of RNA, and reverse transcription. We generated deletions of 10 amino acids in each of the four hydrophilic domains of the protein and found that all four mutations reduced transposition activity. The N-terminal deletion removed a nuclear localization signal and inhibited nuclear import of the transposon. The two mutations in the center of Gag destabilized the protein and resulted in no virus-like particles. The C-terminal deletion caused a defect in RNA packaging and, as a result, low levels of cDNA. The electron microscopy of cells expressing a truncated Tf1 showed that Gag alone was sufficient for the formation of virus-like particles. Taken together, these results indicate that Tf1 encodes a Gag protein that is a functional equivalent of the Gag proteins of retroviruses.
Long terminal repeat (LTR)-containing retrotransposons are a diverse group of elements found in a broad variety of eukaryotic hosts. Their structures and mechanisms of propagation are closely related to those of retroviruses. Both retroviruses and LTR-containing retrotransposons encode Gag, reverse transcriptase (RT), and integrase (IN) proteins. The Gag proteins assemble into the coats of virus and virus-like particles (VLPs), and the formation of these particles is required for reverse transcription. Once RT produces the full-length double-stranded cDNA, IN inserts the cDNA into the genome of a host cell.
Most of what is known about the function of Gag proteins resulted from the study of retroviruses. With an estimated 1,500 Gag molecules per virion in the case of the Rous sarcoma virus (59), Gag constitutes the major component of viral particles. It is well established that Gag is able to form extracellular particles in the absence of the other viral proteins (10, 19, 23, 62). As a result of its role in particle formation, Gag is responsible for the size of virions, the process of budding, and the packaging of the other components of the virus, such as the genomic RNA, RT, IN, and envelope proteins (56).
Once the particles have assembled, their maturation results from the processing of Gag by the viral protease (PR) into at least three major products, matrix (MA), capsid (CA), and nucleocapsid (NC) (33, 64). MA is derived from the amino terminus of Gag and directs the location of particle formation (for a review, see reference 58). CA, the largest cleavage product of Gag, forms the shell surrounding the ribonucleoprotein complex that contains the genomic RNA. CA has several separate functions that contribute to the assembly of particles, the release of particles from infected cells, and the reverse transcription of viral RNA. Numerous mutations in the CA domains of the murine leukemia virus and of human immunodeficiency virus type 1 have been found to block assembly and particle release (11, 24, 43, 67). Certain mutations in the CA sequences of several retroviruses have been found to destroy infectivity without affecting assembly or budding (27, 43, 55, 60). Strong evidence for the role of the CA in postentry replication comes from experiments with the murine leukemia virus that showed that specific mutations inhibit the synthesis of viral cDNA (3). Each of the functions of Gag is thought to require the major homology region (MHR), a motif of CA that is about 20 amino acids long and is highly conserved among Gag proteins (49a, 64). Mutations in the MHR of the Rous sarcoma virus can be deleterious to particle release or have an effect on cDNA synthesis (9, 14).
Finally, the NC protein, located at the carboxy terminus of Gag, is tightly bound to the genomic RNA (18, 21). NC is basic and contains one or more zinc finger motifs involved in the packaging of the viral RNA (13). In biochemical experiments, NC appears to have chaperone activity resulting in the stimulation of template switching by RT (4, 17, 50, 65; for a review, see reference 51).
The spumaretroviruses are replication-competent retroviruses that express a Gag protein that has no sequence similarity to the MA, CA, and NC of other retroviruses (for a review, see references 31 and 41). This dissimilarity includes the lack of an MHR motif in CA and of Zn fingers in NC (44, 52). In place of the Zn fingers, a glycine-arginine-rich motif near the carboxy terminus of the spumaretroviral Gag has been shown to have nucleic acid binding properties (66).
Although less is known about the Gag proteins of LTR-containing retrotransposons, evidence from electron microscopy indicates that they form VLPs, as documented for Ty1 (22) and Ty3 (25) of Saccharomyces cerevisiae as well as for copia (46) of Drosophila melanogaster. Intracytoplasmic particles have also been detected for the endogenous retroviruses of Drosophila, gypsy (32) and ZAM (30), elements closely related to LTR-containing retrotransposons. The retrotransposon Ty3 encodes a CA protein that contains an MHR-like motif (48). However, no homologs of CA and no MHR motifs have been identified in elements such as Ty1 (12) or the Tf1 and Tf2 elements from Schizosaccharomyces pombe (37).
Tf1 and Tf2 are two closely related LTR-containing retrotransposons (37), and their sequences encode single primary products of translation that have homologies to PR, RT, and IN (36, 37, 61). Although the amino acid sequences encoded by Tf1 and Tf2 are virtually identical for RT and IN, the sequences of Gag diverge.
Tf1 and Tf2 have been studied as model retroviruses by using the techniques of yeast genetics. The expression of Tf1 RNA from an inducible promoter results in high levels of transposition (26, 34, 36). Mutations within the conserved residues of PR, RT, and IN greatly reduce the transposition of Tf1 and indicate that these enzymes are required for Tf1 activity (5, 6, 35). However, much less is known about whether a Gag-like protein is required for Tf1 transposition. A protein of 27 kDa, derived from the N terminus of the product of the open reading frame (ORF), cosediments in sucrose gradients with RT, IN, and cDNA of Tf1 (36). This protein also has a nuclear localization signal (NLS) that is required for nuclear localization and, as a result, for transposition activity (16). This evidence is consistent with the possibility that the 27-kDa protein may function as the Gag of Tf1. Nevertheless, its sequence has no similarity to that of any known Gag. It lacks the MHR motif, and the 27-kDa protein is not processed into smaller species, such as MA, CA, and NC. In addition, it is not known whether this putative Gag protein assembles to form the shell of a particle or even if it is required for transposition.
We tested the Gag of Tf1 for functions associated with Gag proteins. We generated deletions of 10 amino acids in each of the four hydrophilic domains of the protein and found that all four mutations reduced transposition activity. The characterization of these mutant transposons revealed that Gag expression was required for packaging of RNA, particle formation, and reverse transcription. These observations were possible because we report here for the first time that Tf1 is able to assemble into intracytoplasmic particles easily visible by electron microscopy. In fact, the expression of Gag alone was sufficient for the formation of VLPs. Taken together, these results indicate that the Gag of Tf1 functions similarly to the Gag proteins of retroviruses.
MATERIALS AND METHODS
Media.
The S. pombe minimal liquid and plate media were composed of Edinburgh minimal medium (EMM). Ten micromolar vitamin B1 (thiamine) was added to EMM when indicated to repress the nmt1 promoter. 5-Fluoroorotic acid (5-FOA; U. S. Biological, Swampscott, Mass.) was used at 1 mg/ml in EMM or in yeast extract plus supplements (YES) with 500 μg of Geneticin (GIBCO)/ml.
Plasmid construction.
Most of the plasmids used for this study were constructed with techniques of PCR cloning. The oligonucleotides and plasmids used are listed in Tables 1 and 2.
TABLE 1.
Oligonucleotides
| Oligonucleotide | Description |
|---|---|
| HL38 | 5′-GGAAGAGGAATCCTGGC-3′: 5′-flanking primer, upstream of XhoI site, used to generate ΔA, ΔB, and ΔC mutations |
| HL39 | 5′-CCAATTGCTTCCAGTCTTTG-3′: 3′-flanking primer, downstream of AvrII site, used to generate ΔA, ΔB, and ΔC mutations |
| HL87 | 5′-CGATAATTGAACGCTACACC-3′: 5′-flanking primer, upstream of AvrII site, used to generate ΔD mutation |
| HL88 | 5′-TTGTTTATTTGACATGTATGGC-3′: 3′-flanking primer, downstream of BsrGI site, used to generate ΔD mutation |
| HL205 | 5′-CAAGTCGTTCTATAGCGTAGCCGGCATCGGAATGTTCACCAAAAAA-3′: bottom-strand primer used to create ΔB deletiona |
| HL206 | 5′-TTTTTTGGTGAACATTCCGATGCCGGCTACGCTATAGAACGACTTG-3′: top-strand primer used to create ΔB deletion |
| HL207 | 5′-CAATATCCACCATTTCCATCGCCGGCCATTCTTGGAATAGCGAGT-3′: bottom-strand primer used to create ΔA deletion |
| HL208 | 5′-ACTCGCTATTCCAAGAATGGCCGGCGATGGAAATGGTGGATATTG-3′: top-strand primer used to create ΔA deletion |
| HL209 | 5′-AAAGTTAAAAATCTTGTCTATATTGCCGGCCTGTTTAATCAGATAGGGTG-3′: bottom-strand primer used to create ΔC deletion |
| HL210 | 5′-CACCCTATCTGATTAAACAGGCCGGCAATATAGACAAGATTTTTAACTTT-3′: top-strand primer used to create ΔC deletion |
| HL211 | 5′-TGAAACATTTGTTTTCTTTGGGCCGGCAAACTTATTGTTTTTCCAATTG-3′: bottom-strand primer used to create ΔD deletion |
| HL212 | 5′-CAATTGGAAAAACAATAAGTTTGCCGGCCCAAAGAAAACAAATGTTTCA-3′: top-strand primer used to create ΔD deletion |
Top- and bottom-strand primers were used to introduce an NgoMI site, as well as to delete amino acids. Deletion constructs were created by fusion PCR, including amplification with HL38 (for bottom strands corresponding to HL205, HL207, and HL209), HL39 (for top strands corresponding to HL206, HL208, and HL210), HL87 (for a bottom strand corresponding to HL211), and HL88 (for a top strand corresponding to HL212).
TABLE 2.
Plasmids
| Plasmid(s) | Description | Source or reference |
|---|---|---|
| pHL167-1 | EcoRI fragment containing Tf1 Gag sequence | 38 |
| pHL449-1 | neoA1-marked version of Tf1 in a ura4 selectable plasmid | 34 |
| pHL490-80 | Tf1-neoAI with frameshift in PR | 6 |
| pHL476-3 | Tf1-neoAI with frameshift in IN | 34 |
| pFL20 | S. pombe vector with autonomous replicating sequence and stabilization fragment | 42 |
| pHL891-19 | neoA1-marked Tf1 in a ura4 selectable plasmid; XhoI-less neo without intron; SpeI site just 3′ downstream of hairpin after Tf1 primer binding site | 40 |
| pHL1260 and pHL1261 | Tf1-neoA1 with deletion of amino acids 2 to 11, (ΔA) | This study |
| pHL1262 and pHL1263 | Tf1-neoA1 with deletion of amino acids 64 to 73 (ΔB) | This study |
| pHL1264 and pHL1265 | Tf1-neoA1 with deletion of amino acids 165 to 174 (ΔC) | This study |
| pHL1258 and pHL1259 | Tf1-neoA1 with deletion of amino acids 215 to 224 (ΔD) | This study |
The plasmids pHL891-19 and pHL449-1 were used to create mutant versions of the Tf1 Gag protein. The mutations corresponded to deletions of 10 amino acids and the additions of the dipeptide alanine-glycine. The sequence of the introduced DNA contained an NgoMI restriction site that was unique within the plasmids. The constructs are hereafter referred to as Tf1-ΔA, Tf1-ΔB, Tf1-ΔC, and Tf1-ΔD, and two independently generated plasmids were created for each, pHL1260 and pHL1261, pHL1262 and pHL1263, pHL1264 and pHL1265, and pHL1258 and pHL1259, respectively (Table 2).
The constructs were created by fusion PCR as described earlier (34). Briefly, two overlapping oligonucleotides containing a specific deletion were used to amplify by PCR two fragments that overlap at the site of the mutation. Each of these fragments extends to a unique restriction site that can be used to subclone the final fusion product. The fusion PCR products were produced by mixing both overlapping PCR products with an oligonucleotide that hybridizes to the extreme end of the top strand of one product and another oligonucleotide that hybridizes to the extreme end of the bottom strand of the other fragment.
The two overlapping oligonucleotides were HL207 and HL208 for Tf1-ΔA, HL205 and HL206 for Tf1-ΔB, HL209 and HL210 for Tf1-ΔC, and HL211 and HL212 for Tf1-ΔD; the flanking oligonucleotides were HL38 and HL39 for the Tf1-ΔA, Tf1-ΔB, and Tf1-ΔC constructs and HL87 and HL88 for the Tf1-ΔD construct (Table 1).
The fusion PCR products were then digested by XhoI and AvrII and cloned into pHL891-19, also digested by XhoI and AvrII, for the Tf1-ΔA, Tf1-ΔB, and Tf1-ΔC constructs or digested by BsrGI and AvrII and cloned into pHL449-1, also digested by BsrGI and AvrII, for the Tf1-ΔD construct.
The neo gene interrupted by an intron was introduced in the transposons with ΔA, ΔB, and ΔC by replacing the NarI/BamHI fragment of 2.0 kb with that of pHL449-1. Although the mutations introduced by PCR were verified by sequencing, the plasmids were created in duplicate from independent PCRs and the properties of each plasmid were studied in parallel. Since no differences were seen between the duplicate plasmids, we present here the results for one duplicate. Plasmids were introduced into S. pombe strains (Table 3) by lithium acetate transformation (47).
TABLE 3.
Yeast strains
| Straina | Plasmid | Source or reference |
|---|---|---|
| YHL 912 | No plasmid | J. Boeke, 21X52 |
| YHL 1282 | pHL449-1 | 34 |
| YHL 1836 | pHL490-80 | 6 |
| YHL 1554 | pHL476-3 | 34 |
| YHL 5889 | pHL1260 | This study |
| YHL 5890 | pHL1261 | This study |
| YHL 5891 | pHL1262 | This study |
| YHL 5892 | pHL1263 | This study |
| YHL 5893 | pHL1264 | This study |
| YHL 5894 | pHL1265 | This study |
| YHL 5826 | pHL1258 | This study |
| YHL 5827 | pHL1259 | This study |
| YHL 6566 | pFL20 | 7 |
All strains had the genotype h− ura4-294 leu1-32 and YHL 912 as the parent strain.
Transposition and homologous recombination assays.
Tf1 transposition and homologous recombination frequencies were determined as described previously (35). Tf1 transposition was monitored by placing a neoAI-marked Tf1 element under the control of an inducible nmt1 promoter. The neo gene allowed cells to grow in the presence of 500 μg of G418/ml. S. pombe strains that contained a Tf1-neoAI plasmid were grown as patches on EMM-uracil dropout agar plates in the absence of thiamine to induce transcription of the nmt1 promoter. After 4 days of incubation at 32°C, the plates were replica printed to medium containing 5-FOA to eliminate the URA3-marked plasmid with Tf1-neoAI (8). Finally, 5-FOA-resistant patches were printed to YES medium containing both 5-FOA and G418 and incubated at 32°C for 2 days to detect strains that became resistant to G418 as the result of insertions of the Tf1-neo element into the genome. Wild-type Tf1 produced confluent patches of G418 resistance, while mutations in the element reduced the growth on the G418 plates.
Homologous recombinations between cDNA and plasmid sequences were assayed by using a protocol similar to the transposition assay (5). The strains with the Tf1-neoAI plasmids were first grown as patches on agar plates that contained EMM (plus 10 μM thiamine and dropout powder) and then replica printed to similar EMM plates that lacked thiamine. After 4 days of incubation at 32°C, the plates were replica printed directly to YES medium that contained 500 μg of G418/ml. cDNA recombination was scored on the G418 plates after 48 h of growth at 32°C.
Preparation and analysis of nucleic acid.
cDNA preparations were performed as described previously (6). Total DNA was phenol extracted from stationary-phase cells grown under induction conditions (absence of thiamine) for 36 h. These preparations were then digested with BstXI prior to analysis by DNA blotting. The probe consisted of a 1-kb BamHI fragment with the sequence of neo (6). The DNA blot analyses were done by using the DIG High Prime DNA labeling and detection kit (Roche).
Protein extraction and detection of Tf1 Gag, IN, and RT by immunoblot analysis.
Total proteins were extracted from cells grown under inducing conditions (absence of thiamine) by a previously published protocol (6). Cultures at the optical densities (ODs) of 1 and 10 were harvested and resuspended in 400 μl of buffer B (15 mM NaCl, 10 mM HEPES-KOH [pH 7.8], 5 mM EDTA). Cells were subjected to a vortex in the presence of glass beads (acid washed) with a diameter of 0.4 mm. Protein extracts were then recovered, and an equal volume of 2× sample buffer (6) was added. The mixture was boiled for 3 min, and 10 μg of total protein from each sample was loaded on sodium dodecyl sulfate-10% polyacrylamide gels for immunoblot analysis. Standard electrophoresis and transfer techniques were used with Immobilon-P membranes (Millipore). The detection method used was the ECL system as described by the manufacturer (Amersham). Horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin was used at a 1:10,000 dilution. The previously described primary polyclonal antisera used for each filter were from production bleeds 660 (anti-Gag), 657 (anti-IN) (36), and 1571 (anti-RT) (26). Other bleeds (63085-4 and 63085-5) were prepared by immunizing rabbits with a peptide containing the first 15 amino acids of the Gag protein (see Fig. 1A). These bleeds were pooled and then affinity purified.
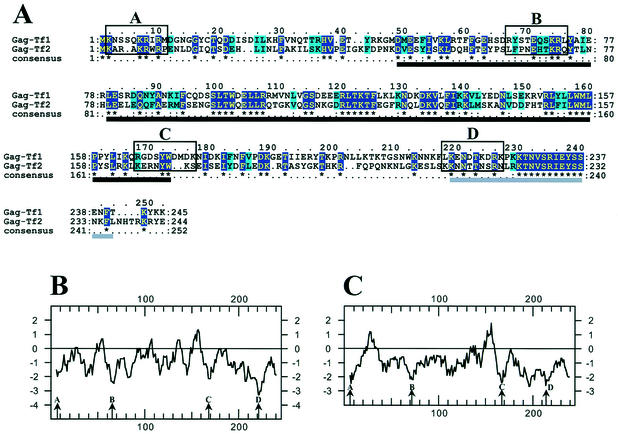
FIG. 1.
Comparative analysis of the Gag proteins of Tf1 and Tf2. (A) Clustal alignment of Gag proteins from Tf1 and Tf2. The dark blue boxes with yellow letters highlight identical amino acids, while the light blue boxes with black letters highlight similar amino acids. The four open boxes (A, B, C, and D) contain the amino acids deleted for this study. The black and gray lines under the alignment indicate the two blocks used to measure the Ks/Ka ratios (see the text). (B and C) Kyte-Doolittle hydropathy profiles of the Tf1 (B) and Tf2 (C) Gag proteins. Arrows indicate the four conserved hydrophilic domains (A, B, C, and D) targeted by the mutagenesis.
Computational analyses of Gag.
ClustalX (default parameters) (57) was used to construct an amino acid alignment of the Gag portions of Tf1 and Tf2 polyproteins. We used Se-Al (courtesy of Andrew Rambant) to construct a DNA alignment of the corresponding codons. DnaSP version 3.51 (53) was used to calculate the Ks (the number of synonymous changes over the number of potential sites of synonymous changes) and Ka (the number of nonsynonymous changes over the number of potential sites of nonsynonymous changes) values. Statistical analysis of negative selection was done by using the Z test as implemented in Mega version 2.1 (29).
Electron microscopy.
After 36 h of culture at 32°C in EMM lacking thiamine to induce Tf1 transposition, 6 OD units of cells was washed once in 10 ml of PEM [0.1 M piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.9), 2 mM EGTA, 1 mM MgCl2] and fixed in 2% glutaraldehyde (Tousimis, Rockville, Md.) in PEM for 2 h at room temperature. Cells were then washed and resuspended in 5 ml of PEM containing 0.2% tannic acid (catalog no. 21710; Electron Microscopy Sciences, Ft. Washington, Pa.) and incubated for 15 min at room temperature. Following two washes in 5 ml of 0.1 M potassium phosphate, pH 7.5, cell walls were digested in 2 ml of potassium phosphate, 0.25 mg of Zymolyase 100T (no. Z1004; U.S. Biological)/ml, and 90 mM β-mercaptoethanol for 60 min at 30°C. Cells were then washed in 0.1 M cacodylate, pH 6.8, and resuspended in 1 ml of 2% osmium peroxide (Electron Microscopy Sciences) in 0.1 M cacodylate for 1 h at room temperature. Cells were then washed in water and resuspended in 1 ml of 2% uranyl acetate for 1 h at room temperature, washed in water, and dehydrated progressively in a series of increasing concentrations of ethanol solution (35, 50, 70, 90, and 95% and three times in 100%) and then in propylene oxide (100%). The cells were infiltrated in equal volumes of 100% propylene oxide and epoxy resin (Electron Microscopy Sciences) overnight and embedded in pure resin on the following day. The epoxy resin was cured in a 60°C oven for 48 h. The cured block was thin-sectioned and stained in uranyl acetate and lead citrate. The cells were examined and imaged with a transmission electron microscope (H7000; Hitachi, Tokyo, Japan) equipped with a charge-coupled device camera (Gatan, Pleasanton, Calif.).
Protection assays of Tf1 RNA.
Cells were inoculated at an OD at 600 nm of 0.05 in 50 ml of dropout medium (EMM minus uracil) and grown for 36 h to an OD at 600 nm of 10.0. Twenty milliliters of cells was harvested, washed, and then broken with glass beads in 1 ml of extraction buffer (15 mM KCl, 10 mM HEPES-KOH [pH 7.8], 5 mM EDTA, 0.2% Triton X-100) containing protease inhibitors (aprotinin [1 μg/ml], leupeptin [0.5 μg/ml], and pepstatin [0.7 μg/ml]), 3 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, and 60 U of RNase inhibitor (RNAguard; Pharmacia Biotech). Supernatants of the cell extracts were recovered after centrifugation at 1,000 × g for 5 min. Fifteen microliters of 10× buffer M (100 mM Tris-HCl [pH 7.5], 100 mM MgCl2, 500 mM NaCl) with or without specified amounts of Benzonase (catalog no. E-8263; Sigma) was added to 135 μl of the supernatant and incubated at room temperature for 6 min. The reactions were stopped by the addition of 5 μl of 0.5 M EDTA. Nucleic acids were extracted immediately by the addition of 300 μl each of the nucleic acid extraction buffer and phenol-chloroform-isoamyl alcohol. After four additional extractions with phenol-chloroform-isoamyl alcohol, the samples were ethanol precipitated, resuspended in 25 μl of H2O, and kept at −20°C until RNA blot analysis. The levels of RNase-resistant RNA were analyzed as described below.
RNA blot analysis.
Five microliters of each RNA sample was loaded on an agarose gel containing formaldehyde by using the NorthernMax kit from Ambion (no. 1940). The RNA was transferred to a BrightStar-Plus membrane (Ambion). The membrane was hybridized with 32P-labeled probes specific for Tf1 Gag, prepared by using the 0.75-kb EcoRI fragment from pHL167-1 (37). The membrane was treated according to the manufacturer's protocol.
RESULTS
Sequences of Gag have been maintained by selective pressure.
Although very little was known about the function of the 27-kDa protein of Tf1, its cosedimentation with components of Tf1 (36) and its position at the N terminus of the product of the Tf1 ORF led to the hypothesis that it was the Gag of Tf1. To test this hypothesis, we investigated whether this putative Gag was required for transposition and whether it contained domains with specific functions commonly associated with Gag proteins. In our initial approach, we examined the sequence of Gag for evidence of a conserved function. We aligned the sequence of the Gag of Tf1 with the sequence of the corresponding protein of Tf2, a closely related and active transposon. The best alignments of the Gags were obtained when the second methionine of the sequence corresponding to the Tf2 ORF was aligned with the first methionine of the sequence corresponding to the Tf1 ORF (Fig. 1A). Based on this alignment, we propose that the second methionine of the Tf2 Gag initiates translation, contrary to what has been previously proposed (61).
Out of the 245 amino acids from the Gag of Tf1 and the 244 amino acids from that of Tf2, 36% were identical. Of the 89 pairs of identical amino acids, 45% corresponded to codons with synonymous nucleotide changes. To test whether the identical amino acids were being maintained due to natural selection, we determined the Ks/Ka ratio. This was calculated independently for two regions where the two sequences align unambiguously. The sequences used were amino acids 46 through 170 for Tf1 Gag and 46 through 170 for Tf2 Gag and, separately, amino acids 216 through 241 for Tf1 Gag and 211 through 236 for Tf2 Gag (Fig. 1A). The Ks/Ka ratios were 2.6 (P = 0.0005) and 3.0 (P = 0.0183), respectively. Both values were well above 1.0, indicating that the amino acid identities between the Gag of Tf1 and that of Tf2 resulted from negative or purifying selection. The evidence for this selective pressure in these regions demonstrates that Gag has recently had an important function that was or is needed for the activity of the transposon.
To examine the sequences of the Tf1 and Tf2 Gags for evidence of domain structure, we compared their hydropathy profiles. A positive value indicates local hydrophobicity and a negative value suggests a water-exposed region on the face of a protein. Figure 1B shows four regions of Tf1 Gag with strong hydrophilic scores. Interestingly, the positions of these hydrophilic domains were conserved in Tf2 Gag, suggesting that these domains possess an important function (Fig. 1C). In addition, there were several amino acid conservations within each hydrophilic domain (Fig. 1A).
Mutations in Gag resulted in reduced frequencies of transposition.
To determine whether Gag was required for transposition and whether domains with independent functions exist, we generated deletions of 10 amino acids in each of the four regions of hydrophilicity (Fig. 1A). For the purposes of future studies, an NgoMI site was introduced at the deletion positions, and these nucleotides resulted in the insertion of an alanine followed by a glycine. Thus, 10 residues of Gag were replaced with alanine-glycine.
We tested the transposition activities of the Tf1 elements with the deletions, referred to as Tf1-ΔA, Tf1-ΔB, Tf1-ΔC, and Tf1-ΔD, by using an assay based on the expression of Tf1 RNA from a strong promoter on a multicopy plasmid (Fig. 2A) (34). The sequence of Tf1 in the plasmid contained a neo gene as an indicator for transposition events. For reasons described below, the neo gene was disrupted by an artificial intron that can be spliced out of the Tf1 RNA. Patches of cells in which transcription of Tf1-neoAI was induced were subsequently exposed to 5-FOA to select for cells that no longer possessed the Tf1-neoAI plasmid (see Materials and Methods). The presence of transposed copies of Tf1-neoAI was identified by replica printing the cells from the 5-FOA medium to medium with G418. Figure 2B (left panel) shows that patches of cells that contained wild-type Tf1-neoA (Tf1-WT) exhibited sufficient transposition to produce confluent growth on the plate containing G418. A strain that contained a copy of Tf1-neoAI that, due to a frameshift mutation, lacked IN expression (Tf1-INfs) exhibited significantly reduced resistance to G418. This low level of resistance to G418 from Tf1-INfs was due to homologous recombination between Tf1 cDNA and endogenous copies of the related element, Tf2 (data not shown). A transposon that lacked PR, RT, and IN, due to a frameshift in PR (Tf1-PRfs), exhibited no residual transposition activity.
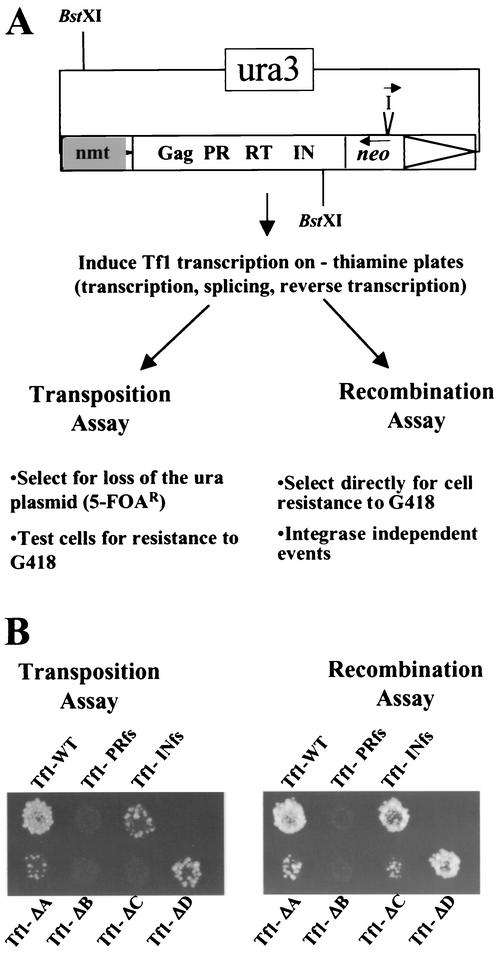
FIG. 2.
Effects of Gag deletions on transposition and recombination activity. (A) Schematic of the genetic assay for measuring the levels of transposition and homologous recombination. The restriction sites (BstXI) used for the cDNA blot are shown. I indicates the artificial intron placed in neo, −thiamine indicates the absence of thiamine, and 5-FOAR indicates resistance to 5-FOA. (B) Results of the transposition and recombination assays for the four transposons containing deletions in Gag (Tf1-ΔA, Tf1-ΔB, Tf1-ΔC, and Tf1-ΔD) as well as for the control strains (Tf1-WT, Tf1-PRfs, and Tf1-INfs). Patches of cells in which the expression of Tf1 was induced were replica printed to plates containing G418 to measure recombination activity (right panel). To measure transposition, cells were replica printed to plates with 5-FOA, then printed to plates with 5-FOA and G418.
The transposition assays showed that Tf1-ΔB and Tf1-ΔC exhibited no transposition at all. Tf1-ΔA generated low levels of G418-resistant papillae that were below those observed for Tf1-INfs. The Tf1-ΔD element also exhibited a significant defect in transposition, albeit less than that of the other constructs.
To identify which aspects of the transposition pathway were affected by the mutations, we employed an assay for homologous recombination that detected the presence of Tf1 cDNA in the nucleus (5). This method used the cDNA produced by the same Tf1 expression plasmid described above. The configuration of the neo gene in reverse orientation to Tf1 and the orientation of the artificial intron allowed the intron to be spliced only from the Tf1 RNA. Once transcription of Tf1-neoAI was induced and without selecting against the expression plasmid, we replica printed cells directly to medium containing G418 (see Materials and Methods). This allowed us to detect homologous recombination events between Tf1 cDNA and copies of the Tf1 plasmid. Reduced levels of homologous recombination would be caused by mutations that either reduced reverse transcription or inhibited the import of Tf1 cDNA into the nucleus. The level of recombination that was generated by a wild-type copy of Tf1-neoAI is shown in Fig. 2B (right panel). The Tf1-INfs element showed that these recombination events were independent of IN. The lack of any G418 resistance generated by the Tf1-PRfs element indicated that reverse transcription was necessary for any homologous recombination to be detected. The mutants Tf1-ΔA, Tf1-ΔB, and Tf1-ΔC showed significant reductions in recombination. While Tf1-ΔB appeared to be totally defective in this assay, Tf1-ΔC showed residual levels of recombination activity. Tf1-ΔA appeared to generate slightly more G418 resistance than Tf1-ΔC, and the Tf1-ΔD element showed more activity than Tf1-ΔA and Tf1-ΔC. These defects in homologous recombination could reflect a reduction in reverse transcription or a defect in the nuclear import pathway of the preintegration complex containing cDNA, IN, and Gag.
Three of the deletion mutations caused reduced levels of cDNA synthesis.
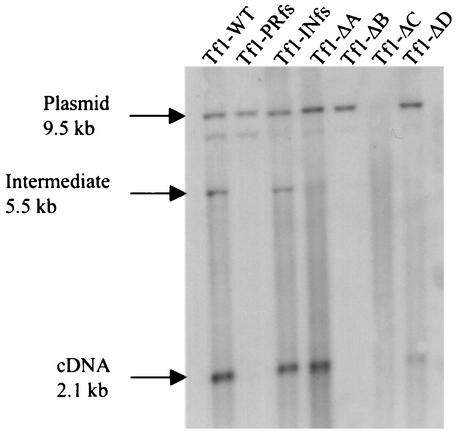
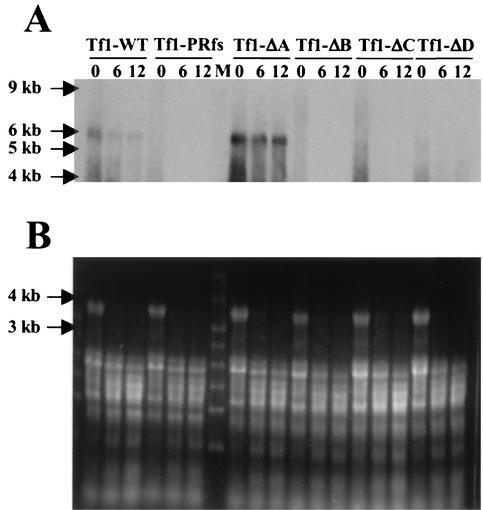
To test whether the deletions in Gag caused defects in the production of cDNA, we used DNA blot analysis (39). The strains of S. pombe expressing the different versions of Tf1-neoAI were grown to stationary phase. Extracts of total DNA were digested with BstXI, subjected to agarose gel electrophoresis, and blotted with a probe specific for the neo sequence. The restriction enzyme BstXI was chosen because there is only one site within the mature cDNA (Fig. 2A), and its use resulted in a 2.1-kb fragment from the cDNA and a 9.5-kb band from the plasmid. The plasmid expressing Tf1-INfs produced wild-type levels of cDNA as expected for an element that expressed RT (39) (Fig. 3). Also as expected, Tf1-PRfs did not produce cDNA because it did not express RT. The DNA blot also showed that Tf1-ΔA generated a wild-type level of cDNA. Tf1-ΔD produced significantly lower levels of cDNA, while repeated examination of Tf1-ΔB and Tf1-ΔC detected no cDNA band. These data indicated that the reduced levels of transposition observed for Tf1-ΔB, Tf1-ΔC, and Tf1-ΔD were due to significantly reduced levels of reverse transcription. On the other hand, the reduced transposition and homologous recombination caused by Tf1-ΔA were not the result of defects in reverse transcription. Instead, the mutation likely reduced transport of Tf1 into the nucleus. This conclusion is based on previous work that identified an NLS within the sequence deleted from Tf1-ΔA (16).
FIG. 3.
Effects of mutations in Gag on the synthesis of cDNA. Total DNA was extracted from S. pombe strains in which Tf1 expression had been induced. The DNA was digested with BstXI (Fig. 2) and probed with a neo-specific sequence. The 9.5-kb band was produced by a plasmid sequence, and the 2.1-kb band was generated by Tf1 cDNA. The additional band is likely derived from single-LTR circles; however, this identification is not definite. The reduced level of the plasmid band from Tf1-ΔC was reproducible with independent transformants and may be due to degradation during extraction that is mediated by RT or IN.
Deletion mutants Tf1-ΔB and Tf1-ΔC produced reduced levels of Gag.
To investigate whether the transposition and recombination defects were due to decreased stability of Tf1 proteins, we performed immunoblot analysis with antibodies raised against the Tf1 Gag, IN, and RT. Total cell proteins were extracted from log-phase and stationary-phase cultures of strains expressing Tf1-WT, Tf1-INfs, Tf1-PRfs, and the four versions of Tf1 with the deletions. From previous results, it is known that the Tf1 Gag and IN proteins are expressed at equal levels within a single primary translation product (Fig. 4A) and that during the log-to-stationary phase transition of the culture, the IN and RT proteins are specifically targeted for degradation (6).
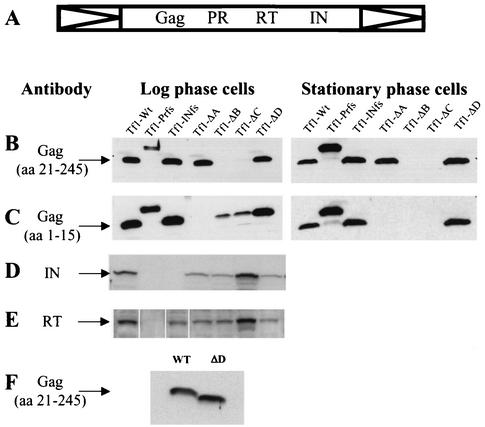
FIG. 4.
Immunoblot of extracts made from S. pombe expressing different mutants of Tf1. (A) Schematic of the Tf1 polyprotein. (B to F) Immunoblots were performed by using polyclonal antibodies directed against Gag peptide amino acids (aa) 20 through 245 (bleed 660) (B and F) and 1 through 15 (bleed 63085-4+5) (C), against IN peptide (bleed 657) (D), and against RT peptide (bleed 1571) (E). The predicted sizes of the proteins are 27, 56, and 60 kDa, respectively, for Gag, IN, and RT. WT, wild type.
In cultures grown to log phase, only Tf1-ΔA and Tf1-ΔD were able to produce normal levels of Gag protein relative to Tf1-WT (Fig. 4B, left panel). No Gag proteins were detected for the Tf1-ΔB and Tf1-ΔC elements by using polyclonal antibodies raised against residues 21 through 245 of Gag. The Gag proteins produced by Tf1-ΔA and Tf1-ΔD were also stable in stationary-phase cells (Fig. 4B, right panel). To test whether the Gag proteins produced by Tf1-ΔB and Tf1-ΔC were not detected with this antiserum because the deletions removed key epitopes, we raised new antibodies against the first 15 amino acids of the Gag protein and used them for immunoblotting of the same extracts. The results show that in log-phase cells (Fig. 4C, left panel), Tf1-ΔB and Tf1-ΔC expressed unstable Gag proteins which were no longer detectable in stationary-phase cells (Fig. 4C, right panel). The fact that the Gag protein of Tf1-ΔA was not recognized by this set of antibodies was expected since the ΔA deletion removed the majority of the epitope recognized by the antibodies. In log-phase cultures, all the mutants except for Tf1-ΔC produced slightly lower levels of IN than the wild type (Fig. 4D). Tf1-ΔC accumulated wild-type amounts of IN. The extracts from cells grown to stationary phase showed that none of the deletions affected the degradation of IN that was observed with Tf1-WT (data not shown). Antibodies that recognized RT showed that all the mutants except for Tf1-ΔC produced slightly lower levels of the 60-kDa species of RT seen in log-phase cultures (Fig. 4E).
The fact that the Gag protein of Tf1-ΔD showed a faster mobility than the wild type (Fig. 4F) was consistent with the assumption that the residues removed from Tf1-ΔD exist in the Gag protein. This confirmation is important because the position of the PR cleavages in Tf1 are not known but are predicted based on alignments with Ty3, a retrotransposon for which the cleavage sites were sequenced (28).
The particles of Tf1 can be observed by electron microscopy, and expression of Gag is sufficient for their formation.
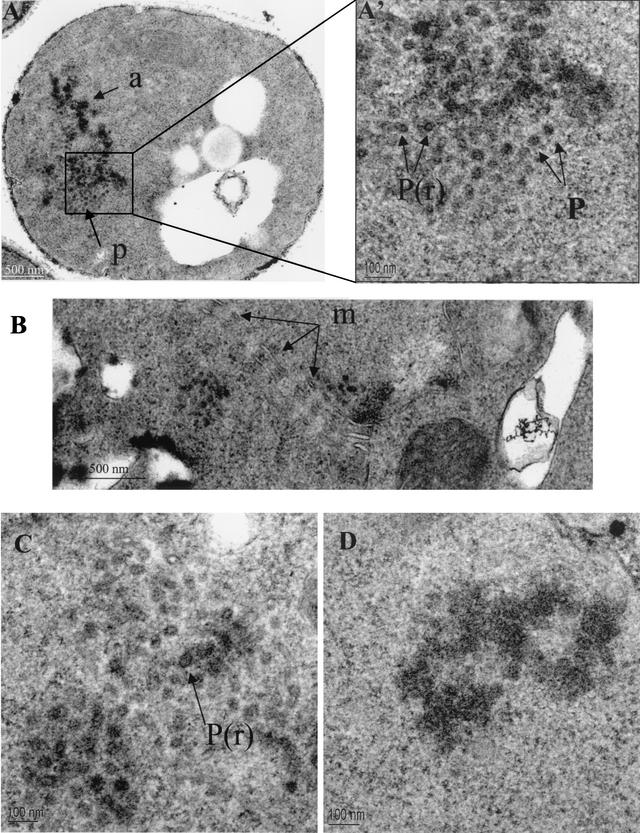
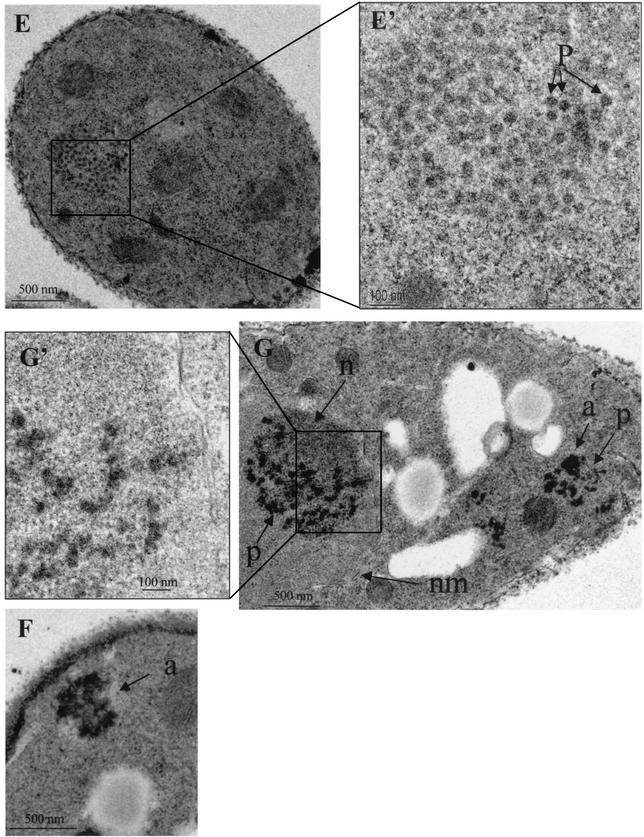
Previous work established that Gag, RT, IN, and cDNA assemble into large macromolecular complexes that cosediment in sucrose gradients (36). Here we tested whether Tf1 assembled spherical VLPs that can be observed with electron microscopy. Figure 5A shows typical structures of particles easily detectable in the cytoplasm of cells expressing the Tf1-WT element. They were observed in 3.7% of 269 cells sampled with a single section and were never observed in sections of 530 cells not expressing Tf1. They appear as clusters of well-defined particles or aggregates. The value 3.7% is not surprising given that the particles were tightly clustered and a single section samples only a small portion of each cell. A fraction of the particles were electron-dense rings with electron-lucent centers, while the majority had dense centers (Fig. 5A, A', and C). Associated with the particles were electron-dense aggregates (Fig. 5A and D). The maximum diameter of these intracytoplasmic particles was 50 nm. Surprisingly, the cytoplasmic particles and dense aggregates were often associated with arrays of membranes of unknown origin (Fig. 5B).
FIG. 5.
Electron micrographs of S. pombe cells expressing Tf1-WT (A, A', B, C, and D), Tf1-PRfs (G and G'), Tf1-ΔA (F), and Tf1-ΔD (E and E'). A', E', and G' are higher magnifications of the images shown in A, E, and G, respectively. a, aggregates of Tf1 material; P(r), particles with dense rings; P, particles without rings; m, membranes; n, heterochromatin; nm, nuclear membrane.
The same techniques of electron microscopy were used to determine whether the forms of Tf1 with deletions in Gag produced spherical particles. No particles or dense aggregates were found in the strains with Tf1-ΔC (data not shown). Since the Gag proteins of Tf1-ΔC were unstable (Fig. 4B), these results indicate that Gag was required for particle formation. In the case of Tf1-ΔA, no spherical VLPs were observed. However, dense aggregates in the cytoplasm were found in 2.0% of 300 cells (Fig. 5F). In the case of Tf1-ΔD (Fig. 5E and E'), the size and the number of particles were similar to those for wild-type Tf1. Of 350 cells, 7.1% contained particles. The presence of cytoplasmic membranes in the vicinity of Tf1-ΔD particles was also seen (data not shown).
The finding that the expression of Gag correlated with the formation of spherical particles is consistent with a role for Gag in particle structure. Many Gag proteins of retroviruses and LTR-containing retrotransposons are able to form particles in the absence of any other retroelement proteins (1, 10, 19, 23, 62). We tested whether the expression of the Tf1 Gag was sufficient for the formation of spherical particles. The version of Tf1 with the frameshift at the beginning of PR, Tf1-PRfs, does not express RT or IN (Fig. 4B, D, and E). The sole protein produced was a version of Gag fused to the first approximately 30 amino acids of PR followed by 20 residues encoded out of frame. The additional sequence results in an increase in molecular weight that can be observed in Fig. 4B. The electron microscopy revealed that a large percentage of cells expressing Tf1-PRfs contained particles and dense aggregates (Fig. 5G). Although the particles were more dense and less uniform than those of the wild type, their shapes and sizes resembled those of normal particles (Fig. 5G'). Of 300 cells, 5.3% contained structures related to particles. Since the fraction of cells with particles was approximately similar to that with Tf1-WT, these data indicate that Tf1 Gag is sufficient for the assembly of spherical particles. However, it was surprising that particles produced by Tf1-PRfs were often found in the nucleus whereas wild-type particles were observed only in the cytoplasm. Even more interesting was the observation that the nuclear particles appeared to associate with heterochromatin (Fig. 5G).
Gag is required for packaging of the Tf1 RNA.
One principal role of Gag proteins is to assemble into particles that package the RNA of the retroelement. We therefore tested whether Tf1 Gag was required for packaging of the Tf1 RNA. Since retrotransposons do not release their VLPs into culture medium, measuring packaged mRNA in purified VLPs has been hindered by the difficulties of isolating particles free of cellular transcripts. The approach taken here to address whether Gag is required for packaging of Tf1 mRNA was to use a previously developed assay that detects the amount of Tf1 RNA that is protected from degradation by the nuclease Benzonase (40). Cellular extracts containing VLPs were subjected to digestion with various concentrations of Benzonase for 6 min. The resistant RNA was isolated from each digested sample by phenol extraction and analyzed by RNA blot analysis. An ethidium bromide stain shows that for each condition, equal amounts of digested RNA were loaded (Fig. 6A). The RNA was transferred to a membrane that was hybridized with a probe for the Tf1 RNA. As previously described, the Tf1 RNA from Tf1-WT shows significant resistance to Benzonase treatment (Fig. 6A) (40). It was also shown previously that this protection from degradation was specific for the Tf1 RNA, since the ribosomal and actin RNAs were degraded. Here too, we found that the ribosomal RNAs were not protected from degradation (Fig. 6B). Although the electron microscopy indicated that Tf1-PRfs formed spherical particles, the RNA was not packaged (Fig. 6A). This suggests that Tf1 Gag is not sufficient for the packaging of Tf1 RNA. The strains expressing Tf1-ΔB and Tf1-ΔC were unable to protect RNA from degradation, and this corresponded to the reduced levels of Gag (Fig. 4) and the lack of particles detected by electron microscopy. The Gag protein of Tf1-ΔD also exhibited a significant defect in RNA packaging. This lack of RNA packaging correlated well with the reduced levels of cDNA produced by Tf1-ΔD (Fig. 3). The packaging of RNA by the Gag protein from Tf1-ΔA was perhaps the most surprising. Since we identified a defect in particle assembly, we expected that the Tf1 RNA would not be packaged. Instead, the result obtained demonstrated that the deletion in Tf1-ΔA significantly increased the protection and stability of the RNA.
FIG. 6.
RNase protection assay to detect packaging of Tf1 RNA in VLPs. (A) Cellular extracts containing VLPs were treated with various amounts of Benzonase (0, 6, or 12 U, lanes 0, 6, and 12, respectively) for 6 min at room temperature. RNA was then transferred to membranes and subjected to RNA blot analysis with a probe specific for the Gag region of Tf1. (B) The total RNA was visualized by ethidium bromide staining of the agarose gel under UV light.
DISCUSSION
In this study, we addressed whether the 27-kDa protein of Tf1 encoded by the segment of the ORF corresponding to the N terminus of the polyprotein was the functional equivalent of the Gag proteins of retroviruses and LTR-containing retrotransposons. Although the position of the Tf1 protein at the N terminus of the polyprotein is similar to that of Gag proteins, very little was known about its function. Our experiments tested the putative Gag of Tf1 for typical functions of Gag, such as roles in reverse transcription, the assembly of spherical particles, and the packaging of RNA.
Our initial characterization of Tf1 Gag was to determine whether the protein was required for transposition. The comparison of sequences from Tf1 and the related element, Tf2, revealed that 36% of the amino acids in the Tf1 and Tf2 Gags were identical. That 45% of the conserved residues were encoded by synonymous codons suggested strongly that the proteins possessed a critical function. This was supported by the high Ks/Ka ratio.
The indications that the Gag of Tf1 possessed an important function were supported by our study of four Tf1 elements with deletions in the conserved hydrophilic domains of Gag. Each of these deletions significantly reduced the ability of Tf1 to transpose and indicated that Gag possessed activity necessary for the process of transposition. The most dramatic defects in transposition were observed in the cases of Tf1-ΔA, Tf1-ΔB, and Tf1-ΔC. Tf1-ΔD showed trace levels of transposition activity above that of Tf1-INfs.
We explored the individual functions of Gag by characterizing the nature of the defects caused by each deletion. For each of the four Tf1 elements with deletions in Gag, we measured the production of Gag, RT, IN, and cDNA (Table 4). Tf1-ΔA produced wild-type levels of cDNA. As indicated above, the defect in transposition despite the wild-type levels of cDNA correlated well with the likely possibility that Tf1-ΔA had a defect in nuclear import due to the deletion of the critical NLS sequence (16). The critical residues of the NLS were KRIR, and when these were removed by a deletion of the first 10 amino acids of Gag, the frequency of transposition was significantly reduced. In addition, it was shown that the deletion of the first 10 residues and the replacement of them with a Flag tag caused a defect in the nuclear localization of Gag (16). As was found with Tf1-ΔA, no reduction in cDNA or Tf1 proteins was observed in this case.
TABLE 4.
Summary of Gag mutant phenotypesa
| Tf1 element | Transposition | cDNA synthesis | Gag | RT | IN | Particle formation | RNA packaging |
|---|---|---|---|---|---|---|---|
| Tf1-WT | +++ | +++ | +++ | +++ | +++ | Cytoplasmic particles and aggregates | +++ |
| Tf1-PRfs | − | − | +++ | − | − | Cytoplasmic and nuclear particles and aggregates | − |
| Tf1-ΔA | − | +++ | +++ | ++ | ++ | Cytoplasmic aggregates | ++++ |
| Tf1-ΔB | − | − | − | ++ | ++ | Not done | − |
| Tf1-ΔC | − | − | − | +++ | +++ | No particles | − |
| Tf1-ΔD | + | + | +++ | ++ | ++ | Cytoplasmic particles and aggregates | − |
+, very little; ++, less than the wild type; +++, wild type; ++++, more than the wild type; −, not detected.
Our study of protein levels identified severe defects in the expression of Gag by Tf1-ΔB and Tf1-ΔC. This lack of Gag correlated with the significant reduction in cDNA levels for Tf1-ΔB and Tf1-ΔC. The elements Tf1-ΔA, Tf1-ΔB, and Tf1-ΔD accumulated slightly smaller amounts of RT and IN (Table 4). The reduced levels of RT were not sufficient to cause the defects in cDNA levels since Tf1-ΔA had reduced RT levels but nevertheless made wild-type amounts of cDNA. In addition, we know that the reduced levels of IN could not have caused defects in reverse transcription since Tf1-INfs produced normal levels of cDNA.
The impact of the reduced levels of protein on particle formation was examined by electron microscopy. A deletion that greatly reduced levels of Gag, ΔC, also resulted in the absence of spherical particles and aggregates, even though IN and RT were expressed at wild-type levels. The results of RNA packaging assays for Tf1-ΔB and Tf1-ΔC indicate that Gag expression was required for particle formation. Tf1-ΔB and Tf1-ΔC produced no cDNA, indicating that reverse transcription was dependent on particle formation. Tf1-ΔD made spherical particles clearly visible by electron microscopy. It was therefore surprising that the particles were unable to package the Tf1 RNA. In their entirety, these data demonstrate that Tf1-ΔB and Tf1-ΔC are defective for transposition because the reduced levels of Gag resulted in the absence of particles. Without particles, the Tf1 RNA could not be packaged or reverse transcribed. Although the proteolytic processing of the Tf1 proteins produced by Tf1-ΔB and Tf1-ΔC suggests that Gag-Gag associations occur, the reduced levels of Gag indicate that any particles that did form were unstable. In addition, the data demonstrate that Tf1-ΔD was defective for transposition not because of a lack of particles but because the deletion in Gag blocked the packaging of the RNA.
The mutation in Tf1-ΔD caused a defect in reverse transcription and in the packaging of RNA. This indicates that Gag plays an important role in reverse transcription by packaging the Tf1 RNA into the particles. The deletions in Tf1-ΔB and Tf1-ΔC resulted in low levels of Gag expression, and taken together with the lack of spherical particles, these data demonstrate that Gag is required for the formation of VLPs. More importantly, the expression of Tf1-PRfs, the element that does not produce RT or IN, revealed that the accumulation of Gag is sufficient for the assembly of spherical particles. Our findings that Gag is required and sufficient for particle formation and that it participates in the packaging of its RNA correlate well with the functions of the Gag proteins of retroviruses. As a result, it is likely that the Gag of Tf1 is a functional equivalent of the Gags encoded by other LTR-containing retroelements.
The characterization of Tf1 with deletions in Gag also contributed to our understanding of how Tf1 enters the nucleus. The Tf1-ΔA protein lacks the N-terminal Gag residues whose replacement with the Flag epitope resulted in a defect in entry into the nucleus (16). The deletion in Tf1-ΔA also caused a defect in transposition, and this too was likely due to a block of import into the nucleus. The DNA blot demonstrated that Tf1-ΔA produced normal levels of cDNA, and the homologous recombination assay indicated that the cDNA was not transported into the nucleus.
The results of the electron microscopy demonstrated that spherical particles produced by Tf1-WT in stationary-phase cells were observed only in the cytoplasm. Previous results of immunofluorescence microscopy revealed that under these conditions, the bulk of Gag is localized in the nucleus (7, 15). These observations are consistent with a process of particle disassembly that obscures the visualization by electron microscopy of Gag in the nucleus. The disassembly of particles could occur before or after transit through the nuclear pores. This is an important question because the size of Tf1 particles, 50 nm, is larger than the maximum diameter of the channel, 20 to 25 nm, as measured by testing nucleoplasmin-coated gold particles of various sizes for the ability to pass through the nuclear pore (20, 45). However, the mutation in Tf1-PRfs resulted in large numbers of spherical and aggregated particles within the nucleus, suggesting that particles were able to pass through the pores. It is therefore possible that the particles reorganize to fit through the pore or that the pore itself has the capability to accommodate the particles. Nevertheless, the presence of particles in the nucleus indicates that Tf1 particles have the ability to pass through the nuclear pore without first being disassembled. The ability of Tf1 particles to pass through the nuclear pore intact is not surprising since evidence indicates that the 50-nm particles of simian virus 40 and the 39-nm particles of human hepatitis B virus pass intact through nuclear pores (49, 63).
The electron micrographs of cells expressing Tf1-PRfs not only showed particles in the nucleus but also revealed a surprising association between the particles in the nucleus and heterochromatin. This association of the particles with heterochromatin correlates well with results of immunofluorescence microscopy that show Gag localized in the nucleolus (M. Kim and H. Levin, unpublished data). Although the biological significance of this is not clear, it indicates that Gag possesses sequences that either target the protein to the nucleolus or cause its retention there. In addition, the recent finding that Tf1 integration exhibits a preference for the third chromosome, which contains the rRNA gene repeats, suggests that the localization of Tf1 protein in the nucleolus may have mechanistic importance (54; N. Bowen, unpublished data).
The deletion in Tf1-ΔA resulted in a surprising combination of effects. This deletion removed the NLS and should have blocked import into the nucleus. The decrease in transposition activity and the drop in homologous recombination between cDNA and plasmid sequences correlate well with a defect in import. Tf1-ΔA produced normal levels of Gag and cDNA, suggesting that the lack of import is the only defect caused by the mutation. However, the electron microscopy demonstrated that cells expressing Tf1-ΔA contained only aggregates and not spherical particles (Table 4). This defect in particle structure may not be the result of removing the NLS but may instead be due to alterations in Gag-Gag and/or Gag-Pol interactions. One unexpected result is that despite the disruption of the particle structure, the Gag of Tf1-ΔA was able to package dramatically higher levels of Tf1 RNA than the wild type (Fig. 6). Although we have no clear explanation for this, one possibility is that the deletion of residues 2 through 11 results in a Gag that assembles with an increase in affinity for RNA. Another hypothesis is that Tf1-ΔA packages more RNA per particle than wild-type Tf1.
The assay for packaging of Tf1 RNA revealed additional information about the role of Gag in protecting the RNA. Although the expression of Tf1-PRfs produced some spherical particles, no packaging was observed. This indicates that the formation of particles is not sufficient for packaging of Tf1 RNA. It also suggests that the expression of Gag alone is not sufficient and that RT and/or IN may be required for packaging to occur. Alternatively, the frameshift mutation in PR caused approximately 50 residues to be fused to the C terminus of Gag, and these could have perturbed the packaging of RNA. More important is the result that Tf1-ΔD allows the formation of spherical particles that are indistinguishable from those of the wild type. Surprisingly, these particles are severely defective for packaging of Tf1 RNA. Thus, the formation of highly ordered particles is not sufficient for packaging of Tf1 RNA. Instead, it is likely that residues of Gag interact with the RNA and protect it from degradation. The residues removed in the Gag of Tf1-ΔD are the first amino acids found to participate in the packaging of the RNA. Given the lack of Zn fingers and NC in Tf1 Gag, the residues removed in Tf1-ΔD are the best candidates for domains of interaction with Tf1 RNA. It is also possible that the deletion in Tf1-ΔD reduced interactions with the Tf1 cDNA. This may explain the surprising result that despite low levels of transposition activity, Tf1-ΔD produced significant levels of homologous recombination between cDNA and plasmid sequences. Reduced association of the cDNA with Gag could increase exposure of the cDNA to the recombination machinery.
The presence of several basic amino acids in the region deleted in Tf1-ΔD and their location at the C terminus are two significant similarities to the unusual nucleic acid binding site in the Gag of spumavirus (66). The presence of basic residues in the C terminus of Gag has also been observed in gypsy, and because this endogenous retrovirus also lacks Zn fingers, these basic residues have been proposed to function in nucleic acid binding (2). It is compelling to think that spumavirus and Tf1 represent two LTR-containing retroelements that have not adopted Zn finger structures to bind RNA but instead may have convergently evolved charged structures that mediate RNA interactions.
The unusual features of Tf1, such as the single ORF, the self-priming mechanism of reverse transcription, and the lack of sequence similarity to the Gags of LTR-containing elements, has led to the speculation that Tf1 may transpose by a pathway significantly different from that of the other well-studied elements in yeast, Ty1 and Ty3. The results of the electron microscopy used here to examine VLPs in S. pombe represent the first report that Tf1 does produce highly ordered, spherical particles. This is strong evidence that Tf1 transposition does indeed occur via intermediates similar to those of other LTR-containing retroelements.
Acknowledgments
We are grateful to Kunio Nagashima for producing the electron micrographs. We are also grateful to Nathan Bowen for his help in the statistical analyses of the Gag proteins. We also thank Nancy Bae, Richard Fekete, Christie Hamilton, Klaus Nielsen, Evelyn Sattleger, and Mark J. Swanson for fruitful discussions.
L.T. was a recipient of fellowships from the ARC (Association pour la Recherche contre le Cancer) and from the NIH Visiting Program for Postdoctoral Research.
REFERENCES
- 1.Adams, S. E., J. Mellor, K. Gull, R. B. Sim, M. F. Tuite, S. M. Kingsman, and A. J. Kingsman. 1987. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian proteins. Cell 49:111-119. [DOI] [PubMed] [Google Scholar]
- 2.Alberola, T. M., and R. de Frutos. 1996. Molecular structure of a gypsy element of Drosophila subobscura (gypsyDs) constituting a degenerate form of insect retroviruses. Nucleic Acids Res. 24:914-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alin, K., and S. P. Goff. 1996. Mutational analysis of interactions between the Gag precursor proteins of murine leukemia viruses. Virology 216:418-424. [DOI] [PubMed] [Google Scholar]
- 4.Allain, B., M. Lapadat-Tapolsky, C. Berlioz, and J. L. Darlix. 1994. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 13:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atwood, A., J. Choi, and H. L. Levin. 1998. The application of a homologous recombination assay revealed amino acid residues in an LTR-retrotransposon that were critical for integration. J. Virol. 72:1324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atwood, A., J. H. Lin, and H. L. Levin. 1996. The retrotransposon Tf1 assembles virus-like particles that contain excess Gag relative to integrase because of a regulated degradation process. Mol. Cell. Biol. 16:338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balasundaram, D., M. J. Benedik, M. Morphew, V. D. Dang, and H. L. Levin. 1999. Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol. Cell. Biol. 19:5768-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 9.Cairns, T. M., and R. C. Craven. 2001. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. J. Virol. 75:242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chazal, N., C. Carriere, B. Gay, and P. Boulanger. 1994. Phenotypic characterization of insertion mutants of the human immunodeficiency virus type 1 Gag precursor expressed in recombinant baculovirus-infected cells. J. Virol. 68:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clare, J., and P. Farabaugh. 1985. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc. Natl. Acad. Sci. USA 82:2829-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covey, S. N. 1986. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 14:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang, V. D., M. J. Benedik, K. Ekwall, J. Choi, R. C. Allshire, and H. L. Levin. 1999. A new member of the Sin3 family of corepressors is essential for cell viability and required for retroelement propagation in fission yeast. Mol. Cell. Biol. 19:2351-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang, V. D., and H. L. Levin. 2000. Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p. Mol. Cell. Biol. 20:7798-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darlix, J. L., A. Vincent, C. Gabus, H. de Rocquigny, and B. Roques. 1993. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C. R. Acad. Sci. Ser. III 316:763-771. [PubMed] [Google Scholar]
- 18.Davis, N. L., and R. R. Rueckert. 1972. Properties of a ribonucleoprotein particle isolated from Nonidet P-40-treated Rous sarcoma virus. J. Virol. 10:1010-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delchambre, M., D. Gheysen, D. Thines, C. Thiriart, E. Jacobs, E. Verdin, M. Horth, A. Burny, and F. Bex. 1989. The GAG precursor of simian immunodeficiency virus assembles into virus like particles. EMBO J. 8:2653-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldherr, C. M., E. Kallenback, and N. Schulz. 1984. Movement of a karyophilic protein through the nuclear pores of oocytes. J. Cell Biol. 99:2216-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleissner, E., and E. Tress. 1973. Isolation of a ribonucleoprotein structure from oncornaviruses. J. Virol. 12:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garfinkel, D. J., J. D. Boeke, and G. R. Fink. 1985. Ty element transposition: reverse transcriptase and virus-like particles. Cell 42:507-517. [DOI] [PubMed] [Google Scholar]
- 23.Gheysen, D., E. Jacobs, F. de Foresta, T. Thiriart, M. Francotte, D. Thines, and M. deWilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 24.Goff, S. P., and L. I. Lobel. 1987. Mutants of murine leukemia viruses and retroviral replication. Biochim. Biophys. Acta 907:93-123. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, L. J., D. L. Chalker, K. J. Orlinsky, and S. B. Sandmeyer. 1992. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J. Virol. 66:1414-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoff, E. F., H. L. Levin, and J. D. Boeke. 1998. Schizosaccharomyces pombe retrotransposon Tf2 mobilizes primarily through homologous cDNA recombination. Mol. Cell. Biol. 18:6839-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu, H.-W., P. Schwartzberg, and S. P. Goff. 1985. Point mutations in the P30 domain of the gag gene of moloney murine leukemia virus. Virology 142:211-214. [DOI] [PubMed] [Google Scholar]
- 28.Kirchner, J., and S. Sandmeyer. 1993. Proteolytic processing of Ty3 proteins is required for transposition. J. Virol. 67:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 30.Leblanc, P., S. Desset, F. Giorgi, A. R. Taddei, A. M. Fausto, M. Mazzini, B. Dastugue, and C. Vaury. 2000. Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J. Virol. 74:10658-10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lecellier, C. H., and A. Saib. 2000. Foamy viruses: between retroviruses and pararetroviruses. Virology 271:1-8. [DOI] [PubMed] [Google Scholar]
- 32.Lecher, P., A. Bucheton, and A. Pelisson. 1997. Expression of the Drosophila retrovirus gypsy as ultrastructurally detectable particles in the ovaries of flies carrying a permissive flamenco allele. J. Gen. Virol. 78:2379-2388. [DOI] [PubMed] [Google Scholar]
- 33.Leis, J., D. Baltimore, J. M. Bishop, J. Coffin, E. Fleissner, S. P. Goff, S. Oroszlan, H. Robinson, A. M. Skalka, H. M. Temin, and V. Vogt. 1988. Standardized and simplified nomenclature for proteins common to all retroviruses. J. Virol. 62:1808-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levin, H. L. 1995. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol. Cell. Biol. 15:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levin, H. L. 1996. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol. Cell. Biol. 16:5645-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin, H. L., D. C. Weaver, and J. D. Boeke. 1993. Novel gene expression mechanism in a fission yeast retroelement: Tf1 proteins are derived from a single primary translation product. EMBO J. 12:4885-4895. (Erratum, 13:1494, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levin, H. L., D. C. Weaver, and J. D. Boeke. 1990. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol. Cell. Biol. 10:6791-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levin, H. L., D. C. Weaver, and J. D. Boeke. 1990. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol. Cell. Biol. 10:6791-6798. (Erratum, 11:2334, 1991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, J. H., and H. L. Levin. 1997. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes Dev. 11:270-285. [DOI] [PubMed] [Google Scholar]
- 40.Lin, J. H., and H. L. Levin. 1998. Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5-IR stem-loop of retroviruses. Mol. Cell. Biol. 18:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Losson, R., and F. Lacroute. 1983. Plasmids carrying the yeast OMP decarboxylase structural and regulatory genes: transcription regulation in a foreign environment. Cell 32:371-377. [DOI] [PubMed] [Google Scholar]
- 43.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurer, B., H. Bannert, G. Darai, and R. M. Flugel. 1988. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J. Virol. 62:1590-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehlin, H., B. Daneholt, and U. Skoglund. 1992. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell 69:605-613. [DOI] [PubMed] [Google Scholar]
- 46.Miyake, T., N. Mae, T. Shiba, and S. Kondo. 1987. Production of virus-like particles by the transposable genetic element, copia, of Drosophila melanogaster. Mol. Gen. Genet. 207:29-37. [DOI] [PubMed] [Google Scholar]
- 47.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 48.Orlinsky, K. J., J. Gu, M. Hoyt, S. Sandmeyer, and T. M. Menees. 1996. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J. Virol. 70:3440-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pante, N., and M. Kann. 2002. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 13:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Patarca, R., and W. A. Haseltine. 1985. A major retroviral core protein related to EPA and TIMP. Nature 318:390. [DOI] [PubMed] [Google Scholar]
- 50.Peliska, J. A., S. Balasubramanian, D. P. Giedroc, and S. J. Benkovic. 1994. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry 33:13817-13823. [DOI] [PubMed] [Google Scholar]
- 51.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 52.Renne, R., E. Friedl, M. Schweizer, U. Fleps, R. Turek, and D. Neumann-Haefelin. 1992. Genomic organization and expression of simian foamy virus type 3 (SFV-3). Virology 186:597-608. [DOI] [PubMed] [Google Scholar]
- 53.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 54.Singleton, T. L., and H. L. Levin. 2002. A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of insertion. Eukaryot. Cell 1:44-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strambio-de-Castillia, C., and E. Hunter. 1992. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J. Virol. 66:7021-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 296-308. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 57.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogt, V. M. 1997. Retroviral virions and genomes, p. 27-70. In J. M. Coffin (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 59.Vogt, V. M., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, C. T., and E. Barklis. 1993. Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J. Virol. 67:4264-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weaver, D. C., G. V. Shpakovski, E. Caputo, H. L. Levin, and J. D. Boeke. 1993. Sequence analysis of closely related retrotransposon families from fission yeast. Gene 131:135-139. [DOI] [PubMed] [Google Scholar]
- 62.Weldon, R. A., Jr., and J. W. Wills. 1993. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J. Virol. 67:5550-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whittaker, G. R., M. Kann, and A. Helenius. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627-651. [DOI] [PubMed] [Google Scholar]
- 64.Wills, J. W., and R. C. Craven. 1991. Form, function and use of retroviral Gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 65.You, J. C., and C. S. McHenry. 1994. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 269:31491-31495. [PubMed] [Google Scholar]
- 66.Yu, S. F., K. Edelmann, R. K. Strong, A. Moebes, A. Rethwilm, and M. L. Linial. 1996. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 70:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhoa, Y., I. M. Jones, D. J. Hockley, M. V. Nermut, and P. Roy. 1994. Complementation of human immunodeficiency virus (HIV-1) gag particle formation. Virology 199:403-408. [DOI] [PubMed] [Google Scholar]